Interleukins (Cytokines) as Biomarkers in Colorectal Cancer: Progression, Detection, and Monitoring
Abstract
1. Introduction
2. Molecular Pathways and Cytokine Role in CRC
3. Advancements in Cytokine Detection and Monitoring Clinically
4. Cytokines’ Role in CRC
4.1. Forkhead Box P3 (FOXP3)
4.2. Tumor Necrosis Factor-α (TNF-α)
4.3. Interferon-Gamma (IFN-γ)
5. Interleukins in Colorectal Cancer
5.1. Interleukin-1β
5.2. Interleukin-17
5.3. Interleukin-22
5.4. Interleukin-6
5.5. Interleukin-23
5.6. Interleukin-33
5.7. Interleukin-15
5.8. Interleukin-18
5.9. Interleukin-13
5.10. Interleukin-4
5.11. Interleukin-8
5.12. Interleukin-11
6. Discussion
7. Future Perspective
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Safarzadeh, E.; Sandoghchian, S.; Baradaran, B. Herbal medicine as inducers of apoptosis in cancer treatment. Adv. Pharm. Bull. 2014, 4, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Shewach, D.S.; Kuchta, R.D. Introduction to cancer chemotherapeutics. Chem. Rev. 2009, 109, 2859–2861. [Google Scholar] [CrossRef] [PubMed]
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global cancer statistics. CA Cancer J. Clin. 2011, 61, 69–90. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Keum, N.; Giovannucci, E. Global burden of colorectal cancer: Emerging trends.; risk factors and prevention strategies. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 713–732. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.; White, P.; Nieto, J.; Vieira, D.; Francois, F.; Hamilton, F. Colorectal Cancer in African Americans: An Update. Clin. Transl. Gastroenterol. 2016, 7, e185. [Google Scholar] [CrossRef]
- Jess, T.; Rungoe, C.; Peyrin-Biroulet, L. Risk of colorectal cancer in patients with ulcerative colitis: A meta-analysis of population-based cohort studies. Clin. Gastroenterol. Hepatol. 2012, 10, 639–645. [Google Scholar] [CrossRef]
- Marventano, S.; Forjaz, M.J.; Grosso, G.; Mistretta, A.; Giorgianni, G.; Platania, A.; Gangi, S.; Basile, F.; Biondi, A. Health related quality of life in colorectal cancer patients: State of the art. BMC Surg. 2013, 13, S15. [Google Scholar] [CrossRef]
- Vacante, M.; Borzì, A.M.; Basile, F.; Biondi, A. Biomarkers in colorectal cancer: Current clinical utility and future perspectives. World J. Clin. Cases 2018, 6, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Mager, L.F.; Wasmer, M.H.; Rau, T.T.; Krebs, P. Cytokine-Induced Modulation of Colorectal Cancer. Front. Oncol. 2016, 6, 96. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Margolin, K. Cytokines in cancer immunotherapy. Cancers 2011, 3, 3856–3893. [Google Scholar] [CrossRef] [PubMed]
- Koltsova, E.K.; Grivennikov, S.I. IL-22 Gets to the Stem of Colorectal Cancer. Immunity 2014, 40, 639–641. [Google Scholar] [CrossRef]
- Wang, J.; Li, D.; Cang, H.; Guo, B. Crosstalk between cancer and immune cells: Role of tumor-associated macrophages in the tumor microenvironment. Cancer Med. 2019, 8, 4709–4721. [Google Scholar] [CrossRef] [PubMed]
- Kawanishi, S.; Ohnishi, S.; Ma, N.; Hiraku, Y.; Murata, M. Crosstalk between DNA Damage and Inflammation in the Multiple Steps of Carcinogenesis. Int. J. Mol. Sci. 2017, 18, 1808. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, L.; Zhao, H.; Yan, Y.; Lu, J. The Role of Interleukins in Colorectal Cancer. Int. J. Biol. Sci. 2020, 16, 2323–2339. [Google Scholar] [CrossRef]
- Veziant, J.; Villéger, R.; Barnich, N.; Bonnet, M. Gut Microbiota as Potential Biomarker and/or Therapeutic Target to Improve the Management of Cancer: Focus on Colibactin-Producing Escherichia coli in Colorectal Cancer. Cancers 2021, 13, 2215. [Google Scholar] [CrossRef]
- Kim, J.; Lee, H.K. Potential Role of the Gut Microbiome In Colorectal Cancer Progression. Front. Immunol. 2022, 12, 807648. [Google Scholar] [CrossRef]
- Olovo, C.V.; Huang, X.; Zheng, X.; Xu, M. Faecal microbial biomarkers in early diagnosis of colorectal cancer. J. Cell. Mol. Med. 2021, 25, 10783–10797. [Google Scholar] [CrossRef]
- Rye, M.S.; Garrett, K.L.; Holt, R.A.; Platell, C.F.; McCoy, M.J. Fusobacterium nucleatum and Bacteroides fragilis detection in colorectal tumours: Optimal target site and correlation with total bacterial load. PLoS ONE 2022, 17, e0262416. [Google Scholar] [CrossRef] [PubMed]
- Eklöf, V.; Löfgren-Burström, A.; Zingmark, C.; Edin, S.; Larsson, P.; Karling, P.; Alexeyev, O.; Rutegård, J.; Wikberg, M.L.; Palmqvist, R. Cancer-associated fecal microbial markers in colorectal cancer detection. Int. J. Cancer 2017, 141, 2528–2536. [Google Scholar] [CrossRef] [PubMed]
- Nassar, F.J.; Msheik, Z.S.; Itani, M.M.; Helou, R.E.; Hadla, R.; Kreidieh, F.; Bejjany, R.; Mukherji, D.; Shamseddine, A.; Nasr, R.R.; et al. Circulating miRNA as Biomarkers for Colorectal Cancer Diagnosis and Liver Metastasis. Diagnostics 2021, 11, 341. [Google Scholar] [CrossRef]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [PubMed]
- Staiteieh, S.A.; Akil, L.; Al Khansa, R.; Nasr, R.; Sagheer, A.Z.; Houshaymi, B.; Merhi, R.A. Study of microRNA expression profiling as biomarkers for colorectal cancer patients in Lebanon. Mol. Clin. Oncol. 2022, 16, 39. [Google Scholar] [CrossRef] [PubMed]
- Sur, D.; Advani, S.; Braithwaite, D. MicroRNA panels as diagnostic biomarkers for colorectal cancer: A systematic review and meta-analysis. Front. Med. 2022, 9, 915226. [Google Scholar] [CrossRef]
- Precazzini, F.; Detassis, S.; Imperatori, A.S.; Denti, M.A.; Campomenosi, P. Measurements Methods for the Development of MicroRNA-Based Tests for Cancer Diagnosis. Int. J. Mol. Sci. 2021, 22, 1176. [Google Scholar] [CrossRef]
- Kaur, J.; Preethi, M.; Srivastava, R.; Borse, V. Role of IL-6 and IL-8 biomarkers for optical and electrochemical based point-of-care detection of oral cancer. Biosens. Bioelectron. X 2022, 11, 100212. [Google Scholar] [CrossRef]
- Liu, G.; Jiang, C.; Lin, X.; Yang, Y. Point-of-care detection of cytokines in cytokine storm management and beyond: Significance and challenges. View 2021, 2, 20210003. [Google Scholar] [CrossRef]
- Chaudhry, H.; Zhou, J.; Zhong, Y.; Ali, M.M.; McGuire, F.; Nagarkatti, P.S.; Nagarkatti, M. Role of cytokines as a double-edged sword in sepsis. In Vivo 2013, 27, 669–684. [Google Scholar]
- Febbo, P.G.; Ladanyi, M.; Aldape, K.D.; De Marzo, A.M.; Hammond, M.E.; Hayes, D.F.; Lafrate, A.J.; Kelley, R.K.; Marcucci, G.; Ogino, S.; et al. NCCN Task Force report: Evaluating the clinical utility of tumor markers in oncology. J. Natl. Compr. Cancer Netw. 2011, 9, S1–S32. [Google Scholar] [CrossRef]
- Kartikasari, A.E.R.; Huertas, C.S.; Mitchell, A.; Plebanski, M. Tumor-Induced Inflammatory Cytokines and the Emerging Diagnostic Devices for Cancer Detection and Prognosis. Front. Oncol. 2021, 11, 692142. [Google Scholar] [CrossRef] [PubMed]
- Capone, F.; Guerriero, E.; Sorice, A.; Colonna, G.; Ciliberto, G.; Costantini, S. Serum Cytokinome Profile Evaluation: A Tool to Define New Diagnostic and Prognostic Markers of Cancer Using Multiplexed Bead-Based Immunoassays. Mediat. Inflamm. 2016, 2016, 3064643. [Google Scholar] [CrossRef] [PubMed]
- Schett, G.; Elewaut, D.; McInnes, I.B.; Dayer, J.M.; Neurath, M.F. How cytokine networks fuel inflammation: Toward a cytokine-based disease taxonomy. Nat. Med. 2013, 19, 822–824. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Jia, D.; Li, L.; Andrew, S.; Allan, D.; Li, X.; Lee, J.; Ji, Z.; Yao, Z.; Gadde, S.; Figeys, D.; et al. An autocrine inflammatory forward-feedback loop after chemotherapy withdrawal facilitates the repopulation of drug-resistant breast cancer cells. Cell Death Dis. 2017, 8, e2932. [Google Scholar] [CrossRef]
- Nengroo, M.A.; Verma, A.; Datta, D. Cytokine chemokine network in tumor microenvironment: Impact on CSC properties and therapeutic applications. Cytokine 2022, 156, 155916. [Google Scholar] [CrossRef]
- Furman, D.; Campisi, J.; Verdin, E.; Carrera-Bastos, P.; Targ, S.; Franceschi, C.; Ferrucci, L.; Gilroy, D.W.; Fasano, A.; Miller, G.W.; et al. Chronic inflammation in the etiology of disease across the life span. Nat. Med. 2019, 25, 1822–1832. [Google Scholar] [CrossRef]
- Greten, F.R.; Grivennikov, S.I. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity 2019, 51, 27–41. [Google Scholar] [CrossRef]
- Sokolova, O.; Naumann, M. Crosstalk between DNA Damage and Inflammation in the Multiple Steps of Gastric Carcinogenesis. Curr. Top. Microbiol. Immunol. 2019, 421, 107–137. [Google Scholar] [CrossRef]
- Yang, Z.H.; Dang, Y.Q.; Ji. G. Role of epigenetics in transformation of inflammation into colorectal cancer. World J. Gastroenterol. 2019, 25, 2863–2877. [Google Scholar] [CrossRef] [PubMed]
- Martin, B.; Märkl, B. Immunologic Biomarkers and Biomarkers for Immunotherapies in Gastrointestinal Cancer. Visc. Med. 2019, 35, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Rhea, J.M.; Molinaro, R.J. Cancer biomarkers: Surviving the journey from bench to bedside. MLO Med. Lab. Obs. 2011, 43, 10–12. [Google Scholar] [PubMed]
- Nagata, H.; Ishihara, S.; Hata, K.; Murono, K.; Kaneko, M.; Yasuda, K.; Otani, K.; Watanabe, T. Survival and Prognostic Factors for Metachronous Peritoneal Metastasis in Patients with Colon Cancer. Ann. Surg. Oncol. 2017, 24, 1269–1280. [Google Scholar] [CrossRef] [PubMed]
- Goossens, N.; Nakagawa, S.; Sun, X.; Hoshida, Y. Cancer biomarker discovery and validation. Transl. Cancer Res. 2015, 4, 256. [Google Scholar] [PubMed]
- Sarhadi, V.K.; Armengol, G. Molecular Biomarkers in Cancer. Biomolecules 2022, 12, 1021. [Google Scholar] [CrossRef]
- Herceg, Z.; Hainaut, P. Genetic and epigenetic alterations as biomarkers for cancer detection, diagnosis and prognosis. Mol. Oncol. 2007, 1, 26–41. [Google Scholar] [CrossRef]
- Harbaum, L.; Pollheimer, M.J.; Kornprat, P.; Lindtner, R.A.; Bokemeyer, C.; Langner, C. Peritumoral eosinophils predict recurrence in colorectal cancer. Mod. Pathol. 2015, 28, 403–413. [Google Scholar] [CrossRef]
- HSU, C.-P.; Chung, Y.-C. Influence of Interleukin-6 on the Invasiveness of Human Colorectal Carcinoma. Anticancer. Res. 2006, 26, 4607–4614. Available online: https://ar.iiarjournals.org/content/anticanres/26/6B/4607.full.pdf (accessed on 17 March 2023).
- Waniczek, D.; Lorenc, Z.; Śnietura, M.; Wesecki, M.; Kopec, A.; Muc-Wierzgoń, M. Tumor-Associated Macrophages and Regulatory T Cells Infiltration and the Clinical Outcome in Colorectal Cancer. Arch. Immunol. Ther. Exp. 2017, 65, 445–454. [Google Scholar] [CrossRef]
- Wikberg, M.L.; Ling, A.; Li, X.; Öberg, Å.; Edin, S.; Palmqvist, R. Neutrophil infiltration is a favorable prognostic factor in early stages of colon cancer. Hum. Pathol. 2017, 68, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Terzić, J.; Grivennikov, S.; Karin, E.; Karin. M. Inflammation and colon cancer. Gastroenterology 2010, 138, 2101–2114.e5. [Google Scholar] [CrossRef]
- Calon, A.; Espinet, E.; Palomo-Ponce, S.; Tauriello, D.V.; Iglesias, M.; Céspedes, M.V. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell 2012, 22, 571–584. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Deng, C.X. Effect of Stromal Cells in Tumor Microenvironment on Metastasis Initiation. Int. J. Biol. Sci. 2018, 14, 2083–2093. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.H.; Goel, A.; Chung, D.C. Pathways of Colorectal Carcinogenesis. Gastroenterology 2020, 158, 291–302. [Google Scholar] [CrossRef]
- Pino, M.S.; Chung, D.C. The chromosomal instability pathway in colon cancer. Gastroenterology 2010, 138, 2059–2072. [Google Scholar] [CrossRef]
- Powell, D.W.; Adegboyega, P.A.; Di Mari, J.F.; Mifflin, R.C. Epithelial cells and their neighbors I. Role of intestinal myofibroblasts in development repair and cancer. Am. J. Physiol. Gastrointest. Liver Physiol. 2005, 289, G2–G7. [Google Scholar] [CrossRef]
- Koliaraki, V.; Pallangyo, C.K.; Greten, F.R.; Kollias, G. Mesenchymal Cells in Colon Cancer. Gastroenterology 2017, 152, 964–979. [Google Scholar] [CrossRef]
- Kryczek, I.; Lin, Y.; Nagarsheth, N.; Peng, D.; Zhao, L.; Zhao, E.; Vatan, L.; Szeliga, W.; Dou, Y.; Owens, S.; et al. IL-22(+) CD4(+) T cells promote colorectal cancer stemness via STAT3 transcription factor activation and induction of the methyltransferase DOT1L. Immunity 2014, 40, 772–784. [Google Scholar] [CrossRef]
- Todaro, M.; Gaggianesi, M.; Catalano, V.; Benfante, A.; Iovino, F.; Biffoni, M.; Apuzzo, T.; Sperduti, I.; Volpe, S.; Cocorullo, G.; et al. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell 2014, 14, 342–356. [Google Scholar] [CrossRef]
- Lotti, F.; Jarrar, A.M.; Pai, R.K.; Hitomi, M.; Lathia, J.; Mace, A.; Rich, J.N. Chemotherapy activates cancer-associated fibroblasts to maintain colorectal cancer-initiating cells by IL-17A. J. Exp. Med. 2013, 210, 2851–2872. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Liu, Q.; Tsang, L.L.; Ye, Q.; Chan, H.C.; Sun, Y.; Jiang, X. Human MSCs promotes colorectal cancer epithelial–mesenchymal transition and progression via CCL5/β-catenin/Slug pathway. Cell Death Dis. 2017, 8, e2819. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen, L.; De Sousa, E.M.F.; van der Heijden, M.; Cameron, K.; de Jong, J.H.; Borovski, T.; Tuynman, J.B.; Todaro, M.; Merz, C.; Rodermond, H.; et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat. Cell Biol. 2010, 12, 468–476. [Google Scholar] [CrossRef] [PubMed]
- Schulte, W.; Jürgen, B.; Richard, B. Cytokines in sepsis: Potent immunoregulators and potential therapeutic targets—An updated view. Mediat. Inflamm. 2013, 2013, 165974. [Google Scholar] [CrossRef] [PubMed]
- Sprague, A.H.; Khalil, R.A. Inflammatory cytokines in vascular dysfunction and vascular disease. Biochem. Pharmacol. 2009, 78, 539–552. [Google Scholar] [CrossRef] [PubMed]
- Rea, I.M.; Gibson, D.S.; McGilligan, V.; McNerlan, S.E. Alexander HD, Ross OA. Age and Age-Related Diseases: Role of Inflammation Triggers and Cytokines. Front. Immunol. 2018, 9, 586. [Google Scholar] [CrossRef]
- Nedoszytko, B.; Sokołowska-Wojdyło, M.; Ruckemann-Dziurdzińska, K.; Roszkiewicz, J.; Nowicki, R.J. Chemokines and cytokines network in the pathogenesis of the inflammatory skin diseases: Atopic dermatitis, psoriasis and skin mastocytosis. Postępy Dermatol. I Alergol. 2014, 31, 84–91. [Google Scholar] [CrossRef]
- Kabel, A.M. Relationship between cancer and cytokines. J. Cancer Res. Treat. 2014, 2, 41–43. [Google Scholar]
- Comella-Del-Barrio, P.; Abellana, R.; Villar-Hernández, R.; Jean Coute, M.D.; Sallés Mingels, B.; Canales Aliaga, L.; Narcisse, M.; Gautier, J.; Ascaso, C.; Latorre, I.; et al. A model based on the combination of IFN-γ, IP-10, ferritin and 25-hydroxyvitamin D for discriminating latent from active tuberculosis in children. Front. Microbiol. 2019, 10, 1855. [Google Scholar] [CrossRef]
- Pugliese, N.R.; Fabiani, I.; Conte, L.; Nesti, L.; Masi, S.; Natali, A.; Colombo, P.C.; Pedrinelli, R.; Dini, F.L. Persistent congestion, renal dysfunction and inflammatory cytokines in acute heart failure: A prognosis study. J. Cardiovasc. Med. 2020, 21, 494–502. [Google Scholar] [CrossRef]
- Shimabukuro-Vornhagen, A.; Gödel, P.; Subklewe, M.; Stemmler, H.J.; Schlößer, H.A.; Schlaak, M.; Kochanek, M.; Böll, B.; von Bergwelt-Baildon, M.S. Cytokine release syndrome. J. Immunother. Cancer 2018, 6, 56. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.B.; Carl, H.J. Cytokine release syndrome in severe COVID-19. Science 2020, 368, 473–474. [Google Scholar] [CrossRef] [PubMed]
- Simpson, S.; Kaislasuo, J.; Guller, S.; Pal, L. Thermal stability of cytokines: A review. Cytokine 2020, 125, 154829. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Qi, M.; Hutchinson, M.R.; Yang, G.; Goldys, E.M. Recent advances in cytokine detection by immunosensing. Biosens. Bioelectron. 2016, 79, 810–821. [Google Scholar] [CrossRef] [PubMed]
- Monastero, R.N.; Srinivas, P. Cytokines as biomarkers and their respective clinical cutoff levels. Int. J. Inflam. 2017, 2017, 4309485. [Google Scholar] [CrossRef] [PubMed]
- Kouwenhoven, M.; Ozenci, V.; Teleshova, N.; Hussein, Y.; Huang, Y.M.; Eusebio, A.; Link, H. Enzyme-linked immunospot assays provide a sensitive tool for detection of cytokine secretion by monocytes. Clin. Diagn. Lab. Immunol. 2001, 8, 1248–1257. [Google Scholar] [CrossRef] [PubMed]
- Stenken, J.A.; Poschenrieder, A.J. Bioanalytical chemistry of cytokines–a review. Anal. Chim. Acta 2015, 853, 95–115. [Google Scholar] [CrossRef]
- Zhang, K.; Baratta, M.V.; Liu, G.; Frank, M.G.; Leslie, N.R.; Watkins, L.R.; Maier, S.F.; Hutchinson, M.R.; Goldys, E.M. A novel platform for in vivo detection of cytokine release within discrete brain regions. Brain Behav. Immun. 2018, 71, 18–22. [Google Scholar] [CrossRef]
- Arman, A.; Deng, F.; Goldys, E.M.; Liu, G.; Hutchinson, M.R. In vivo intrathecal IL-1β quantification in rats: Monitoring the molecular signals of neuropathic pain. Brain Behav. Immun. 2020, 88, 442–450. [Google Scholar] [CrossRef]
- Tertis, M.; Leva, P.I.; Bogdan, D.; Suciu, M.; Graur, F.; Cristea, C. Impedimetric aptasensor for the label-free and selective detection of Interleukin-6 for colorectal cancer screening. Biosens. Bioelectron. 2019, 137, 123–132. [Google Scholar] [CrossRef]
- Aldo, P.; Marusov, G.; Svancara, D.; David, J.; Mor, G. Simple Plex™: A novel multi-analyte, automated microfluidic immunoassay platform for the detection of human and mouse cytokines and chemokines. Am. J. Reprod. Immunol. 2016, 75, 6. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Tang, X.; Li, J.; Lan, Q.; Min, L.; Ren, C.; Hu, X.; Torrente, R.M.; Gao, W.; Yang, Z. A nanozyme tag enabled chemiluminescence imaging immunoassay for multiplexed cytokine monitoring. Chem. Commun. 2018, 54, 13813–13816. [Google Scholar] [CrossRef] [PubMed]
- Emoto, S.; Ishigami, H.; Yamashita, H.; Yamaguchi, H.; Kaisaki, S.; Kitayama, J. Clinical significance of CA125 and CA72-4 in gastric cancer with peritoneal dissemination. Gastric Cancer 2012, 15, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Wang, W.; Fang, C.; Raj, S.S.; Hu, W.M.; Li, Q.W.; Zhou, Z.W. Clinical significance and diagnostic value of serum CEA, CA19-9 and CA72-4 in patients with gastric cancer. Oncotarget 2016, 7, 49565–49573. [Google Scholar] [CrossRef] [PubMed]
- Ulusoy, H.; Kangalgil, M.; Küçük, A.O.; Özdemir, A.; Karahan, S.C.; Yaman, S.Ö.; Yavuz, H.B.; Ok, Ü. Effects of different lipid emulsions on serum adipokines, inflammatory markers and mortality in critically ill patients with sepsis: A prospective observational cohort study. Clin. Nutr. 2021, 40, 4569–4578. [Google Scholar] [CrossRef]
- He, R.; Zhu, Q.; Wang, Y.; Chen, G.; Chen, S.; Wang, Y. Influence of respiratory function training under the mode of mutual-assisted patients on postoperative pulmonary infection and immune function on lung cancer. Am. J. Transl. Res. 2021, 13, 9260–9268. [Google Scholar]
- Diamandis, E.P.; Hoffman, B.R.; Sturgeon, C.M. National academy of clinical biochemistry laboratory medicine practice guidelines for the use of tumor markers. Clin. Chem. 2008, 54, 1935–1939. [Google Scholar] [CrossRef]
- Hing, J.X.; Mok, C.W.; Tan, P.T.; Sudhakar, S.S.; Seah, C.M.; Lee, W.P.; Tan, S.M. Clinical utility of tumour marker velocity of cancer antigen 15-3 (CA 15-3) and carcinoembryonic antigen (CEA) in breast cancer surveillance. Breast 2020, 52, 95–101. [Google Scholar] [CrossRef]
- Diaz, C.B.; Marono, S.H.; Olivas, M.L.G.; Sosa, M.A.; Soto, R.G.; Arrieta, O. Association of neurologic manifestations and CEA levels with the diagnosis of brain metastases in lung cancer patients. Clin. Transl. Oncol. 2019, 21, 1538–1542. [Google Scholar] [CrossRef]
- Song, X.; Liang, B.; Wang, C.; Shi, S. Clinical value of color Doppler ultrasound combined with serum CA153, CEA and TSGF detection in the diagnosis of breast cancer. Exp. Ther. Med. 2020, 20, 1822–1828. [Google Scholar] [CrossRef]
- Sakamoto, T.; Saito, H.; Amisaki, M.; Tokuyasu, N.; Honjo, S.; Fujiwara, Y. Combined preoperative platelet-to-lymphocyte ratio and serum carbohydrate antigen 19-9 level as a prognostic factor in patients with resected pancreatic cancer. Hepatobiliary Pancreat. Dis. Int. 2019, 18, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.X.; Yang, L.; He, B.S.; Pan, Y.Q.; Ying, H.Q.; Sun, H.L.; Lin, K.; Hu, X.X.; Xu, T.; Wang, S.K. Combination of preoperative NLR, PLR and CEA could increase the diagnostic efficacy for I-III stage CRC. J. Clin. Lab. Anal. 2017, 31, e22075. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Han, L.; Zhao, R.; Fatima, S.; Zhao, L.; Gao, F. Prognosis value of IL-6, IL-8, and IL-1β in serum of patients with lung cancer: A fresh look at interleukins as a biomarker. Heliyon 2022, 8, e09953. [Google Scholar] [CrossRef] [PubMed]
- Xi, C.; Zhang, G.Q.; Sun, Z.K.; Song, H.J.; Shen, C.T.; Chen, X.Y.; Sun, J.W.; Qiu, Z.L.; Luo, Q.Y. Interleukins in Thyroid Cancer: From Basic Researches to Applications in Clinical Practice. Front. Immunol. 2020, 11, 1124. [Google Scholar] [CrossRef]
- Provatopoulou, X.; Georgiadou, D.; Sergentanis, T.N.; Kalogera, E.; Spyridakis, J.; Gounaris, A.; Zografos, G.N. Interleukins as markers of inflammation in malignant and benign thyroid disease. Inflamm. Res. 2014, 63, 667–674. [Google Scholar] [CrossRef]
- Rébé, C.; Ghiringhelli, F. Interleukin-1β and Cancer. Cancers 2020, 12, 1791. [Google Scholar] [CrossRef]
- Valeri, M.; Raffatellu, M. Cytokines IL-17 and IL-22 in the host response to infection. Pathog. Dis. 2016, 74, ftw111. [Google Scholar] [CrossRef]
- Anestakis, D.; Petanidis, S.; Kalyvas, S.; Nday, C.M.; Tsave, O.; Kioseoglou, E.; Salifoglou, A. Mechanisms and Applications of Ιnterleukins in Cancer Immunotherapy. Int. J. Mol. Sci. 2015, 16, 1691–1710. [Google Scholar] [CrossRef]
- Zarogoulidis, P.; Lampaki, S.; Yarmus, L.; Kioumis, I.; Pitsiou, G.; Katsikogiannis, N.; Hohenforst-Schmidt, W.; Li, Q.; Huang, H.; Sakkas, A. Interleukin-7 and interleukin-15 for cancer. J. Cancer 2014, 5, 765–773. [Google Scholar] [CrossRef]
- Todoric, J.; Antonucci, L.; Karin, M. Targeting Inflammation in Cancer Prevention and Therapy. Cancer Prev. Res. 2016, 9, 895. [Google Scholar] [CrossRef]
- Coussens, L.M.; Werb, Z. Inflammation and cancer. Nature 2002, 420, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Allavena, P.; Sica, A.; Balkwill, F. Cancer-related inflammation. Nature 2008, 454, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Ilyin, S.E.; Belkowski, S.M.; Plata-Salamán, C.R. Biomarker discovery and validation: Technologies and integrative approaches. Trends Biotechnol. 2004, 22, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Brunkow, M.E.; Jeffery, E.W.; Hjerrild, K.A.; Paeper, B.; Clark, L.B.; Yasayko, S.A.; Wilkinson, J.E.; Galas, D.; Ziegler, S.F.; Ramsdell, F. Disruption of a new forkhead/winged-helix protein.; scurfin.; results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 2001, 27, 68–73. [Google Scholar] [CrossRef]
- Katoh, M.; Igarashi, M.; Fukuda, H.; Nakagama, H.; Katoh, M. Cancer genetics and genomics of human FOX family genes. Cancer Lett. 2013, 328, 198–206. [Google Scholar] [CrossRef]
- Tian, T.; Wang, M.; Zheng, Y.; Yang, T.; Zhu, W.; Li, H.; Lin, S.; Liu, K.; Xu, P.; Deng, Y.; et al. Association of two FOXP3 polymorphisms with breast cancer susceptibility in Chinese Han women. Cancer Manag. Res. 2018, 10, 867–872. [Google Scholar] [CrossRef]
- Luo, Q.; Zhang, S.; Wei, H.; Pang, X.; Zhang, H. Roles of Foxp3 in the occurrence and development of cervical cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 8717–8730. [Google Scholar]
- Sun, X.; Feng, Z.; Wang, Y.; Qu, Y.; Gai, Y. Expression of Foxp3 and its prognostic significance in colorectal cancer. Int. J. Immunopathol. Pharmacol. 2017, 30, 201–206. [Google Scholar] [CrossRef]
- Liu, Y.; Lan, Q.; Lu, L.; Chen, M.; Xia, Z.; Ma, J.; Wang, J.; Fan, H.; Shen, Y.; Ryffel, B. Phenotypic and functional characteristic of a newly identified CD8+ Foxp3− CD103+ regulatory T cells. J. Mol. Cell Biol. 2014, 6, 81–92. [Google Scholar] [CrossRef]
- Kim, M.; Grimmig, T.; Grimm, M.; Lazariotou, M.; Meier, E.; Rosenwald, A.; Tsaur, I.; Blaheta, R.; Heemann, U.; Germer, C.T. Expression of Foxp3 in colorectal cancer but not in Treg cells correlates with disease progression in patients with colorectal cancer. PLoS ONE 2013, 8, e53630. [Google Scholar] [CrossRef]
- Kuwahara, T.; Hazama, S.; Suzuki, N.; Yoshida, S.; Tomochika, S.; Nakagami, Y.; Matsui, H.; Shindo, Y.; Kanekiyo, S.; Tokumitsu, Y. Intratumoural-infiltrating CD4 + and FOXP3 + T cells as strong positive predictive markers for the prognosis of resectable colorectal cancer. Br. J. Cancer 2019, 121, 659–665. [Google Scholar] [CrossRef] [PubMed]
- Grimmig, T.; Kim, M.; Germer, C.-T.; Gasser, M.; Maria Waaga-Gasser, A. The role of FOXP3 in disease progression in colorectal cancer patients. Oncoimmunology 2013, 2, e24521. [Google Scholar] [CrossRef] [PubMed]
- Bessis, N.; Chiocchia, G.; Kollias, G.; Minty, A.; Fournier, C.; Fradelizi, D.; Boissier, M.C. Modulation of proinflammatory cytokine production in tumour necrosis factor-alpha (TNF-alpha)-transgenic mice by treatment with cells engineered to secrete IL-4.; IL-10 or IL-13. Clin. Exp. Immunol. 1998, 111, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Bates, R.C.; Mercurio, A.M. Tumor necrosis factor-alpha stimulates the epithelial-to-mesenchymal transition of human colonic organoids. Mol. Biol. Cell. 2003, 14, 1790–1800. [Google Scholar] [CrossRef] [PubMed]
- Ou, B.; Zhao, J.; Guan, S.; Feng, H.; Wangpu, X.; Zhu, C.; Zong, Y.; Ma, J.; Sun, J.; Shen, X.; et al. CCR4 promotes metastasis via ERK/NF-κB/MMP13 pathway and acts downstream of TNF-α in colorectal cancer. Oncotarget 2016, 7, 47637. [Google Scholar] [CrossRef] [PubMed]
- Jovanovic, D.V.; Di Battista, J.A.; Martel-Pelletier, J.; Jolicoeur, F.C.; He, Y.; Zhang, M.; Mineau, F.; Pelletier, J.P. IL-17 stimulates the production and expression of proinflammatory cytokines.; IL-beta and TNF-alpha. by human macrophages. J. Immunol. 1998, 160, 3513–3521. [Google Scholar] [CrossRef]
- Hunter, K. Host genetics influence tumour metastasis. Nat. Rev. Cancer 2006, 6, 141–146. [Google Scholar] [CrossRef]
- Al Obeed, O.A.; Alkhayal, K.A.; Al Sheikh, A.; Zubaidi, A.M.; Vaali-Mohammed, M.A.; Boushey, R.; Mckerrow, J.H.; Abdulla, M.H. Increased expression of tumor necrosis factor-α is associated with advanced colorectal cancer stages. World J. Gastroenterol. 2014, 20, 18390–18396. [Google Scholar] [CrossRef]
- Grimm, M.; Kim, M.; Rosenwald, A.; von Raden, B.H.; Tsaur, I.; Meier, E.; Heemann, U.; Germer, C.T.; Gasser, M.; Waaga-Gasser, A.M. Tumour-mediated TRAIL-Receptor expression indicates effective apoptotic depletion of infiltrating CD8+ immune cells in clinical colorectal cancer. Eur. J. Cancer 2010, 46, 2314–2323. [Google Scholar] [CrossRef]
- Stanilov, N.; Miteva, L.; Dobreva, Z.; Stanilova, S. Colorectal cancer severity and survival in correlation with tumour necrosis factor-alpha. Biotechnol. Biotechnol. Equip. 2014, 28, 911–917. [Google Scholar] [CrossRef]
- Warsinggih; Limanu, F.; Labeda, I.; Lusikooy, R.E.; Mappincara; Faruk, M. The relationship of tumor necrosis factor alpha levels in plasma toward the stage and differentiation degree in colorectal cancer. Med. Clínica Práctica 2021, 4, 100224. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Y.; Song, Z.; Chu, J.; Qu, X. Deficiency of interferon-gamma or its receptor promotes colorectal cancer development. J. Interferon Cytokine Res. 2015, 35, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Kosmidis, C.; Sapalidis, K.; Koletsa, T.; Kosmidou, M.; Efthimiadis, C.; Anthimidis, G.; Varsamis, N.; Michalopoulos, N.; Koulouris, C.; Atmatzidis, S.; et al. Interferon-γ and Colorectal Cancer: An up-to date. J. Cancer 2018, 9, 232–238. [Google Scholar] [CrossRef]
- Liu, S.; Yu, X.; Wang, Q.; Liu, Z.; Xiao, Q.; Hou, P.; Hu, Y.; Hou, W.; Yang, Z.; Guo, D.; et al. Specific expression of interferon-γ induced by synergistic activation mediator-derived systems activates innate immunity and inhibits tumorigenesis. J. Microbiol. Biotechnol. 2017, 27, 1855–1866. [Google Scholar] [CrossRef] [PubMed]
- Schroder, K.; Hertzog, P.J.; Ravasi, T.; Hume, D.A. Interferon-gamma: An overview of signals.; mechanisms and functions. J. Leukoc. Biol. 2004, 75, 163–189. [Google Scholar] [CrossRef]
- Jorgovanovic, D.; Song, M.; Wang, L.; Zhang, Y. Roles of IFN-γ in tumor progression and regression: A review. Biomark. Res. 2020, 8, 49. [Google Scholar] [CrossRef]
- Bellucci, R.; Martin, A.; Bommarito, D.; Wang, K.; Hansen, S.H.; Freeman, G.J.; Ritz, J. Interferon-γ-induced activation of JAK1 and JAK2 suppresses tumor cell susceptibility to NK cells through upregulation of PD-L1 expression. Oncoimmunology 2015, 4, e1008824. [Google Scholar] [CrossRef]
- Mimura, K.; The, J.L.; Okayama, H.; Shiraishi, K.; Kua, L.F.; Koh, V.; Smoot, D.T.; Ashktorab, H.; Oike, T.; Suzuki, Y.; et al. PD-L1 expression is mainly regulated by interferon gamma associated with JAK-STAT pathway in gastric cancer. Cancer Sci. 2018, 109, 43–53. [Google Scholar] [CrossRef]
- Zhao, T.; Li, Y.; Zhang, J.; Zhang, B. PD-L1 expression increased by IFN-γ via JAK2-STAT1 signaling and predicts a poor survival in colorectal cancer. Oncol. Lett. 2020, 20, 1127–1134. [Google Scholar] [CrossRef]
- Calik, I.; Calik, M.; Turken, G.; Ozercan, I.H.; Dagli, A.F.; Artas, G.; Sarikaya, B. Intratumoral Cytotoxic T-Lymphocyte Density and PD-L1 Expression Are Prognostic Biomarkers for Patients with Colorectal Cancer. Medicina 2019, 55, 723. [Google Scholar] [CrossRef]
- Carson, W.E.; Dierksheide, J.E.; Jabbour, S.; Anghelina, M.; Bouchard, P.; Ku, G.; Yu, H.; Baumann, H.; Shah, M.H.; Cooper, M.A.; et al. Coadministration of interleukin-18 and interleukin-12 induces a fatal inflammatory response in mice: Critical role of natural killer cell interferon-γ production and STAT-mediated signal transduction. Blood 2000, 96, 1465–1473. [Google Scholar] [CrossRef]
- Chaisavaneeyakorn, S.; Othoro, C.; Shi, Y.P.; Otieno, J.; Chaiyaroj, S.C.; Lal, A.A.; Udhayakumar, V. Relationship between plasma Interleukin-12 (IL-12) and IL-18 levels and severe malarial anemia in an area of holoendemicity in western Kenya. Clin. Diagn. Lab. Immunol. 2003, 10, 362–366. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Zhang, Z.; Sun, P.; Song, G.; Wang, L.; Sun, Z.; Yuan, N.; Wang, Q.; Lun, L. Interleukin-18 Is a Prognostic Marker and Plays a Tumor Suppressive Role in Colon Cancer. Disease Markers 2020, 2020, 6439614. [Google Scholar] [CrossRef] [PubMed]
- Braumüller, H.; Mauerer, B.; Andris, J.; Berlin, C.; Wieder, T.; Kesselring, R. The Cytokine Network in Colorectal Cancer: Implications for New Treatment Strategies. Cells 2023, 12, 138. [Google Scholar] [CrossRef] [PubMed]
- Briukhovetska, D.; Dörr, J.; Endres, S.; Libby, P.; Dinarello, C.A.; Kobold, S. Interleukins in cancer: From biology to therapy. Nat. Rev. Cancer 2021, 21, 481–499. [Google Scholar] [CrossRef]
- Chen, X.W.; Zhou, S.F. Inflammation, cytokines, the IL-17/IL-6/STAT3/NF-κB axis, and tumorigenesis. Drug. Des. DevelTher. 2015, 9, 2941–2946. [Google Scholar] [CrossRef]
- Cheng, K.J.; Mejia, M.E.H.; Khong, T.L.; Mohd, Z.S.; Thavagnanam, S.; Ibrahim, Z.A. IL-1α and colorectal cancer pathogenesis: Enthralling candidate for anti-cancer therapy. Crit. Rev. Oncol. Hematol. 2021, 163, 103398. [Google Scholar] [CrossRef]
- Li, Y.; Wang, L.; Pappan, L. IL-1β promotes stemness and invasiveness of colon cancer cells through Zeb1 activation. Mol. Cancer 2012, 11, 87. [Google Scholar] [CrossRef]
- Lubberink, M.; Golla, S.S.; Jonasson, M.; Rubin, K.; Glimelius, B.; Sörensen, J.; Nygren, P. (15)O-Water PET Study of the Effect of Imatinib.; a Selective Platelet-Derived Growth Factor Receptor Inhibitor.; Versus Anakinra.; an IL-1R Antagonist.; on Water-Perfusable Tissue Fraction in Colorectal Cancer Metastases. J. Nucl. Med. 2015, 56, 1144–1149. [Google Scholar] [CrossRef]
- Pastille, E.; Wasmer, M.H.; Adamczyk, A.; Vu, V.P.; Mager, L.F.; Phuong, N.N.T.; Palmieri, V.; Simillion, C.; Hansen, W.; Kasper, S.; et al. The IL-33/ST2 pathway shapes the regulatory T cell phenotype to promote intestinal cancer. Mucosal Immunol. 2019, 12, 990–1003. [Google Scholar] [CrossRef]
- Bergman, M.; Levin, G.S.; Bessler, H.; Djaldetti, M.; Salman, H. Resveratrol affects the cross talk between immune and colon cancer cells. Biomed. Pharmacother. 2013, 67, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Idris, A.; Ghazali, N.B.; Koh, D. Interleukin 1β—A Potential Salivary Biomarker for Cancer Progression? Biomark. Cancer 2015, 7, BIC.S25375. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Wang, F.; Ma, C.; Hao, T.; Geng, L.; Jiang, H. Predictive value of IL-18 and IL-10 in the prognosis of patients with colorectal cancer. Oncol. Lett. 2019, 18, 713–719. [Google Scholar] [CrossRef] [PubMed]
- Nikiteas, N.; Yannopoulos, A.; Chatzitheofylaktou, A.; Tsigris, C. Heterozygosity for Interleukin-18 -607 A/C Polymorphism is Associated with Risk for Colorectal Cancer. Anticancer Res. 2007, 27, 3849–3853. Available online: https://ar.iiarjournals.org/content/anticanres/27/6B/3849.full.pdf (accessed on 17 March 2023). [PubMed]
- Misa, B.I.; Diakowska, D.; Korpacka, K.M. Local and Systemic IL-7 Concentration in Gastrointestinal-Tract Cancers. Medicina 2019, 55, 262. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Gong, C.; Mao, H.; Li, Z.; Fang, Z.; Chen, Q.; Lin, M.; Jiang, X.; Hu, Y.; Wang, W.; et al. E2F1/SP3/STAT6 axis is required for IL-4-induced epithelial-mesenchymal transition of colorectal cancer cells. Int. J. Oncol. 2018, 53, 567–578. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhu, L.; Lu, X.; Bian, H.; Wu, X.; Yang, W.; Qin, Q. IL-33/ST2 pathway contributes to metastasis of human colorectal cancer. Biochem. Biophys. Res. Commun. 2014, 453, 486–492. [Google Scholar] [CrossRef]
- Rocca, Y.S.; Roberti, M.P.; Juliá, E.P.; Pampena, M.B.; Bruno, L.; Rivero, S.; Huertas, E.; Sánchez Loria, F.; Pairola, A.; Caignard, A.; et al. Phenotypic and Functional Dysregulated Blood NK Cells in Colorectal Cancer Patients Can Be Activated by Cetuximab Plus IL-2 or IL-15. Front. Immunol. 2016, 7, 413. [Google Scholar] [CrossRef]
- Wang, C.; Lu, Y.; Chen, L.; Gao, T.; Yang, Q.; Zhu, C.; Chen, Y. Th9 cells are subjected to PD-1/PD-L1-mediated inhibition and are capable of promoting CD8 T cell expansion through IL-9R in colorectal cancer. Int. Immunopharmacol. 2020, 78, 106019. [Google Scholar] [CrossRef]
- Fukushima, Y.; Iinuma, H.; Tsukamoto, M.; Matsuda, K.; Hashiguchi, Y. Clinical significance of microRNA-21 as a biomarker in each Dukes’ stage of colorectal cancer. Oncol. Rep. 2015, 33, 573–582. [Google Scholar] [CrossRef]
- Steele, N.; Anthony, A.; Saunders, M.; Esmarck, B.; Ehrnrooth, E.; Kristjansen, P.E.; Nihlén, A.; Hansen, L.T.; Cassidy, J. A phase 1 trial of recombinant human IL-21 in combination with cetuximab in patients with metastatic colorectal cancer. Br. J. Cancer 2012, 106, 793–798. [Google Scholar] [CrossRef] [PubMed]
- Stolfi, C.; Rizzo, A.; Franzè, E.; Rotondi, A.; Fantini, M.C.; Sarra, M.; Caruso, R.; Monteleone, I.; Sileri, P.; Franceschilli, L. Involvement of interleukin-21 in the regulation of colitis-associated colon cancer. J. Exp. Med. 2011, 208, 2279–2290. [Google Scholar] [CrossRef] [PubMed]
- Belluco, C.; Nitti, D.; Frantz, M.; Toppan, P.; Basso, D.; Plebani, M.; Lise, M.; Jessup, J.M. Interleukin-6 Blood Level Is Associated with Circulating Carcinoembryonic Antigen and Prognosis in Patients with Colorectal Cancer. Ann. Surg. Oncol. 2000, 7, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Knüpfer, H.; Preiss, R. Serum interleukin-6 levels in colorectal cancer patients—A summary of published results. Int. J. Color. Dis. 2010, 25, 135–140. [Google Scholar] [CrossRef]
- St John, M.A.; Li, Y.; Zhou, X.; Denny, P.; Ho, C.M.; Montemagno, C.; Shi, W.; Qi, F.; Wu, B.; Sinha, U.; et al. Interleukin 6 and Interleukin 8 as Potential Biomarkers for Oral Cavity and Oropharyngeal Squamous Cell Carcinoma. Arch. Otolaryngol. Head Neck Surg. 2004, 130, 929–935. [Google Scholar] [CrossRef] [PubMed]
- Vainer, N.; Dehlendorff, C.; Johansen, J.S. Systematic literature review of IL-6 as a biomarker or treatment target in patients with gastric, bile duct, pancreatic and colorectal cancer. Oncotarget 2018, 9, 29820–29841. [Google Scholar] [CrossRef]
- Huynh, J.; Chand, A.; Ernst, M. IL-11 as a therapeutic target to treat colorectal cancer. Cancer Res. 2018, 78, 5733. [Google Scholar] [CrossRef]
- Putoczki, T.L.; Thiem, S.; Loving, A.; Busuttil, R.A.; Wilson, N.J.; Ziegler, P.K.; Nguyen, P.M.; Preaudet, A.; Farid, R.; Edwards, K.M.; et al. Interleukin-11 is the dominant IL-6 family cytokine during gastrointestinal tumorigenesis and can be targeted therapeutically. Cancer Cell 2013, 24, 257–271. [Google Scholar] [CrossRef]
- Yoshizaki, A.; Nakayama, T.; Yamazumi, K.; Yakata, Y.; Taba, M.; Sekine, I. Expression of interleukin (IL)-11 and IL-11 receptor in human colorectal adenocarcinoma: IL-11 up-regulation of the invasive and proliferative activity of human colorectal carcinoma cells. Int. J. Oncol. 2006, 29, 869–876. [Google Scholar] [CrossRef]
- Sun, Q.; Sun, F.; Wang, B.; Liu, S.; Niu, W.; Liu, E.; Peng, C.; Wang, J.; Gao, H.; Liang, B.; et al. Interleukin-8 promotes cell migration through integrin αvβ6 upregulation in colorectal cancer. Cancer Lett. 2014, 354, 245–253. [Google Scholar] [CrossRef]
- Shimizu, M.; Tanaka, N. IL-8-induced O-GlcNAc modification via GLUT3 and GFAT regulates cancer stem cell-like properties in colon and lung cancer cells. Oncogene 2019, 38, 1520–1533. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.; Chen, W.; Zhang, Z.; Wu, D.; Wu, P.; Chen, Z.; Li, C.; Huang, J. Prognostic value, clinicopathologic features and diagnostic accuracy of interleukin-8 in colorectal cancer: A meta-analysis. PLoS ONE 2015, 10, e0123484. [Google Scholar] [CrossRef] [PubMed]
- Miteva, L.D.; Stanilov, N.S.; Deliysky, T.S.; Stanilova, S.A. Significance of−1082A/G polymorphism of IL10 gene for progression of colorectal cancer and IL-10 expression. Tumor Biol. 2014, 35, 12655–12664. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, R.J.; Greenman, J.; MacDonald, A.W.; Gaskell, K.M.; Topping, K.P.; Duthie, G.S.; Kerin, M.J.; Lee, P.W.; Monson, J.R.J. Advanced colorectal cancer is associated with impaired interleukin 12 and enhanced interleukin 10 production. Clin. Cancer Res. 1998, 4, 1943–1948. Available online: https://clincancerres.aacrjournals.org/content/clincanres/4/8/1943.full.pdf (accessed on 17 March 2023). [PubMed]
- Shouval, D.S.; Biswas, A.; Goettel, J.A.; McCann, K.; Conaway, E.; Redhu, N.S.; Mascanfroni, I.D.; Al Adham, Z.; Lavoie, S.; Ibourk, M.; et al. Interleukin-10 receptor signaling in innate immune cells regulates mucosal immune tolerance and anti-inflammatory macrophage function. Immunity 2014, 40, 706–719. [Google Scholar] [CrossRef] [PubMed]
- Doulabi, H.; Rastin, M.; Shabahangh, H.; Maddah, G.; Abdollahi, A.; Nosratabadi, R.; Esmaeili, S.A.; Mahmoudi, M. Analysis of Th22.; Th17 and CD4(+)cells co-producing IL-17/IL-22 at different stages of human colon cancer. Biomed. Pharmacother. 2018, 103, 1101–1106. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.Z.; Wang, H.Y.; Yang, S.P.; Yuan, Z.P.; Xu, F.Y.; Sun, C.; Shi, R.H. Expression of interleukin-22/STAT3 signaling pathway in ulcerative colitis and related carcinogenesis. World J. Gastroenterol. WJG 2013, 19, 2638. [Google Scholar] [CrossRef]
- Wang, C.; Gong, G.; Sheh, A.; Muthupalani, S.; Bryant, E.M.; Puglisi, D.A.; Holcombe, H.; Conaway, E.A.; Parry, N.A.P.; Bakthavatchalu, V.; et al. Interleukin-22 drives nitric oxide-dependent DNA damage and dysplasia in a murine model of colitis-associated cancer. Mucosal Immunol. 2017, 10, 1504–1517. [Google Scholar] [CrossRef]
- Moseley, T.A.; Haudenschild, D.R.; Rose, L.; Reddi, A.H. Interleukin-17 family and IL-17 receptors. Cytokine Growth Factor Rev. 2013, 14, 155–174. [Google Scholar] [CrossRef]
- Razi, S.; Baradaran, N.B.; Keshavarz-Fathi, M.; Rezaei, N. IL-17 and colorectal cancer: From carcinogenesis to treatment. Cytokine 2019, 116, 7–12. [Google Scholar] [CrossRef]
- Alinejad, V.; Dolati, S.; Motallebnezhad, M.; Yousefi, M. The role of IL17B-IL17RB signaling pathway in breast cancer. Biomed. Pharmacother. 2017, 88, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Shamoun, L.; Skarstedt, M.; Andersson, R.E.; Wågsäter, D.; Dimberg, J. Association study on IL-4, IL-4Rα and IL-13 genetic polymorphisms in Swedish patients with colorectal cancer. Clin. Chim. Acta 2018, 487, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Neurath, M.F. IL-23 in inflammatory bowel diseases and colon cancer. Cytokine Growth Factor Rev. 2019, 45, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, J.; Li, L.; Zhang, J.; Wang, X.; Yang, C.; Li, Y.; Lan, F.; Lin, P. IL-23 selectively promotes the metastasis of colorectal carcinoma cells with impaired Socs3 expression via the STAT5 pathway. Carcinogenesis 2014, 35, 1330–1340. [Google Scholar] [CrossRef]
- Suzuki, H.; Ogawa, H.; Miura, K.; Haneda, S.; Watanabe, K.; Ohnuma, S.; Sasaki, H.; Sase, T.; Kimura, S.; Kajiwara, T.; et al. IL-23 directly enhances the proliferative and invasive activities of colorectal carcinoma. Oncol. Lett. 2012, 4, 199–204. [Google Scholar] [CrossRef]
- Hu, W.H.; Chang, C.D.; Liu, T.T.; Chen, H.H.; Hsiao, C.C.; Kang, H.Y.; Chuang, J.H. Association of sarcopenia and expression of interleukin-23 in colorectal cancer survival. Clin. Nutr. 2021, 40, 5322–5326. [Google Scholar] [CrossRef]
- Voronov, E.; Apte, R.N. IL-1 in Colon Inflammation.; Colon Carcinogenesis and Invasiveness of Colon Cancer. Cancer Microenviron. 2015, 8, 187–200. [Google Scholar] [CrossRef]
- Cohen, I.; Rider, P.; Carmi, Y.; Braiman, A.; Dotan, S.; White, M.R.; Voronov, E.; Martin, M.U.; Dinarello, C.A.; Apte, R.N. Differential release of chromatin-bound IL-1α discriminates between necrotic and apoptotic cell death by the ability to induce sterile inflammation. Proc. Natl. Acad. Sci. USA 2010, 107, 2574–2579. [Google Scholar] [CrossRef]
- Rider, P.; Carmi, Y.; Guttman, O.; Braiman, A.; Cohen, I.; Voronov, E.; White, M.R.; Dinarello, C.A.; Apte, R.N. IL-1alpha and IL-1beta recruit different myeloid cells and promote different stages of sterile inflammation. J. Immunol. 2011, 187, 4835–4843. [Google Scholar] [CrossRef]
- Voronov, E.; Yaron, C.; Ron, N.A. The role IL-1 in tumor-mediated angiogenesis. Front. Physiol. 2014, 5, 114. [Google Scholar] [CrossRef]
- Gunter, M.J.; Canzian, F.; Landi, S.; Chanock, S.J.; Sinha, R.; Rothman, N. Inflammation-related gene polymorphisms and colorectal adenoma. Cancer Epidemiol. Biomark. Prev. 2006, 15, 1126–1131. [Google Scholar] [CrossRef] [PubMed]
- Graziano, F.; Ruzzo, A.; Canestrari, E.; Loupakis, F.; Santini, D.; Rulli, E.; Humar, B.; Galluccio, N.; Bisonni, R.; Floriani, I.; et al. Variations in the interleukin-1 receptor antagonist gene impact on survival of patients with advanced colorectal cancer. Pharm. J. 2009, 9, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Maeda, S.; Hsu, L.C.; Liu, H.; Bankston, L.A.; Iimura, M.; Kagnoff, M.F.; Eckmann, L.; Karin, M. Nod2 mutation in Crohn’s disease potentiates NF-kappaB activity and IL-1beta processing. Science 2005, 307, 734–738. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-P.; Philip, S. Proinflammatory cytokines tumor necrosis factor-alpha and IL-6, but not IL-1, down-regulate the osteocalcin gene promoter. J. Immunol. 1992, 148, 788–794. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, Z.; Deng, B.; Wu, D.; Quan, Y.; Min, Z. Interleukin 1β/1RA axis in colorectal cancer regulates tumor invasion, proliferation and apoptosis via autophagy. Oncol. Rep. 2020, 43, 908–918. [Google Scholar] [CrossRef]
- Kolls, J.K.; Anders, L. Interleukin-17 family members and inflammation. Immunity 2004, 21, 467–476. [Google Scholar] [CrossRef]
- Feng, H.; Ying, R.; Chai, T.; Chen, H.; Ju, H. The association between IL-17 gene variants and risk of colorectal cancer in a Chinese population: A case-control study. Biosci. Rep. 2019, 39, BSR20190013. [Google Scholar] [CrossRef]
- Wu, D.; Wu, P.; Huang, Q.; Liu, Y.; Ye, J.; Huang, J. Interleukin-17: A promoter in colorectal cancer progression. Clin. Dev. Immunol. 2013, 2013, 436307. [Google Scholar] [CrossRef]
- Nemati, K.; Golmoghaddam, H.; Hosseini, S.V.; Ghaderi, A.; Doroudchi, M. Interleukin-17FT7488 allele is associated with a decreased risk of colorectal cancer and tumor progression. Gene 2015, 561, 88–94. [Google Scholar] [CrossRef]
- Tseng, J.Y.; Yang, C.Y.; Liang, S.C.; Liu, R.S.; Yang, S.H.; Lin, J.K.; Chen, Y.M.; Wu, Y.C.; Jiang, J.K.; Lin, C.H. Interleukin-17A modulates circulating tumor cells in tumor draining vein of colorectal cancers and affects metastases. Clin. Cancer Res. 2014, 20, 2885–2897. [Google Scholar] [CrossRef]
- Cui, G.; Yuan, A.; Goll, R.; Florholmen, J. IL-17A in the tumor microenvironment of the human colorectal adenoma-carcinoma sequence. Scand. J. Gastroenterol. 2012, 47, 1304–1312. [Google Scholar] [CrossRef] [PubMed]
- Straus, D.S. TNFα and IL-17 cooperatively stimulate glucose metabolism and growth factor production in human colorectal cancer cells. Mol. Cancer 2013, 12, 78. [Google Scholar] [CrossRef] [PubMed]
- Chae, W.J.; Gibson, T.F.; Zelterman, D.; Hao, L.; Henegariu, O.; Bothwell, A.L. Ablation of IL-17A abrogates progression of spontaneous intestinal tumorigenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 5540–5544. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Duan, Y.; Cheng, X.; Chen, X.; Xie, W.; Long, H.; Lin, Z.; Zhu, B. IL-17 is associated with poor prognosis and promotes angiogenesis via stimulating VEGF production of cancer cells in colorectal carcinoma. Biochem. Res. Commun. 2011, 407, 348–354. [Google Scholar] [CrossRef]
- Sobhani, I.; Tap, J.; Roudot-Thoraval, F.; Roperch, J.P.; Letulle, S.; Langella, P.; Corthier, G.; Van Nhieu, J.T.; Furet, J.P. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS ONE 2011, 6, e16393. [Google Scholar] [CrossRef]
- Radosavljevic, G.; Ljujic, B.; Jovanovic, I.; Srzentic, Z.; Pavlovic, S.; Zdravkovic, N.; Milovanovic, M.; Bankovic, D.; Knezevic, M.; Acimovic, L.; et al. Interleukin-17 may be a valuable serum tumor marker in patients with colorectal carcinoma. Neoplasma 2010, 57, 135–144. [Google Scholar] [CrossRef]
- Al Obeed, O.A.; Vaali-Mohamed, M.A.; Alkhayal, K.A.; Bin Traiki, T.A.; Zubaidi, A.M.; Arafah, M.; Harris, R.A.; Khan, Z.; Abdulla, M.H. IL-17 and colorectal cancer risk in the Middle East: Gene polymorphisms and expression. Cancer Manag. Res. 2018, 10, 2653. [Google Scholar] [CrossRef]
- Le Gouvello, S.; Bastuji-Garin, S.; Aloulou, N.; Mansour, H.; Chaumette, M.T.; Berrehar, F.; Seikour, A.; Charachon, A.; Karoui, M.; Leroy, K.; et al. High prevalence of Foxp3 and IL17 in MMR-proficient colorectal carcinomas. Gut 2008, 57, 772–779. [Google Scholar] [CrossRef]
- Huang, Y.H.; Cao, Y.F.; Jiang, Z.Y.; Zhang, S.; Gao, F. Th22 cell accumulation is associated with colorectal cancer development. World J. Gastroenterol. 2015, 21, 4216–4224. [Google Scholar] [CrossRef]
- Wu, T.; Wang, Z.; Liu, Y.; Mei, Z.; Wang, G.; Liang, Z.; Cui, A.; Hu, X.; Cui, L.; Yang, Y.; et al. Interleukin 22 protects colorectal cancer cells from chemotherapy by activating the STAT3 pathway and inducing autocrine expression of interleukin 8. Clin. Immunol. 2014, 154, 116–126. [Google Scholar] [CrossRef]
- Harrison, C. IL-22: Linking inflammation and cancer. Nat. Rev. Drug. Discov. 2013, 12, 505. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.L.; Plummer, S.J.; Tucker, T.C.; Casey, G.; Li, L. Interleukin-22 genetic polymorphisms and risk of colon cancer. Cancer Causes Control 2010, 21, 1165–1170. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Wang, H.; Deng, L.; Hou, J.; Shi, R.; Yao, M.; Gao, Y.; Yao, A.; Wang, X.; Yu, L.; et al. IL-22 is related to development of human colon cancer by activation of STAT3. BMC Cancer 2013, 13, 59. [Google Scholar] [CrossRef] [PubMed]
- Bobe, G.; Albert, P.S.; Sansbury, L.B.; Lanza, E.; Schatzkin, A.; Colburn, N.H.; Cross, A.J. Interleukin-6 as a Potential Indicator for Prevention of High-Risk Adenoma Recurrence by Dietary Flavonols in the Polyp Prevention Trial. Cancer Prev. Res. 2010, 3, 764–775. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Keku, T.O.; Martin, C.; Galanko, J.; Woosley, J.T.; Schroeder, J.C.; Satia, J.A.; Halabi, S.; Sandler, R.S. Circulating Levels of Inflammatory Cytokines and Risk of Colorectal Adenomas. Cancer Res. 2008, 68, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Xu, F.; Lu, T.; Duan, Z.; Zhang, Z. Interleukin-6 signaling pathway in targeted therapy for cancer. Cancer Treat. Rev. 2012, 38, 904–910. [Google Scholar] [CrossRef]
- Waetzig, G.H.; Rose-John, S. Hitting a complex target: An update on interleukin-6 trans-signalling. Expert Opin. Ther. Targets 2012, 16, 225–236. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, P.; Wu, D.; Zhang, Z.; Hu, G.; Zhao, S.; Lai, Y.; Huang, J. Prognostic and clinicopathological significance of serum interleukin-6 expression in colorectal cancer: A systematic review and meta-analysis. OncoTargets Ther. 2015, 8, 3793–3801. [Google Scholar] [CrossRef]
- Waldner, M.J.; Foersch, S.; Neurath, M.F. Interleukin-6--a key regulator of colorectal cancer development. Int. J. Biol. Sci. 2012, 8, 1248–1253. [Google Scholar] [CrossRef]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef]
- Ham, I.H.; Oh, H.J.; Jin, H.; Bae, C.A.; Jeon, S.M.; Choi, K.S.; Son, S.Y.; Han, S.U.; Brekken, R.A.; Lee, D. Targeting interleukin-6 as a strategy to overcome stroma-induced resistance to chemotherapy in gastric cancer. Mol. Cancer 2019, 18, 68. [Google Scholar] [CrossRef] [PubMed]
- Hue, S.; Ahern, P.; Buonocore, S.; Kullberg, M.C.; Cua, D.J.; McKenzie, B.S.; Powrie, F.; Maloy, K.J. Interleukin-23 drives innate and T cell-mediated intestinal inflammation. J. Exp. Med. 2006, 203, 2473–2483. [Google Scholar] [CrossRef] [PubMed]
- Oppmann, B.; Lesley, R.; Blom, B.; Timans, J.C.; Xu, Y.; Hunte, B.; Vega, F.; Yu, N.; Wang, J.; Singh, K.; et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity 2000, 13, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Langowski, J.L.; Zhang, X.; Wu, L.; Mattson, J.D.; Chen, T.; Smith, K.; Basham, B.; McClanahan, T.; Kastelein, R.A.; Oft, M. IL-23 promotes tumour incidence and growth. Nature 2006, 442, 461–465. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, J.Y.; Yang, Y.J.; Chang, K.; Wang, Q.F.; Kong, Y.Y.; Dai, B. High IL-23+ cells infiltration correlates with worse clinical outcomes and abiraterone effectiveness in patients with prostate cancer. Asian J. Androl. 2022, 24, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Kortylewski, M.; Xin, H.; Kujawski, M.; Lee, H.; Liu, Y.; Harris, T.; Drake, C.; Pardoll, D.; Yu, H. Regulation of the IL-23 and IL-12 Balance by Stat3 Signaling in the Tumor Microenvironment. Cancer Cell 2009, 15, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Grivennikov, S.I.; Wang, K.; Mucida, D.; Stewart, C.A.; Schnabl, B.; Jauch, D.; Taniguchi, K.; Yu, G.Y.; Osterreicher, C.H.; Hung, K.E.; et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 2012, 491, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.C.; Tan, X.Y.; Luxenberg, D.P.; Karim, R.; Dunussi-Joannopoulos, K.; Collins, M.; Fouser, L.A. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J. Exp. Med. 2006, 203, 2271–2279. [Google Scholar] [CrossRef] [PubMed]
- Buonocore, S.; Ahern, P.P.; Uhlig, H.H.; Ivanov, I.I.; Littman, D.R.; Maloy, K.J.; Powrie, F. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature 2010, 464, 1371–1375. [Google Scholar] [CrossRef] [PubMed]
- McGovern, D.; Powrie, F. The IL23 axis plays a key role in the pathogenesis of IBD. Gut 2007, 56, 1333–1336. [Google Scholar] [CrossRef] [PubMed]
- Lan, F.; Zhang, L.; Wu, J.; Zhang, J.; Zhang, S.; Li, K.; Qi, Y.; Lin, P. IL-23/IL-23R: Potential mediator of intestinal tumor progression from adenomatous polyps to colorectal carcinoma. Int. J. Color. Dis. 2011, 26, 1511–1518. [Google Scholar] [CrossRef] [PubMed]
- Lopetuso, L.R.; Scaldaferri, F.; Pizarro, T.T. Emerging role of the interleukin (IL)-33/ST2 axis in gut mucosal wound healing and fibrosis. Fibrogenesis Tissue Repair 2012, 5, 18. [Google Scholar] [CrossRef] [PubMed]
- Garlanda, C.; Dinarello, C.A.; Mantovani, A. The interleukin-1 family: Back to the future. Immunity 2013, 39, 1003–1018. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, J.; Owyang, A.; Oldham, E.; Song, Y.; Murphy, E.; McClanahan, T.K.; Zurawski, G.; Moshrefi, M.; Qin, J.; Li, X.; et al. IL-33.; an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity 2005, 23, 479–490. [Google Scholar] [CrossRef]
- Wood, I.S.; Wang, B.; Trayhurn, P. IL-33.; a recently identified interleukin-1 gene family member.; is expressed in human adipocytes. Biochem. Biophys. Res. Commun. 2009, 384, 105–109. [Google Scholar] [CrossRef]
- Yangngam, S.; Thongchot, S.; Pongpaibul, A.; Vaeteewoottacharn, K.; Pinlaor, S.; Thuwajit, P.; Okada, S.; Hermoso, M.A.; Thuwajit, C. High level of interleukin-33 in cancer cells and cancer-associated fibroblasts correlates with good prognosis and suppressed migration in cholangiocarcinoma. J. Cancer 2020, 11, 6571–6581. [Google Scholar] [CrossRef]
- Fang, K.-M.; Yang, C.-S.; Lin, T.-C.; Chan, T.-C.; Tzeng, S.-F. Induced interleukin-33 expression enhances the tumorigenic activity of rat glioma cells. Neuro-Oncology 2014, 16, 552–566. [Google Scholar] [CrossRef]
- Gao, X.; Wang, X.; Yang, Q.; Zhao, X.; Wen, W.; Li, G.; Lu, J.; Qin, W.; Qi, Y.; Xie, F.; et al. Tumoral expression of IL-33 inhibits tumor growth and modifies the tumor microenvironment through CD8+ T and NK cells. J. Immunol. 2015, 194, 438–445. [Google Scholar] [CrossRef]
- Cui, G.; Qi, H.; Gundersen, M.D.; Yang, H.; Christiansen, I.; Sørbye, S.W.; Goll, R.; Florholmen, J. Dynamics of the IL-33/ST2 network in the progression of human colorectal adenoma to sporadic colorectal cancer. Cancer Immunol. Immunother. 2015, 64, 181–190. [Google Scholar] [CrossRef]
- O’Donnell, C.; Mahmoud, A.; Keane, J.; Murphy, C.; White, D.; Carey, S.; O’Riordain, M.; Bennett, M.W.; Brint, E.; Houston, A. An antitumorigenic role for the IL-33 receptor, ST2L, in colon cancer. Br. J. Cancer. 2016, 114, 37–43. [Google Scholar] [CrossRef]
- Lucarini, V.; Ziccheddu, G.; Macchia, I.; La Sorsa, V.; Peschiaroli, F.; Buccione, C.; Sistigu, A.; Sanchez, M.; Andreone, S.; D’Urso, M.T.; et al. IL-33 restricts tumor growth and inhibits pulmonary metastasis in melanoma-bearing mice through eosinophils. Oncoimmunology 2017, 6, e1317420. [Google Scholar] [CrossRef] [PubMed]
- Santana Carrero, R.M.; Beceren-Braun, F.; Rivas, S.C.; Hegde, S.M.; Gangadharan, A.; Plote, D.; Pham, G.; Anthony, S.M.; Schluns, K.S. IL-15 is a component of the inflammatory milieu in the tumor microenvironment promoting antitumor responses. Proc. Natl. Acad. Sci. USA 2019, 116, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Klebanoff, C.A.; Finkelstein, S.E.; Surman, D.R.; Lichtman, M.K.; Gattinoni, L.; Theoret, M.R.; Grewal, N.; Spiess, P.J.; Antony, P.A.; Palmer, D.C. IL-15 enhances the in vivo antitumor activity of tumor-reactive CD8+ T cells. Proc. Natl. Acad. Sci. USA 2004, 101, 1969–1974. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Bamford, R.N.; Waldmann, T.A. IL-15-dependent CD8+ CD122+ T cells ameliorate experimental autoimmune encephalomyelitis by modulating IL-17 production by CD4+ T cells. Eur. J. Immunol. 2014, 44, 3330–3341. [Google Scholar] [CrossRef] [PubMed]
- Mlecnik, B.; Bindea, G.; Angell, H.K.; Sasso, M.S.; Obenauf, A.C.; Fredriksen, T.; Lafontaine, L.; Bilocq, A.M.; Kirilovsky, A.; Tosolini, M.; et al. Functional network pipeline reveals genetic determinants associated with in situ lymphocyte proliferation and survival of cancer patients. Sci. Transl. Med. 2014, 6, 228ra237. [Google Scholar] [CrossRef] [PubMed]
- Curran, M.A.; Geiger, T.L.; Montalvo, W.; Kim, M.; Reiner, S.L.; Al-Shamkhani, A.; Sun, J.C.; Allison, J.P. Systemic 4-1BB activation induces a novel T cell phenotype driven by high expression of Eomesodermin. J. Exp. Med. 2013, 210, 743–755. [Google Scholar] [CrossRef]
- Kuniyasu, H.; Oue, N.; Nakae, D.; Tsutsumi, M.; Denda, A.; Tsujiuchi, T.; Yokozaki, H.; Yasui, W. Interleukin-15 expression is associated with malignant potential in colon cancer cells. Pathobiology 2001, 69, 86–95. [Google Scholar] [CrossRef]
- Chen, J.; Li, Z.Y.; Huang, S.Y.; Petersen, E.; Song, H.Q.; Zhou, D.H.; Zhu, X.Q. Protective efficacy of Toxoplasma gondiicalcium-dependent protein kinase 1 (TgCDPK1) adjuvated with recombinant IL-15 and IL-21 against experimental toxoplasmosis in mice. BMC Infect. Dis. 2014, 14, 487. [Google Scholar] [CrossRef]
- Berger, C.; Berger, M.; Hackman, R.C.; Gough, M.; Elliott, C.; Jensen, M.C.; Riddell, S.R. Safety and immunologic effects of IL-15 administration in nonhuman primates. Blood 2009, 114, 2417–2426. [Google Scholar] [CrossRef]
- Pilipow, K.; Roberto, A.; Roederer, M.; Waldmann, T.A.; Mavilio, D.; Lugli, E. IL15 and T-cell Stemness in T-cell-Based Cancer Immunotherapy. Cancer Res. 2015, 75, 5187–5193. [Google Scholar] [CrossRef]
- Lusty, E.; Poznanski, S.M.; Kwofie, K.; Mandur, T.S.; Lee, D.A.; Richards, C.D.; Ashkar, A.A. IL-18/IL-15/IL-12 synergy induces elevated and prolonged IFN-γ production by ex vivo expanded NK cells which is not due to enhanced STAT4 activation. Mol. Immunol. 2017, 88, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, K.; Nakanishi, K.; and Tsutsui, H. Interleukin-18 in Health and Disease. Int. J. Mol. Sci. 2019, 20, 649. [Google Scholar] [CrossRef] [PubMed]
- Gosmann, C.; Frazer, I.H.; Mattarollo, S.R.; Blumenthal, A. IL-18, but not IL-12, induces production of IFN-γ in the immunosuppressive environment of HPV16 E7 transgenic hyperplastic skin. J. Investig. Dermatol. 2014, 134, 2562–2569. [Google Scholar] [CrossRef]
- Formentini, A.; Braun, P.; Fricke, H.; Link, K.-H.; Henne-Bruns, D.; Kornmann, M. Expression of interleukin-4 and interleukin-13 and their receptors in colorectal cancer. Int. J. Color. Dis. 2012, 27, 1369–1376. [Google Scholar] [CrossRef]
- Shi, J.; Song, X.; Traub, B.; Luxenhofer, M.; Kornmann, M. Involvement of IL-4, IL-13 and their receptors in pancreatic cancer. Int. J. Mol. Sci. 2021, 22, 2998. [Google Scholar] [CrossRef]
- Barderas, R.; Bartolomé, R.A.; Fernandez-Acenero, M.J.; Torres, S.; Casal, J.I. High Expression of IL-13 Receptor α2 in Colorectal Cancer Is Associated with Invasion.; Liver Metastasis.; and Poor PrognosisRole of IL-13Rα2 in Colorectal Cancer Metastasis. Cancer Res. 2012, 72, 2780–2790. [Google Scholar] [CrossRef] [PubMed]
- Jin, R.; Hu, S.; Liu, X.; Guan, R.; Lu, L.; Lin, R. Intranasal instillation of miR-410 targeting IL-4/IL-13 attenuates airway inflammation in OVA-induced asthmatic mice. Mol. Med. Rep. 2019, 19, 895–900. [Google Scholar] [CrossRef]
- Wang, H.P.; Wang, Y.Y.; Pan, J.; Cen, R.; Cai, Y.K. Evaluation of specific fecal protein biochips for the diagnosis of colorectal cancer. World J. Gastroenterol. 2014, 20, 1332–1339. [Google Scholar] [CrossRef]
- Cho, Y.A.; Kim, J. Association of IL4, IL13, and IL4R polymorphisms with gastrointestinal cancer risk: A meta-analysis. J. Epidemiol. 2017, 27, 215–220. [Google Scholar] [CrossRef]
- Song, X.; Traub, B.; Shi, J.; Kornmann, M. Possible Roles of Interleukin-4 and -13 and Their Receptors in Gastric and Colon Cancer. Int. J. Mol. Sci. 2021, 22, 727. [Google Scholar] [CrossRef]
- Terada, H.; Urano, T.; Konno, H. Association of interleukin-8 and plasminogen activator system in the progression of colorectal cancer. Eur. Surg. Res. 2005, 37, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Baggiolini, M.; Clark-Lewis, I. Interleukin-8, a chemotactic and inflammatory cytokine. FEBS Lett. 1992, 307, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Ning, Y.; Lenz, H.-J. Targeting IL-8 in colorectal cancer. Expert Opin. Ther. Targets. 2012, 16, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.J.; Xu, J.M.; Xu, W.L.; Gu, D.H.; Li, P.W. Diagnostic value of interleukin-8 in colorectal cancer: A case-control study and meta-analysis. World J. Gastroenterol. 2014, 20, 16334. [Google Scholar] [CrossRef]
- Itoh, Y.; Joh, T.; Tanida, S.; Sasaki, M.; Kataoka, H.; Itoh, K.; Oshima, T.; Ogasawara, N.; Togawa, S.; Wada, T.; et al. IL-8 promotes cell proliferation and migration through metalloproteinase-cleavage proHB-EGF in human colon carcinoma cells. Cytokine 2005, 29, 275–282. [Google Scholar] [CrossRef]
- Rubie, C.; Frick, V.O.; Pfeil, S.; Wagner, M.; Kollmar, O.; Kopp, B.; Graber, S.; Rau, B.M.; Schilling, M.K. Correlation of IL-8 with induction.; progression and metastatic potential of colorectal cancer. World J. Gastroenterol. 2007, 13, 4996. [Google Scholar] [CrossRef]
- Galffy, G.; Mohammed, K.A.; Dowling, P.A.; Nasreen, N.; Ward, M.J.; Antony, V.B. Interleukin 8: An autocrine growth factor for malignant mesothelioma. Cancer Res. 1999, 59, 367–371. [Google Scholar]
- Brew, R.; Erikson, J.S.; West, D.C.; Kinsella, A.R.; Slavin, J.; Christmas, S.E. Interleukin-8 as an autocrine growth factor for human colon carcinoma cells in vitro. Cytokine 2000, 12, 78–85. [Google Scholar] [CrossRef]
- Haqqani, A.S.; Sandhu, J.K.; Birnboim, H.C. Expression of interleukin-8 promotes neutrophil infiltration and genetic instability in mutatect tumors. Neoplasia 2000, 2, 561–568. [Google Scholar] [CrossRef]
- Bazzichetto, C.; Milella, M.; Zampiva, I.; Simionato, F.; Amoreo, C.A.; Buglioni, S.; Pacelli, C.; Le Pera, L.; Colombo, T.; Bria, E.; et al. Interleukin-8 in Colorectal Cancer: A Systematic Review and Meta-Analysis of Its Potential Role as a Prognostic Biomarker. Biomedicines 2022, 10, 2631. [Google Scholar] [CrossRef]
- Lee, Y.S.; Choi, I.; Ning, Y.; Kim, N.Y.; Khatchadourian, V.; Yang, D.; Chung, H.K.; Choi, D.; LaBonte, M.J.; Ladner, R.D.; et al. Interleukin-8 and its receptor CXCR2 in the tumour microenvironment promote colon cancer growth.; progression and metastasis. Br. J. Cancer 2012, 106, 1833–1841. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, P.J.; Gallagher, R.; Seaton, A.; Wilson, C.; Scullin, P.; Pettigrew, J.; Stratford, I.J.; Williams, K.J.; Johnston, P.G.; Waugh, D.J. HIF-1 and NF-κB-mediated upregulation of CXCR1 and CXCR2 expression promotes cell survival in hypoxic prostate cancer cells. Oncogene 2007, 26, 7333–7345. [Google Scholar] [CrossRef]
- Moreno-Guerrero, S.S.; Ramírez-Pacheco, A.; Rocha-Ramírez, L.M.; Hernández-Pliego, G.; Eguía-Aguilar, P.; Escobar-Sánchez, M.A.; Reyes-López, A.; Juárez-Villegas, L.E.; Sienra-Monge, J.J.L. Association of Genetic Polymorphisms and Serum Levels of IL-6 and IL-8 with the Prognosis in Children with Neuroblastoma. Cancers 2021, 13, 529. [Google Scholar] [CrossRef] [PubMed]
- Gordon, M.A.; Gil, J.; Lu, B.; Zhang, W.; Yang, D.; Yun, J.; Schneider, S.; Groshen, S.; Iqbal, S.; Press, O.A.; et al. Genomic profiling associated with recurrence in patients with rectal cancer treated with chemoradiation. Pharmacogenomics 2006, 7, 67–88. [Google Scholar] [CrossRef] [PubMed]
- Ning, Y.; Manegold, P.C.; Hong, Y.K.; Zhang, W.; Pohl, A.; Lurje, G.; Winder, T.; Yang, D.; LaBonte, M.J.; Wilson, P.M.; et al. Interleukin-8 is vivo in colon cancer cell line models. Int. J. Cancer 2011, 128, 2038–2049. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, A.; Lafferton, B.; Plotz, G.; Zeuzem, S.; Brieger, A. Expression and secretion of the pro-inflammatory cytokine IL-8 is increased in colorectal cancer cells following the knockdown of non-erythroid spectrin αII. Int. J. Oncol. 2020, 56, 1551–1564. [Google Scholar] [CrossRef]
- Howlett, M.; Giraud, A.S.; Lescesen, H.; Jackson, C.B.; Kalantzis, A.; Van Driel, I.R.; Robb, L.; Van der Hoek, M.; Ernst, M.; Minamoto, T.; et al. The interleukin-6 family cytokine interleukin-11 regulates homeostatic epithelial cell turnover and promotes gastric tumor development. Gastroenterology 2009, 136, 967–977. [Google Scholar] [CrossRef]
- Jones, S.A.; Jenkins, B.J. Recent insights into targeting the IL-6 cytokine family in inflammatory diseases and cancer. Nat. Rev. Immunol. 2018, 18, 773–789. [Google Scholar] [CrossRef]
- Yamazumi, K.; Nakayama, T.; Kusaba, T.; Wen, C.Y.; Yoshizaki, A.; Yakata, Y.; Nagayasu, T.; Sekine, I. Expression of interleukin-11 and interleukin-11 receptor alpha in human colorectal adenocarcinoma; immunohistochemical analyses and correlation with clinicopathological factors. World J. Gastroenterol. 2006, 12, 317–321. [Google Scholar] [CrossRef]
- West, N.R. Coordination of immune-stroma crosstalk by IL-6 family cytokines. Front. Immunol. 2019, 10, 1093. [Google Scholar] [CrossRef]
- Kiessling, S.; Muller-Newen, G.; Leeb, S.N.; Hausmann, M.; Rath, H.C.; Strater, J.; Spottl, T.; Schlottmann, K.; Grossmann, J.; Montero-Julian, F.A.; et al. Functional expression of the interleukin-11 receptor α-chain and evidence of antiapoptotic effects in human colonic epithelial cells. J. Biol. Chem. 2004, 279, 10304–10315. [Google Scholar] [CrossRef] [PubMed]
- Nishina, T.; Deguchi, Y.; Ohshima, D.; Wakami, T.; Masato, O.; Shigeyuki, S.; Saroshi, U.; Soh, Y.; Mika, K.; Eri, N.; et al. Interleukin-11-expressing fibroblasts have a unique gene signature correlated with poor prognosis of colorectal cancer. Nat. Commun. 2021, 12, 2281. [Google Scholar] [CrossRef] [PubMed]
- Andoh, A.; Zhang, Z.; Inatomi, O.; Fujino, S.; Deguchi, Y.; Araki, Y.; Tsujikawa, T.; Kitoh, K.; Kim-Mitsuyama, S.; Takayanagi, A.; et al. Interleukin-22.; a member of the IL-10 subfamily.; induces inflammatory responses in colonic subepithelial myofibroblasts. Gastroenterology 2005, 129, 969–984. [Google Scholar] [CrossRef]
- Bamba, S.; Andoh, A.; Yasui, H.; Makino, J.; Kim, S.; Fujiyama, Y. Regulation of IL-11 expression in intestinal myofibroblasts: Role of c-Jun AP-1-and MAPK-dependent pathways. Am. J. Physiol. Gastrointest. Liver Physiol. 2003, 285, G529–G538. [Google Scholar] [CrossRef] [PubMed]
- Putoczki, T.; Ernst, M. More than a sidekick: The IL-6 family cytokine IL-11 links inflammation to cancer. J. Leukoc. Biol. 2010, 88, 1109–1117. [Google Scholar] [CrossRef]
- Klein, W.; Tromm, A.; Griga, T.; Fricke, H.; Folwaczny, C.; Hocke, M.; Eitner, K.; Marx, M.; Duerig, N.; Epplen, J.T. A polymorphism in the IL11 gene is associated with ulcerative colitis. Genes Immun. 2002, 3, 494–496. [Google Scholar] [CrossRef]
- Sabzevary-Ghahfarokhi, M.; Shohan, M.; Shirzad, H.; Rahimian, G.; Bagheri, N.; Soltani, A.; Deris, F.; Ghatreh-Samani, M.; Razmara, E. The expression analysis of Fra-1 gene and IL-11 protein in Iranian patients with ulcerative colitis. BMC Immunol. 2018, 19, 17. [Google Scholar] [CrossRef]
- Nishina, T.; Deguchi, Y.; Ohshima, D.; Takeda, W.; Ohtsuka, M.; Shichino, S.; Ueha, S.; Yamazaki, S.; Kawauchi, M.; Nakamura, E.; et al. Interleukin-11 is a Marker for Both Cancer-and Inflammation-Associated Fibroblasts that Contribute to Colorectal Cancer Progression. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ault, P.; Kantarjian, H.; Welch, M.A.; Giles, F.; Rios, M.B.; Cortes, J. Interleukin 11 may improve thrombocytopenia associated with imatinib mesylate therapy in chronic myelogenous leukemia. Leuk. Res. 2004, 28, 613–618. [Google Scholar] [CrossRef]
- Espinosa-Cotton, M.; Rodman Iii, S.N.; Ross, K.A.; Jensen, I.J.; Sangodeyi-Miller, K.; McLaren, A.J.; Dahl, R.A.; Gibson-Corley, K.N.; Koch, A.T.; Fu, Y.X.; et al. Interleukin-1 alpha increases anti-tumor efficacy of cetuximab in head and neck squamous cell carcinoma. J. Immunother. Cancer 2019, 7, 79. [Google Scholar] [CrossRef]
- Malik, A.; Sharma, D.; Zhu, Q.; Karki, R.; Guy, C.S.; Vogel, P.; Kanneganti, T.D. IL-33 regulates the IgA-microbiota axis to restrain IL-1α-dependent colitis and tumorigenesis. J. Clin. Investig. 2016, 126, 4469–4481. [Google Scholar] [CrossRef] [PubMed]
- Maywald, R.L.; Doerner, S.K.; Pastorelli, L.; De, S.C.; Benton, S.M.; Dawson, E.P.; Lanza, D.G.; Berger, N.A.; Markowitz, S.D.; Lenz, H.J.; et al. IL-33 activates tumor stroma to promote intestinal polyposis. Proc. Natl. Acad. Sci. USA 2015, 112, E2487–E2496. [Google Scholar] [CrossRef] [PubMed]
- Schiering, C.; Krausgruber, T.; Chomka, A.; Fröhlich, A.; Adelmann, K.; Wohlfert, E.A.; Pott, J.; Griseri, T.; Bollrath, J.; Hegazy, A.N.; et al. The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature 2014, 513, 564–568. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, S.; Elhance, A.; Van Duzer, A.; Kumar, S.; Leitenberger, J.J.; Oshimori, N. Tumor-initiating cells establish an IL-33-TGF-β niche signaling loop to promote cancer progression. Science 2020, 369, eaay1813. [Google Scholar] [CrossRef]
- Herzyk, D.J.; Bugelski, P.J.; Hart, T.K.; Wier, P.J. Preclinical safety of recombinant human interleukin-18. Toxicol. Pathol. 2003, 31, 554–561. [Google Scholar] [CrossRef]
- Zhu, B.; Luo, J.; Jiang, Y.; Yu, L.; Liu, M.; Fu, J. Prognostic significance of nomograms integrating IL-37 expression, neutrophil level, and MMR status in patients with colorectal cancer. Cancer Med. 2018, 7, 3682–3694. [Google Scholar] [CrossRef]
- Weinstein, A.M.; Giraldo, N.A.; Petitprez, F.; Julie, C.; Lacroix, L.; Peschaud, F.; Emile, J.F.; Marisa, L.; Fridman, W.H.; Storkus, W.J.; et al. Association of IL-36γ with tertiary lymphoid structures and inflammatory immune infiltrates in human colorectal cancer. Cancer Immunol Immunother. 2019, 68, 109–120. [Google Scholar] [CrossRef]
- Kinoshita, F.; Tagawa, T.; Akamine, T.; Takada, K.; Yamada, Y.; Oku, Y.; Kosai, K.; Ono, Y.; Tanaka, K.; Wakasu, S.; et al. Interleukin-38 promotes tumor growth through regulation of CD8+ tumor-infiltrating lymphocytes in lung cancer tumor microenvironment. Cancer Immunol. Immunother. 2021, 70, 123–135. [Google Scholar] [CrossRef]
- van de Veerdonk, F.L.; de Graaf, D.M.; Joosten, L.A.; Dinarello, C.A. Biology of IL-38 and its role in disease. Immunol. Rev. 2018, 281, 191–196. [Google Scholar] [CrossRef]
- Damoiseaux, J. The IL-2–IL-2 receptor pathway in health and disease: The role of the soluble IL-2 receptor. Clin. Immunol. 2020, 218, 108515. [Google Scholar] [CrossRef]
- Suzuki, A.; Leland, P.; Joshi, B.H.; Puri, R.K. Targeting of IL-4 and IL-13 receptors for cancer therapy. Cytokine 2015, 75, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Krzystek-Korpacka, M.; Zawadzki, M.; Neubauer, K.; Bednarz-Misa, I.; Górska, S.; Wiśniewski, J.; Witkiewicz, W.; Gamian, A. Elevated systemic interleukin-7 in patients with colorectal cancer and individuals at high risk of cancer: Association with lymph node involvement and tumor location in the right colon. Cancer Immunol. Immunother. 2017, 66, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Zhu, Z.; Xiao, H.; Wakefield, M.R.; Ding, V.A.; Bai, Q.; Fang, Y. The role of IL-7 in Immunity and Cancer. Anticancer Res. 2017, 37, 963–967. [Google Scholar] [PubMed]
- Angkasekwinai, P.; Chen, D. IL-9-producing T cells: Potential players in allergy and cancer. Nat. Rev. Immunol. 2021, 21, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Song, L.; Wei, J.; Courtney, A.N.; Gao, X.; Marinova, E.; Guo, L.; Heczey, A.; Asgharzadeh, S.; Kim, E.; et al. IL-15 protects NKT cells from inhibition by tumor-associated macrophages and enhances antimetastatic activity. J. Clin. Investig. 2012, 122, 2221–2233. [Google Scholar] [CrossRef] [PubMed]
- Waldmann, T.A. Cytokines in cancer immunotherapy. Cold Spring Harb. Perspect. Biol. 2018, 10, a028472. [Google Scholar] [CrossRef] [PubMed]
- Dolgin, E. First CD123-targeted drug approved after wowing in rare cancer. Nat. Biotechnol. 2019, 37, 202–204. [Google Scholar] [CrossRef]
- Testa, U.; Elvira, P.; Germana, C. CD123 as a therapeutic target in the treatment of hematological malignancies. Cancers 2019, 11, 1358. [Google Scholar] [CrossRef]
- Grisaru-Tal, S.; Itan, M.; Klion, A.D.; Munitz, A. A new dawn for eosinophils in the tumour microenvironment. Nat. Rev. Cancer 2020, 20, 594–607. [Google Scholar] [CrossRef]
- Hirano, T. IL-6 in inflammation, autoimmunity and cancer. Int. Immunol. 2021, 33, 127–148. [Google Scholar] [CrossRef]
- Cui, G.; Yuan, A.; Sun, Z.; Zheng, W.; Pang, Z. IL-1β/IL-6 network in the tumor microenvironment of human colorectal cancer. Pathol. Res. Pract. 2018, 214, 986–992. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; He, Z.; Ye, J.; Liu, Z.; She, X.; Gao, X.; Liang, R. Progress in Understanding the IL-6/STAT3 Pathway in Colorectal Cancer. OncoTargets Ther. 2020, 13, 13023–13032. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Ye, Y.; Zhang, H.; Szmitkowski, M.; Mäkinen, M.J.; Li, P.; Xia, D.; Yang, J.; Wu, Y.; Wu, H. Diagnostic and Prognostic Value of Serum Interleukin-6 in Colorectal Cancer. Medicine 2016, 95, e2502. [Google Scholar] [CrossRef]
- Grivennikov, S.I. IL-11: A prominent pro-tumorigenic member of the IL-6 family. Cancer Cell 2013, 24, 145–147. [Google Scholar] [CrossRef]
- Naing, A.; Infante, J.R.; Papadopoulos, K.P.; Chan, I.H.; Shen, C.; Ratti, N.P.; Rojo, B.; Autio, K.A.; Wong, D.J.; Patel, M.R.; et al. PEGylated IL-10 (Pegilodecakin) induces systemic immune activation, CD8+ T cell invigoration and polyclonal T cell expansion in cancer patients. Cancer Cell 2018, 34, 775–791. [Google Scholar] [CrossRef]
- Ouyang, W.; O’Garra, A. IL-10 family cytokines IL-10 and IL-22: From basic science to clinical translation. Immunity 2019, 50, 871–891. [Google Scholar] [CrossRef]
- Dumoutier, L.; Lejeune, D.; Colau, D.; Renauld, J.C. Cloning and characterization of IL-22 binding protein.; a natural antagonist of IL-10-related T cell-derived inducible factor/IL-22. J. Immunol. 2001, 166, 7090–7095. [Google Scholar] [CrossRef] [PubMed]
- Markota, A.; Stefan, E.; Sebastian, K. Targeting interleukin-22 for cancer therapy. Hum. Vaccines Immunother. 2018, 14, 2012–2015. [Google Scholar] [CrossRef]
- Hu, H.J.; Liang, X.; Li, H.L.; Du, C.M.; Hao, J.L.; Wang, H.Y.; Gu, J.F.; Ni, A.M.; Sun, L.Y.; Xiao, J.; et al. The armed oncolytic adenovirus ZD55-IL-24 eradicates melanoma by turning the tumor cells from the self-state into the nonself-state besides direct killing. Cell Death Dis. 2020, 11, 1022. [Google Scholar] [CrossRef]
- Menezes, M.E.; Bhatia, S.; Bhoopathi, P.; Das, S.K.; Emdad, L.; Dasgupta, S.; Dent, P.; Wang, X.Y.; Sarkar, D.; Fisher, P.B. MDA-7/IL-24: Multifunctional cancer killing cytokine. Anticancer Genes 2014, 818, 127–153. [Google Scholar]
- Larochette, V.; Miot, C.; Poli, C.; Beaumont, E.; Roingeard, P.; Fickenscher, H.; Jeannin, P.; Delneste, Y. IL-26, a cytokine with roles in extracellular DNA-induced inflammation and microbial defense. Front. Immunol. 2019, 10, 204. [Google Scholar] [CrossRef] [PubMed]
- Trotter, T.N.; Shuptrine, C.W.; Tsao, L.C.; Marek, R.D.; Acharya, C.; Wei, J.P.; Yang, X.Y.; Lei, G.; Wang, T.; Lyerly, H.K.; et al. IL26, a noncanonical mediator of DNA inflammatory stimulation, promotes TNBC engraftment and progression in association with neutrophils. Cancer Res. 2020, 80, 3088–3100. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Smyth, M.J.; Teng, M.W.L. Interleukin (IL)-12 and IL-23 and their conflicting roles in cancer. Cold Spring Harb. Perspect. Biol. 2018, 10, a028530. [Google Scholar] [CrossRef]
- Hewitt, S.L.; Bai, A.; Bailey, D.; Ichikawa, K.; Zielinski, J.; Karp, R.; Apte, A.; Arnold, K.; Zacharek, S.J.; Iliou, M.S.; et al. Durable anticancer immunity from intratumoral administration of IL-23, IL-36γ, and OX40L mRNAs. Sci. Transl. Med. 2019, 11, eaat9143. [Google Scholar] [CrossRef] [PubMed]
- Elessawi, D.F.; Alkady, M.M.; Ibrahim, I.M. Diagnostic and prognostic value of serum IL-23 in colorectal cancer. Arab J. Gastroenterol. 2019, 20, 65–68. [Google Scholar] [CrossRef] [PubMed]
- Chihara, N. Dysregulated T cells in multiple sclerosis. Clin. Exp. Neuroimmunol. 2018, 9, 20–29. [Google Scholar] [CrossRef]
- Figueiredo, M.L.; Figueiredo Neto, M.; Salameh, J.W.; Decker, R.E.; Letteri, R.; Chan-Seng, D.; Emrick, T. Ligand-mediated targeting of cytokine Interleukin-27 enhances its bioactivity in vivo. Mol. Ther. Methods Clin. Dev. 2020, 17, 739–751. [Google Scholar] [CrossRef] [PubMed]
- Kourko, O.; Seaver, K.; Odoardi, N.; Basta, S.; Gee, K. IL-27, IL-30, and IL-35: A cytokine triumvirate in cancer. Front. Oncol. 2019, 9, 969. [Google Scholar] [CrossRef]
- Park, Y.J.; Ryu, H.; Choi, G.; Kim, B.S.; Hwang, E.S.; Kim, H.S.; Chung, Y. IL-27 confers a protumorigenic activity of regulatory T cells via CD39. Proc. Natl. Acad. Sci. USA 2019, 116, 3106–3111. [Google Scholar] [CrossRef]
- Li, X.; Shao, Y.; Sha, X.; Fang, P.; Kuo, Y.M.; Andrews, A.J.; Li, Y.; Yang, W.Y.; Maddaloni, M.; Pascual, D.W.; et al. IL-35 (interleukin-35) suppresses endothelial cell activation by inhibiting mitochondrial reactive oxygen species-mediated site-specific acetylation of H3K14 (histone 3 lysine 14). Arterioscler. Thromb. Vasc. Biol. 2018, 38, 599–609. [Google Scholar] [CrossRef]
- Sawant, D.V.; Yano, H.; Chikina, M.; Zhang, Q.; Liao, M.; Liu, C.; Callahan, D.J.; Sun, Z.; Sun, T.; Tabib, T.; et al. Adaptive plasticity of IL-10+ and IL-35+ Treg cells cooperatively promotes tumor T cell exhaustion. Nat. Immunol. 2019, 20, 724–735. [Google Scholar] [CrossRef] [PubMed]
- Vitiello, G.A.; George, M. Targeting the interleukin-17 immune axis for cancer immunotherapy. J. Exp. Med. 2020, 217, e20190456. [Google Scholar] [CrossRef] [PubMed]
- Bie, Q.; Jin, C.; Zhang, B.; Dong, H. IL-17B: A new area of study in the IL-17 family. Mol. Immunol. 2017, 90, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Jungnickel, C.; Schmidt, L.H.; Bittigkoffer, L.; Wolf, L.; Wolf, A.; Ritzmann, F.; Kamyschnikow, A.; Herr, C.; Menger, M.D.; Spieker, T.; et al. IL-17C mediates the recruitment of tumor-associated neutrophils and lung tumor growth. Oncogene 2017, 36, 4182–4190. [Google Scholar] [CrossRef]
- Liu, R.; Lauridsen, H.M.; Amezquita, R.A.; Pierce, R.W.; Jane-Wit, D.; Fang, C.; Pellowe, A.S.; Kirkiles-Smith, N.C.; Gonzalez, A.L.; Pober, J.S. IL-17 promotes neutrophil-mediated immunity by activating microvascular pericytes and not endothelium. J. Immunol. 2016, 197, 2400–2408. [Google Scholar] [CrossRef] [PubMed]
- Tong, Z.; Yang, X.O.; Yan, H.; Liu, W.; Niu, X.; Shi, Y.; Fang, W.; Xiong, B.; Wan, Y.; Dong, C. A protective role by interleukin-17F in colon tumorigenesis. PLoS ONE 2012, 7, e34959. [Google Scholar] [CrossRef] [PubMed]
- Lazear, H.M.; Schoggins, J.W.; Diamond, M.S. Shared and distinct functions of type I and type III interferons. Immunity 2019, 50, 907–923. [Google Scholar] [CrossRef]
- Bakouny, Z.; Toni, K.C. IL-8 and cancer prognosis on immunotherapy. Nat. Med. 2020, 26, 650–651. [Google Scholar] [CrossRef]
- Ford, R.; Tamayo, A.; Martin, B.; Niu, K.; Claypool, K.; Cabanillas, F.; Ambrus, J., Jr. Identification of B-cell growth factors (interleukin-14; high molecular weight-B-cell growth factors) in effusion fluids from patients with aggressive B-cell lymphomas. Blood 1995, 86, 283–293. [Google Scholar] [CrossRef]
- Richmond, J.; Tuzova, M.; Cruikshank, W.; Center, D. Regulation of cellular processes by interleukin-16 in homeostasis and cancer. J. Cell. Physiol. 2014, 229, 139–147. [Google Scholar] [CrossRef]
- Sloot, Y.J.E.; Smit, J.W.; Joosten, L.A.B.; Netea-Maier, R.T. Insights into the role of IL-32 in cancer. Semin. Immunol. 2018, 38, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Baghdadi, M.; Endo, H.; Takano, A.; Ishikawa, K.; Kameda, Y.; Wada, H.; Miyagi, Y.; Yokose, T.; Ito, H.; Nakayama, H.; et al. High co-expression of IL-34 and M-CSF correlates with tumor progression and poor survival in lung cancers. Sci. Rep. 2018, 8, 418. [Google Scholar] [CrossRef] [PubMed]
- Franzè, E.; Laudisi, F.; Di Grazia, A.; Marônek, M.; Bellato, V.; Sica, G.; Monteleone, G. Macrophages produce and functionally respond to interleukin-34 in colon cancer. Cell Death Discov. 2020, 6, 117. [Google Scholar] [CrossRef] [PubMed]
- Mármol, I.; Sánchez-de-Diego, C.; Pradilla Dieste, A.; Cerrada, E.; Rodriguez Yoldi, M.J. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int. J. Mol. Sci. 2017, 18, 197. [Google Scholar] [CrossRef]
- Bardhan, K.; Liu, K. Epigenetics and colorectal cancer pathogenesis. Cancers 2013, 5, 676–713. [Google Scholar] [CrossRef]
- Waldner, M.; Schimanski, C.C.; Neurath, M.F. Colon cancer and the immune system: The role of tumor invading T cells. World J. Gastroenterol. 2006, 12, 7233–7238. [Google Scholar] [CrossRef]
- Klintrup, K.; Mäkinen, J.M.; Kauppila, S.; Väre, P.O.; Melkko, J.; Tuominen, H.; Tuppurainen, K.; Mäkelä, J.; Karttunen, T.J.; Mäkinen, M.J. Inflammation and prognosis in colorectal cancer. Eur. J. Cancer 2005, 41, 2645–2654. [Google Scholar] [CrossRef]
- Levin, R.; Brown, M.J.; Kashtock, M.E.; Jacobs, D.E.; Whelan, E.A.; Rodman, J.; Schock, M.R.; Padilla, A.; Sinks, T. Lead exposures in US children, 2008: Implications for prevention. Environ. Health Perspect. 2008, 116, 1285–1293. [Google Scholar] [CrossRef]
- Zhao, H.; Wu, L.; Yan, G.; Chen, Y.; Zhou, M.; Wu, Y.; Li, Y. Inflammation and tumor progression: Signaling pathways and targeted intervention. Signal Transduct. Target. Ther. 2021, 6, 263. [Google Scholar] [CrossRef]
- Saber, S.; Alomar, S.Y.; Yahya, G. Blocking prostanoid receptors switches on multiple immune responses and cascades of inflammatory signaling against larval stages in snail fever. Environ. Sci. Pollut. Res. 2022, 29, 43546–43555. [Google Scholar] [CrossRef]
- Youssef, M.E.; El-Azab, M.F.; Abdel-Dayem, M.A.; Yahya, G.; Alanazi, I.S.; Saber, S. Electrocardiographic and histopathological characterizations of diabetic cardiomyopathy in rats. Env. Sci. Pollut. Res. Int. 2022, 29, 25723–25732. [Google Scholar] [CrossRef]
- Saber, S.; Yahya, G.; Gobba, N.A.; Sharaf, H.; Alshaman, R.; Alattar, A.; Amin, N.A.; El-Shedody, R.; Aboutouk, F.H.; Abd El-Galeel, Y.; et al. The Supportive Role of NSC328382, a P2X7R Antagonist, in Enhancing the Inhibitory Effect of CRID3 on NLRP3 Inflammasome Activation in Rats with Dextran Sodium Sulfate-Induced Colitis. J. Inflamm. Res. 2021, 14, 3443–3463. [Google Scholar] [CrossRef] [PubMed]
- Saber, S.; Abd El-Fattah, E.E.; Yahya, G.; Gobba, N.A.; Maghmomeh, A.O.; Khodir, A.E.; Mourad, A.A.E.; Saad, A.S.; Mohammed, H.G.; Nouh, N.A.; et al. A Novel Combination Therapy Using Rosuvastatin and Lactobacillus Combats Dextran Sodium Sulfate-Induced Colitis in High-Fat Diet-Fed Rats by Targeting the TXNIP/NLRP3 Interaction and Influencing Gut Microbiome Composition. Pharmaceuticals 2021, 14, 341. [Google Scholar] [CrossRef] [PubMed]
- Youssef, M.E.; Abdel-Reheim, M.A.; Morsy, M.A.; El-Daly, M.; Atwa, G.M.K.; Yahya, G.; Cavalu, S.; Saber, S.; Ahmed Gaafar, A.G. Ameliorative Effect of Dabigatran on CFA-Induced Rheumatoid Arthritis via Modulating Kallikrein-Kinin System in Rats. Int. J. Mol. Sci. 2022, 23, 10297. [Google Scholar] [CrossRef]
- Zohny, M.H.; Cavalu, S.; Youssef, M.E.; Kaddah, M.M.Y.; Mourad, A.A.E.; Gaafar, A.G.A.; El-Ahwany, E.; Amin, N.A.; Arakeep, H.M.; Shata, A.; et al. Coomassie brilliant blue G-250 dye attenuates bleomycin-induced lung fibrosis by regulating the NF-κB and NLRP3 crosstalk: A novel approach for filling an unmet medical need. Biomed. Pharmacother. 2022, 148, 112723. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Fattah, E.E.; Saber, S.; Mourad, A.A.E.; El-Ahwany, E.; Amin, N.A.; Cavalu, S.; Yahya, G.; Saad, A.S.; Alsharidah, M.; Shata, A.; et al. The dynamic interplay between AMPK/NFκB signaling and NLRP3 is a new therapeutic target in inflammation: Emerging role of dapagliflozin in overcoming lipopolysaccharide-mediated lung injury. Biomed. Pharmacother. 2022, 147, 112628. [Google Scholar] [CrossRef] [PubMed]
- Dranoff, G. Cytokines in cancer pathogenesis and cancer therapy. Nat. Rev. Cancer 2004, 4, 11–22. [Google Scholar] [CrossRef]
- Balkwill, F.; Mantovani, A. Inflammation and cancer: Back to Virchow? Lancet 2001, 357, 539–545. [Google Scholar] [CrossRef] [PubMed]
- Ogunwobi, O.O.; Mahmood, F.; Akingboye, A. Biomarkers in Colorectal Cancer: Current Research and Future Prospects. Int. J. Mol. Sci. 2020, 21, 5311. [Google Scholar] [CrossRef]
- Louie, A.D.; Huntington, K.; Carlsen, L.; Zhou, L.; El-Deiry, W.S. Integrating Molecular Biomarker Inputs Into Development and Use of Clinical Cancer Therapeutics. Front. Pharmacol. 2021, 12, 747194. [Google Scholar] [CrossRef]
- Huang, Z.; Yang, M. Molecular Network of Colorectal Cancer and Current Therapeutic Options. Front. Oncol. 2022, 12, 852927. [Google Scholar] [CrossRef]
- Czajka-Francuz, P.; Francuz, T.; Cisoń-Jurek, S.; Czajka, A.; Fajkis, M.; Szymczak, B.; Kozaczka, M.; Malinowski, K.P.; Zasada, W.; Wojnar, J.; et al. Serum cytokine profile as a potential prognostic tool in colorectal cancer patients-one center study. Rep. Pract. Oncol. Radiother. 2020, 25, 867–875. [Google Scholar] [CrossRef] [PubMed]
- West, N.R.; McCuaig, S.; Franchini, F.; Powrie, F. Emerging cytokine networks in colorectal cancer. Nat. Rev. Immunol. 2015, 15, 615–629. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Zhang, Y.; Yang, Z.; Sha, Y.; Pan, Y.; Chen, Y.; Cai, L. Analysis of the role of the interleukins in colon cancer. Biol. Res. 2020, 53, 20. [Google Scholar] [CrossRef] [PubMed]
- Lindebjerg, J.; Merete, O.; Claus, B. Colorectal cancers detected through screening are associated with lower stages and improved survival. Dan. Med. J. 2014, 61, A4758. [Google Scholar] [PubMed]
- Vogelaar, L.I.; van Ballegooijen, M.; Zauber, A.G.; Habbema, J.D.; Kuipers, E.J. Effect of rising chemotherapy costs on the cost savings of colorectal cancer screening. J. Natl. Cancer Inst. 2009, 101, 1412–1422. [Google Scholar] [CrossRef]
- Wu, Z.; Li, Y.; Zhang, Y.; Hu, H.; Wu, T.; Liu, S.; Chen, W.; Xie, S.; Lu, Z. Colorectal Cancer Screening Methods and Molecular Markers for Early Detection. Technol. Cancer Res. Treat. 2020, 19, 1533033820980426. [Google Scholar] [CrossRef]
- Wu, N.; Yang, X.; Zhang, R.; Li, J.; Xiao, X.; Hu, Y.; Chen, Y.; Yang, F.; Lu, N.; Wang, Z.; et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb. Ecol. 2013, 66, 462–470. [Google Scholar] [CrossRef]
- Rahman, M.M.; Islam, M.R.; Shohag, S.; Ahasan, M.T.; Sarkar, N.; Khan, H.; Hasan, A.M.; Cavalu, S.; Rauf, A. Microbiome in cancer: Role in carcinogenesis and impact in therapeutic strategies. Biomed. Pharmacother. 2022, 149, 112898. [Google Scholar] [CrossRef]
- Cavalu, S.; Popa, A.; Bratu, I.; Borodi, G.; Maghiar, A. New Evidences of Key Factors Involved in Silent Stones Etiopathogenesis and Trace Elements: Microscopic, Spectroscopic, and Biochemical Approach. Biol. Trace Elem. Res. 2015, 168, 311–320. [Google Scholar] [CrossRef]
- Kappen, J.; Skorupa, M.; Krukiewicz, K. Conducting Polymers as Versatile Tools for the Electrochemical Detection of Cancer Biomarkers. Biosensors 2022, 13, 31. [Google Scholar] [CrossRef] [PubMed]
- Burney, I.A.; Lakhtakia, R. Precision medicine: Where have we reached and where are we headed? Sultan Qaboos Univ. Med. J. 2017, 17, e255–e258. [Google Scholar] [CrossRef] [PubMed]
- Maciejko, L.; Smalley, M.; Goldman, A. Cancer immunotherapy and personalized medicine: Emerging technologies and biomarker-based approaches. J. Mol. Biomark. Diagn. 2017, 8, 350. [Google Scholar] [CrossRef] [PubMed]
- Verma, M. Personalized medicine and cancer. J. Pers. Med. 2012, 2, 1–14. [Google Scholar] [CrossRef] [PubMed]

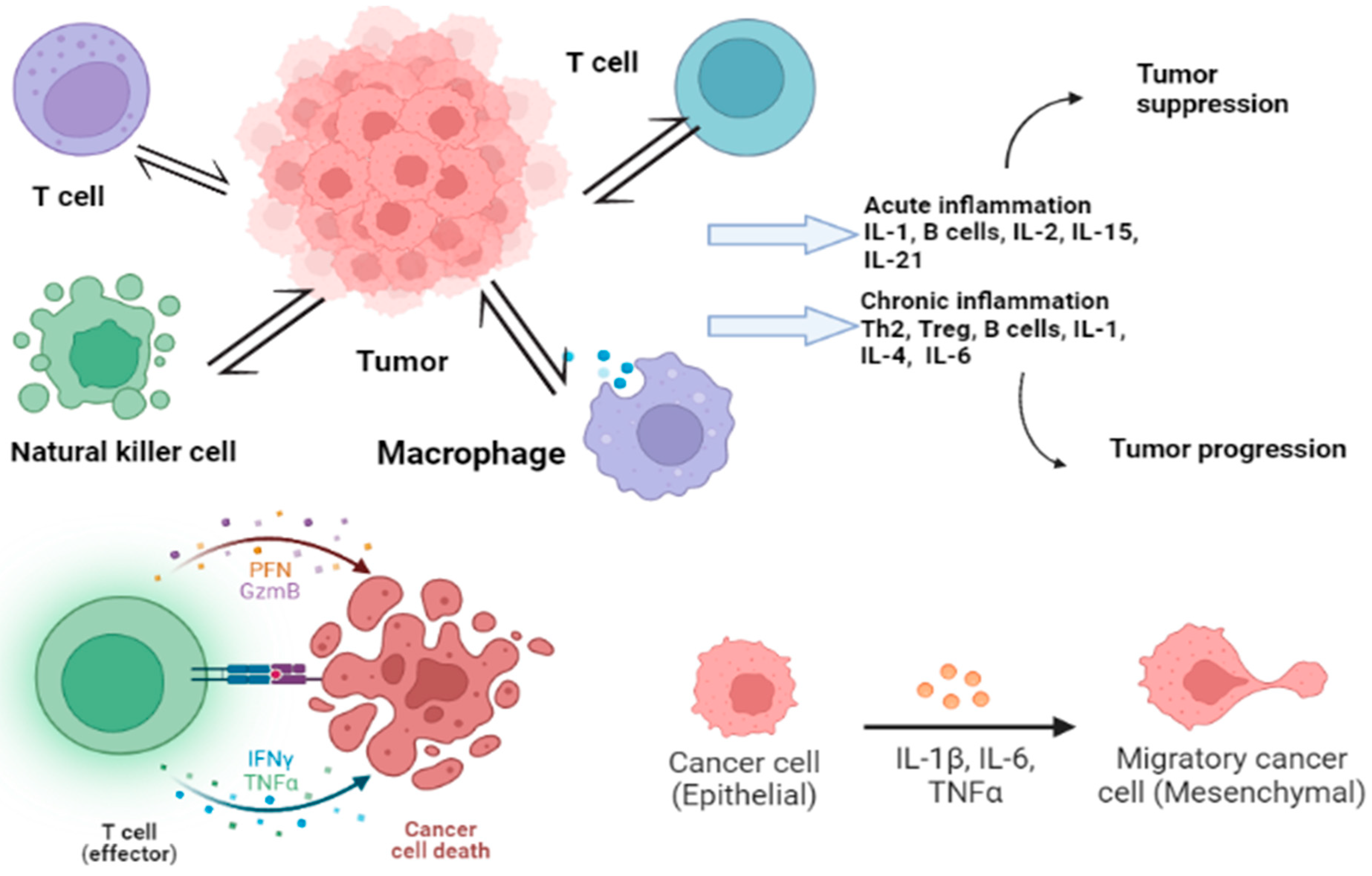
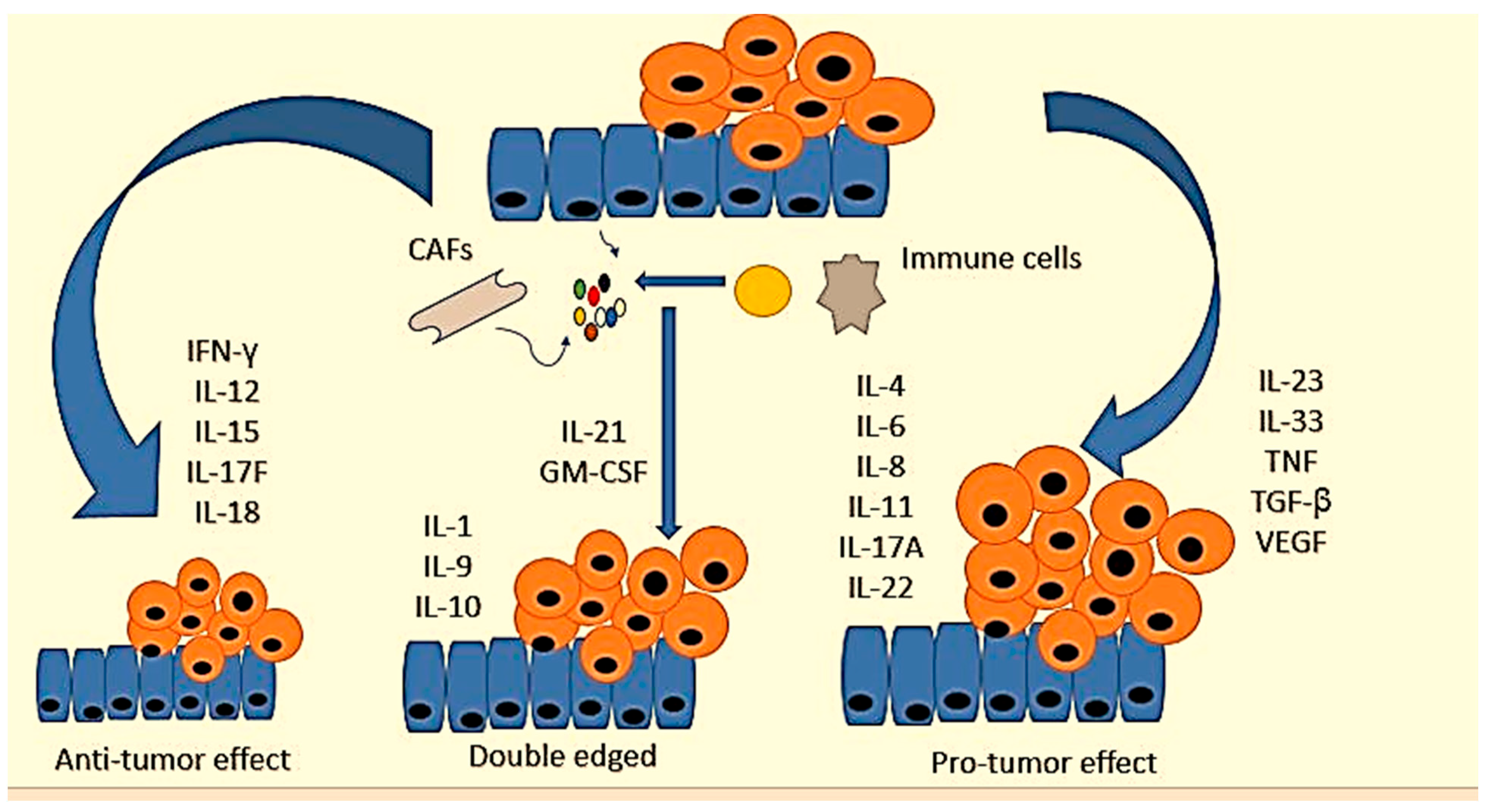
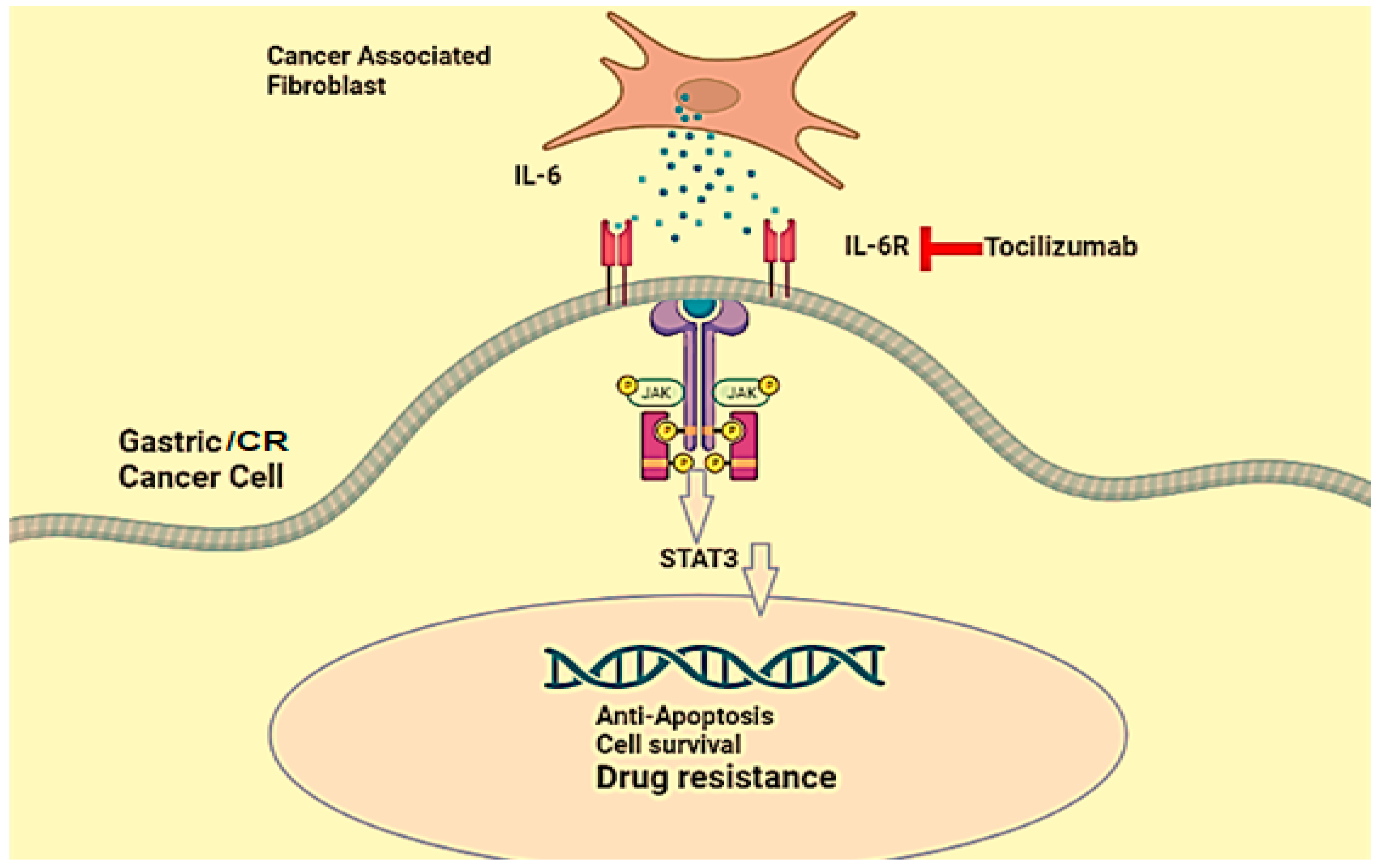
| TME Cells | Soluble Factors | Target Cells | Signaling Pathway | Biological Effects | Reference |
|---|---|---|---|---|---|
| CD4+ | IL-22 | CRC | STAT3/DOT1L (Signal transducer and activator of transcription)/(Disruptor of telomeric silencing 1-like) | Stemness gene regulation | [59] |
| CAF (Cancer-associated fibroblasts) | HGF/SDF1 (Hepatocyte growth factor)/Stromal cell-derived factor-1 | Cancer stem cells (CSC) | Wnt/β-catenin | Clonogenic activity and expression of CD44v6 | [60] |
| Endothelial cells | JAG1 (Jagged) | CRC | Notch | CD133 expression, tumorigenicity and chemoresistance | [61] |
| MSC (Mesenchymal stem cells) | PGE2 (Prostaglandin E2) | CRC | Wnt/β-catenin | EMT (Epithelial-to-mesenchymal transition) and invasion | [62] |
| Myofibroblasts | HGF | CSC | Wnt/β-catenin | Clonogenicity | [63] |
| Cytokine | Functional Effect in CRC | Expression Patterns | Reference |
|---|---|---|---|
| IL-1α | Promotes metastasis and the chemosensitivity |  | [136,137] |
| IL-1β | Promotes the proliferation of colon cancer cells, tumorigenesis, and alters the tumor microenvironment |  | [137,138,139] |
| IL-18 | Antitumorigenic properties and release of other signals |  | [140,141] |
| IL-2, IL-7, IL-9, IL-15 | Antitumor activity, promote EMT, proliferation, invasion, and metastasis | IL-4, IL-7 upregulated, IL-9 downregulated, IL-2 in between | [142,143,144,145,146,147] |
| IL-21 | Activation of immune response biomarkers |  (Potential for biomarker) | [148,149,150] |
| IL-6 | Promotes mitosis, proliferation, metastasis, migration, and angiogenesis |  | [151,152,153,154] |
| IL-11 | Facilitates the proliferation of CRC |  | [155,156,157] |
| IL-8 | Promotes cell proliferation, angiogenesis, cancer metastasis, chemoresistance, antianoikis, maintains CCSC properties |  | [158,159,160] |
| IL-10 | Pathogenesis and progression |  | [141,161,162,163] |
| IL-22 | Dominant role in CRC tumorigenesis, antiapoptosis, and cell proliferation |  | [164,165,166] |
| IL-17a | Promotes cell cycle progression and angiogenesis |  | [167] |
| IL-17b | Promotes tumor |  | [168,169,170,171] |
| IL-4 | Overexpressed in early CRC, tumor development |  | [172] |
| IL-23 | Overexpressed in CRC tissue and predictive for CRC metastasis |  | [173,174,175,176] |
| Interleukin Family | Receptors | Cytokine | Potential Effect in CRC | Therapeutic Strategy | Reference |
|---|---|---|---|---|---|
| IL-1R1 IL-1R2 | IL-1α | Metastasis promotion with chemosensitivity. Promotes antitumor immunity and carcinogenesis (inflammatory) | Therapeutic neutralization in order to tackle severe illness in clinical trials | [102,139,140,280] | |
| IL-2R1 IL-1R2 ILR3 sIL-IR2 sIL-IR3 | IL-1β | Proliferation of cancer cells of colon and promotion of tumorigenesis. Altering the tumor microenvironment | Therapeutic neutralization to manage cytokine release syndrome (CRS) in clotting time and the prevention of cancer | [102,140,141] | |
| IL-1 super family | IL-1R4 (ST2) | IL-33 | Protumor, maintenance of intestinal microbiota, tumor microenvironment change, TH2 polarization, Treg cell function, promotion of Angiogenesis and enhancement of colon cancer cell stemness, maintain intestinal microbiota. | Preclinical neutralization | [15,281,282,283,284] |
| IL-18 subfamily | IL-1R5–IL-1R7 IL-18BP IL-1R5 (IL-18Rα IL-18Rβ) | IL-18 | Antitumor, activates lymphocytes to produce IFN-γ, improve the integrity of intestinal barrier and induces apoptosis to act on NK cells | Preclinical engineered rIL-18 or by combined with activated clotting time, hindered by IL-18BP | [140,285] |
| IL-1R8–IL-1R5 | IL-37 | Antitumor attributes, inhibit the colon cancer cell development by stopping β-catenin. | Not explored | [135,286] | |
| IL-1R6 | IL-36α | Antitumor | [287] | ||
| IL-36 subfamily | IL-1R6 | IL-36γ | Antitumor, inflammatory immune infiltrates promotion, promote inflammation (TH1-type) inhibited by IL-36Ra | Preclinical rIL-36γ as an alternative to IL-1 | [287] |
| IL-38 | IL-1R6–IL-1R9 | Immunosuppressive | Not explored | [288,289] | |
| IL-2Rα, IL-2Rβ/IL-2Rγ, IL-2Rα/IL-2Rβ/IL-2Rγ sIL-2RαIL-2/IL-15Rβ–γc IL-2Rα–IL-2/IL-15Rβ–γc | IL-2 | Antitumor, NK and T cell growth factor, inhibit T cell responses by maintaining Treg cells and AICD induction | rIL-2 approved for monotherapy. Engineered to avoid side effects and to be used in ACT | [147,148,290] | |
| Type (IL-4Rα/γc) and Type (IL-4Rα/IL-13Rα1) | IL-4 | Promote epithelial-to-mesenchymal transition (EMT), metastasis and invasion. Promotes inflammation of TH2-type and polarization of TH9. Promotes cancer cell growth upon overexpression of IL-4R. | IL-4R- targeting to bear cancer cells and block signaling. Antitumor TH9 cells production for ACT | [146,291] | |
| IL-2 family | IL-7R (IL-7Rα/γc) | IL-7 | Metastasis promotion Antitumoural: NK growth factor and T cell production | rIL-7 in combination with interleukins or ACT | [145,292,293] |
| IL-9R (IL-9Rα/γc) | IL-9 | Antitumor action Pleiotropic | Preclinical TH9 cells in ACT | [15,109,294] | |
| IL-15R (IL-15Rα/IL-15Rβ/γc) | IL-15 | Proliferation and angiogenesis inhibition, Antitumor activity by activating lymphocytes to produce IFN γ, Promote apoptosis | rIL-15 or analogues in combination with interleukins or ACT | [295,296] | |
| IL-21R–γc heterodimers of IL-21R and γc | IL-21 | Enhances cytotoxicity of CTLs, Antitumor activity | Combination therapies with rIL-21 in clinical trials | [296] | |
| IL-3 family | IL-3Rα–βc | IL-3 | Promotes malignancy as a haematopoietic factor | Target CD123-bearing cells by fused to toxins | [297,298,299] |
| IL-6 family | gp130 IL-6R IL-6Rα–gp130 (classic) sIL-6Rα–gp130 (trans) | IL-6 | Mitosis promotion, metastasis, proliferation, making the microenvironment for metastasis, activates tumor outgrowth and carcinogenesis, mediates cytokine release syndrome (CRS), Cachexia promotion | Neutralization to manage CRS in ACT, cachexia | [300,301,302,303] |
| gp130 IL-11Ra IL-11Rα–gp130 (classic) sIL-11Rα–gp130 (trans) | IL-11 | Proliferation of CRC Promotes inflammation by inducing carcinogenesis and cancer progression | Preclinical neutralization and gp130 common receptor blockade | [158,304] | |
| IL-31Rα–OSMRβ | IL-31 | TH2-type cytokine, evidently tumorigenic | Unexplored | [287] | |
| IL-10 family | IL-10RA and IL-10RB | IL-10 | Promotes cytotoxicity, inhibits antitumor responses | rIL-10 to increase cytotoxicity in trials | [163,165,305,306] |
| IL-10RB and IL-22R /IL-22BP | IL-22 | Promote tumorigenesis, antiapoptosis and cell proliferation Peritumoral: promotion of carcinoma progression | Preclinical neutralization | [114,306,307,308] | |
| IL-20Rα–IL-20RβIL-22Rα1–IL-20Rβ | IL-24 | Induces autophagy of cancer and apoptosis | Preclinical rIL-24 combined with oncolytic virus | [309,310] | |
| IL-20Rα–IL-10Rβ | IL-26 | Pro-tumoral through TH17 cells and neutrophils | Preclinical neutralization | [311,312] | |
| IL-12Rβ1–IL-12Rβ2 | IL-12 | Antitumoral: the main driver of TH1-type immunity, amplification and initiation of production | Engineered or combined with other interleukins in trials | [286,313] | |
| IL-12 family | IL-23R–IL-12Rβ1 | IL-23 | Mainly pro-tumoral: direct and indirect effect via TH17 cells and TH22 cells | Neutralization in trials, enhances CAR T cell cytotoxicity | [314,315] |
| IL-27Rα | IL-27 and IL-30 | Pleiotropic: induces cytotoxicity and NK cell yet enhances T cell and Treg cell activity | Neutralization and engineered rIL-27 in trials | [316,317,318,319] | |
| IL-12Rβ2–gp130gp130–gp130IL-27Rα–IL-12Rβ2 | IL-35 | Treg cell-mediated suppression of T cell responses and promotion of metastatic colonization | Preclinical neutralization with checkpoint inhibitors and other therapies | [320,321] | |
| IL-17 family | IL-17RA–IL-17RC | IL-17A/F | Cell cycle progression and angiogenesis, facilitate the development indirectly and change the tissue environment and microbiota of CRC | Neutralization in clinical trials | [169,322] |
| IL-17RIL-17RB | IL-17b | Carcinogenesis, immunosuppression, EMT | [170,323] | ||
| IL-17RA–IL-17RE | IL-17c | Mostly pro-tumoral, but antitumoral properties | Not explored | [324] | |
| Unknown | IL-17 D | [325] | |||
| IL-17R | IL-17e | [170] | |||
| IL-17RA–IL-17RB | IL-25 | ||||
| IL-17R | IL-17f | Tumor suppression effect possibly by inhibiting tumor angiogenesis | [326] | ||
| IFN-γ family | IL-28A and IL-28B IL-28Rα–IL-10Rβ | IL-28Rα IL-10Rβ IL-29 | Antitumoral: induces apoptosis of malignant cells | Preclinical gene therapy using IL-28 and IL-29 | [327] |
| CXCR1, CXCR2ACKR1/DARC | IL-8 | Attracts neutrophils and mediates the suppressive environment | Therapeutic neutralization in clinical trials | [328] | |
| IL-13Rα1–IL-4RαIL-13Rα2 | TH2-type cytokine | Targeting or blocking IL-13R | [291] | ||
| IL-14α and IL-14β | IL-14R | Growth factor in B cell and in lymphoma | Not explored | [329] | |
| CD4 | IL-16 | Pro-tumoral: proliferation of lymphoma and chemoattractant | Scarce preclinical evidence | [330] | |
| Other interleukins | Unknown | IL-32 | Pleiotropic in action but depends on cancer type and isoform. | Preclinical antitumor effects in combination | [331] |
| CSF1R | IL-34 | Pro-tumoral: immune suppression, cancer progression, and resistance of therapy | Preclinical neutralization to lessen the pro-tumor effects | [332,333] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maryam, S.; Krukiewicz, K.; Haq, I.U.; Khan, A.A.; Yahya, G.; Cavalu, S. Interleukins (Cytokines) as Biomarkers in Colorectal Cancer: Progression, Detection, and Monitoring. J. Clin. Med. 2023, 12, 3127. https://doi.org/10.3390/jcm12093127
Maryam S, Krukiewicz K, Haq IU, Khan AA, Yahya G, Cavalu S. Interleukins (Cytokines) as Biomarkers in Colorectal Cancer: Progression, Detection, and Monitoring. Journal of Clinical Medicine. 2023; 12(9):3127. https://doi.org/10.3390/jcm12093127
Chicago/Turabian StyleMaryam, Sajida, Katarzyna Krukiewicz, Ihtisham Ul Haq, Awal Ayaz Khan, Galal Yahya, and Simona Cavalu. 2023. "Interleukins (Cytokines) as Biomarkers in Colorectal Cancer: Progression, Detection, and Monitoring" Journal of Clinical Medicine 12, no. 9: 3127. https://doi.org/10.3390/jcm12093127
APA StyleMaryam, S., Krukiewicz, K., Haq, I. U., Khan, A. A., Yahya, G., & Cavalu, S. (2023). Interleukins (Cytokines) as Biomarkers in Colorectal Cancer: Progression, Detection, and Monitoring. Journal of Clinical Medicine, 12(9), 3127. https://doi.org/10.3390/jcm12093127







