The Clinical Implications of KRAS Mutations and Variant Allele Frequencies in Pancreatic Ductal Adenocarcinoma
Abstract
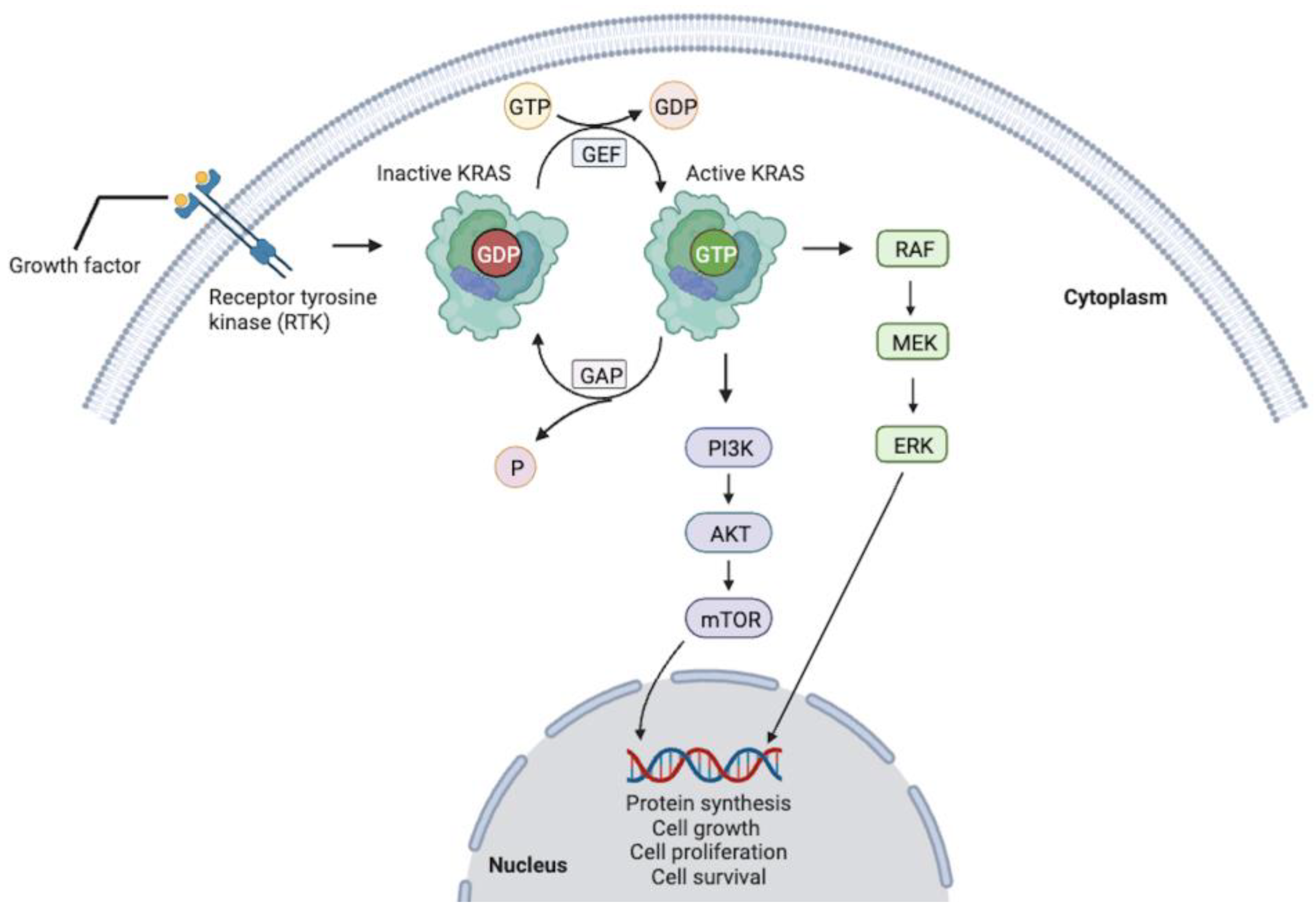
1. Introduction
2. Review Methodology
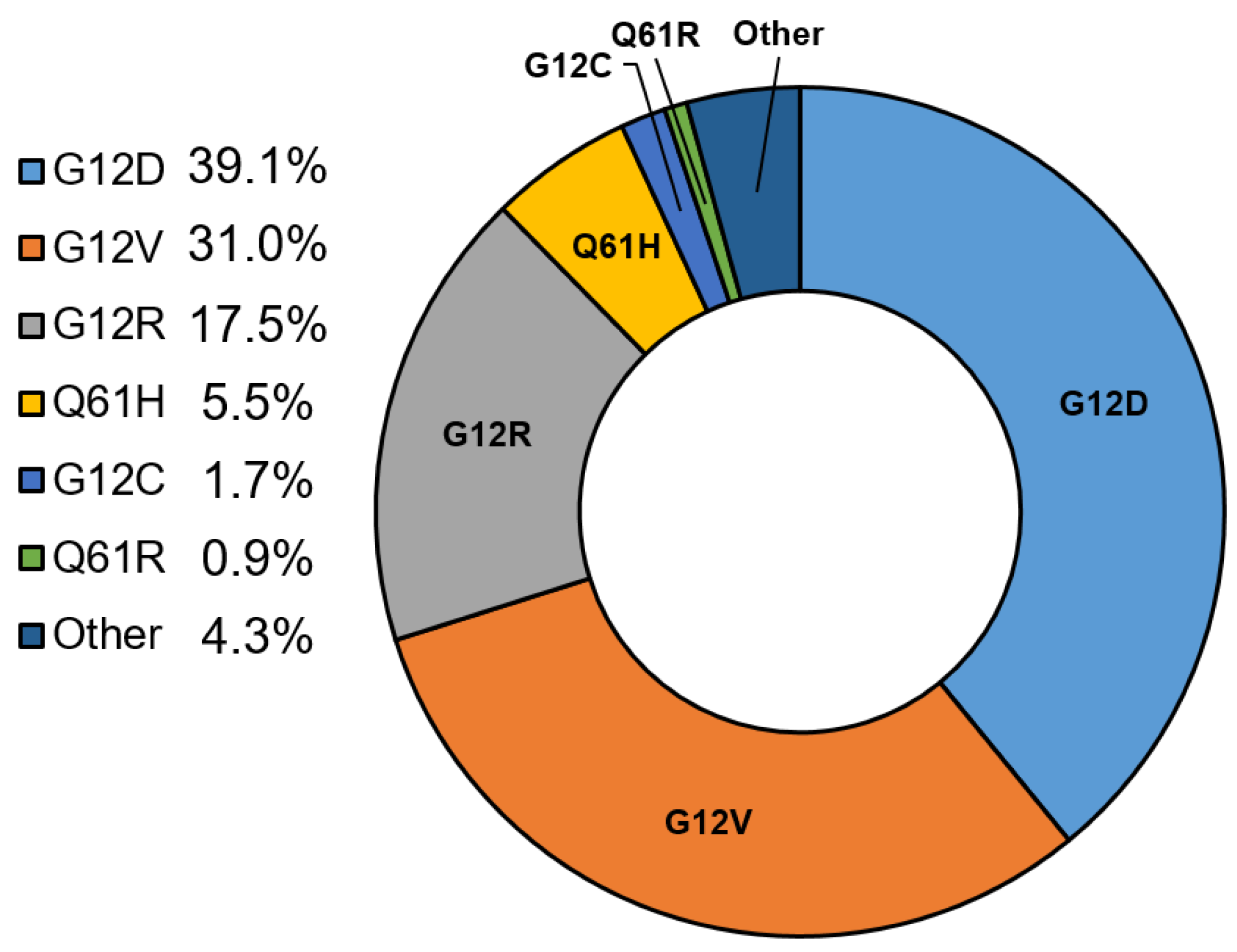
3. Clinical Impact of KRAS Variants
4. Clinical Impact of KRAS Variant Allele Frequency
| Study | Cohort | Findings |
|---|---|---|
| Lennerz et al. [25], Biankin, et al. [29] | Early stage (Stage I–II) | Mutant KRAS VAF ≥ 10% was associated with a trend of worse overall survival compared with VAF < 10% |
| Suzuki et al. [26] | All stages (I–IV) | Both overall and disease-free survival showed a ‘dose-dependent’ impact of mutant KRAS VAF, with VAF < 10% showing the best survival and VAF ≥ 20% showing the worst survival. |
| Nauheim et al. [27] | Early stage (Stage I–II) | Mutant KRAS VAF ≥ 20% patients presented with a more aggressive, advanced-stage disease, and were noted to have worse disease-free survival. |
5. Targeted Therapy
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. Ca Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Waters, A.M.; Der, C.J. KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer. Cold Spring Harb. Perspect. Med. 2018, 8, a031435. [Google Scholar] [CrossRef] [PubMed]
- Klein, A.P. Pancreatic cancer epidemiology: Understanding the role of lifestyle and inherited risk factors. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 493–502. [Google Scholar] [CrossRef]
- Luo, J. KRAS mutation in pancreatic cancer. Semin. Oncol. 2021, 48, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.-M.; Gingras, M.-C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.C.; Quinn, M.C.; et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Hunter, J.C.; Manandhar, A.; Carrasco, M.A.; Gurbani, D.; Gondi, S.; Westover, K.D. Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations. Mol. Cancer Res. 2015, 13, 1325–1335. [Google Scholar] [CrossRef]
- Zhang, X.; Mao, T.; Zhang, B.; Xu, H.; Cui, J.; Jiao, F.; Chen, D.; Wang, Y.; Hu, J.; Xia, Q.; et al. Characterization of the genomic landscape in large-scale Chinese patients with pancreatic cancer. EBioMedicine 2022, 77, 103897. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.A.; Serinelli, S.; Zaccarini, D.J.; Huang, R.; Danziger, N.; Janovitz, T.; Basnet, A.; Sivapiragasam, A.; Graziano, S.; Ross, J.S. Genomic landscape of clinically advanced KRAS wild-type pancreatic ductal adenocarcinoma. Front. Oncol. 2023, 13, 1169586. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.; Jahanzaib, R.; Liao, Y.; Yao, F.; Li, J.; Teng, X.; Chen, K.; Cheng, W. Prognostic value of KRAS subtype in patients with PDAC undergoing radical resection. Front. Oncol. 2022, 12, 1074538. [Google Scholar] [CrossRef]
- Windon, A.L.; Loaiza-Bonilla, A.; Jensen, C.E.; Randall, M.; Morrissette, J.J.D.; Shroff, S.G. A KRAS wild type mutational status confers a survival advantage in pancreatic ductal adenocarcinoma. J. Gastrointest. Oncol. 2018, 9, 1–10. [Google Scholar] [CrossRef]
- Masetti, M.; Acquaviva, G.; Visani, M.; Tallini, G.; Fornelli, A.; Ragazzi, M.; Vasuri, F.; Grifoni, D.; Di Giacomo, S.; Fiorino, S.; et al. Long-term survivors of pancreatic adenocarcinoma show low rates of genetic alterations in KRAS, TP53 and SMAD4. Cancer Biomark. 2018, 21, 323–334. [Google Scholar] [CrossRef]
- Chen, H.; Tu, H.; Meng, Z.; Chen, Z.; Wang, P.; Liu, L. K-ras mutational status predicts poor prognosis in unresectable pancreatic cancer. Eur. J. Surg. Oncol. (EJSO) 2010, 36, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Ogura, T.; Yamao, K.; Hara, K.; Mizuno, N.; Hijioka, S.; Imaoka, H.; Sawaki, A.; Niwa, Y.; Tajika, M.; Kondo, S.; et al. Prognostic value of K-ras mutation status and subtypes in endoscopic ultrasound-guided fine-needle aspiration specimens from patients with unresectable pancreatic cancer. J. Gastroenterol. 2013, 48, 640–646. [Google Scholar] [CrossRef] [PubMed]
- McIntyre, C.A.; Lawrence, S.A.; Richards, A.L.; Chou, J.F.; Wong, W.; Capanu, M.; Berger, M.F.; Donoghue, M.T.A.; Yu, K.H.; Varghese, A.M.; et al. Alterations in driver genes are predictive of survival in patients with resected pancreatic ductal adenocarcinoma. Cancer 2020, 126, 3939–3949. [Google Scholar] [CrossRef]
- Yousef, A.; Yousef, M.; Chowdhury, S.; Abdilleh, K.; Knafl, M.; Edelkamp, P.; Alfaro-Munoz, K.; Chacko, R.; Peterson, J.; Smaglo, B.G.; et al. Impact of KRAS mutations and co-mutations on clinical outcomes in pancreatic ductal adenocarcinoma. NPJ Precis. Oncol. 2024, 8, 27. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Baba, Y.; Kitano, Y.; Miyake, K.; Zhang, X.; Yamamura, K.; Kosumi, K.; Kaida, T.; Arima, K.; Taki, K.; et al. KRAS, BRAF, and PIK3CA mutations, and patient prognosis in 126 pancreatic cancers: Pyrosequencing technology and literature review. Med. Oncol. 2016, 33, 32. [Google Scholar] [CrossRef] [PubMed]
- Bournet, B.; Muscari, F.; Buscail, C.; Assenat, E.; Barthet, M.; Hammel, P.; Selves, J.; Guimbaud, R.; Cordelier, P.; Buscail, L. KRAS G12D Mutation Subtype Is a Prognostic Factor for Advanced Pancreatic Adenocarcinoma. Clin. Transl. Gastroenterol. 2016, 7, e157. [Google Scholar] [CrossRef]
- Diehl, A.C.; Hannan, L.M.; Zhen, D.B.; Coveler, A.L.; King, G.; Cohen, S.A.; Harris, W.P.; Shankaran, V.; Wong, K.M.; Green, S.; et al. KRAS Mutation Variants and Co-occurring PI3K Pathway Alterations Impact Survival for Patients with Pancreatic Ductal Adenocarcinomas. Oncologist 2022, 27, 1025–1033. [Google Scholar] [CrossRef] [PubMed]
- Hendifar, A.E.; Blais, E.M.; Ng, C.; Thach, D.; Gong, J.; Sohal, D.; Chung, V.; Sahai, V.; Fountzilas, C.; Mikhail, S.; et al. Comprehensive analysis of KRAS variants in patients (pts) with pancreatic cancer (PDAC): Clinical/molecular correlations and real-world outcomes across standard therapies. J. Clin. Oncol. 2020, 38, 4641. [Google Scholar] [CrossRef]
- Kawesha, A.; Ghaneh, P.; Andrén-Sandberg, A.; Ögraed, D.; Skar, R.; Dawiskiba, S.; Evans, J.D.; Campbell, F.; Lemoine, N.; Neoptolemos, J.P. K-ras oncogene subtype mutations are associated with survival but not expression of p53, p16INK4A, p21WAF-1, cyclin D1, erbB-2 and erbB-3 in resected pancreatic ductal adenocarcinoma. Int. J. Cancer 2000, 89, 469–474. [Google Scholar] [CrossRef]
- Shen, H.; Lundy, J.; Strickland, A.H.; Harris, M.; Swan, M.; Desmond, C.; Jenkins, B.J.; Croagh, D. KRAS G12D Mutation Subtype in Pancreatic Ductal Adenocarcinoma: Does It Influence Prognosis or Stage of Disease at Presentation? Cells 2022, 11, 3175. [Google Scholar] [CrossRef] [PubMed]
- Shoucair, S.; Habib, J.R.; Pu, N.; Kinny-Köster, B.; van Ooston, A.F.; Javed, A.A.; Lafaro, K.J.; He, J.; Wolfgang, C.L.; Yu, J. Comprehensive Analysis of Somatic Mutations in Driver Genes of Resected Pancreatic Ductal Adenocarcinoma Reveals KRAS G12D and Mutant TP53 Combination as an Independent Predictor of Clinical Outcome. Ann. Surg. Oncol. 2022, 29, 2720–2731. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Shi, X.; Shen, J.; Gao, S.; Wang, H.; Shen, S.; Pan, Y.; Li, B.; Xu, X.; Shao, Z.; et al. Preoperative detection of KRAS G12D mutation in ctDNA is a powerful predictor for early recurrence of resectable PDAC patients. Br. J. Cancer 2020, 122, 857–867. [Google Scholar] [CrossRef] [PubMed]
- Schultz, N.A.; Roslind, A.; Christensen, I.J.; Horn, T.D.; Høgdall, E.; Pedersen, L.N.; Kruhøffer, M.; Burcharth, F.M.; Wøjdemann, M.M.; Johansen, J.S.D. Frequencies and Prognostic Role of KRAS and BRAF Mutations in Patients with Localized Pancreatic and Ampullary Adenocarcinomas. Pancreas 2012, 41, 759–766. [Google Scholar] [CrossRef] [PubMed]
- Lennerz, J.K.; Stenzinger, A. Allelic Ratio of KRAS Mutations in Pancreatic Cancer. Oncologist 2015, 20, e8–e9. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Masugi, Y.; Inoue, Y.; Hamada, T.; Tanaka, M.; Takamatsu, M.; Arita, J.; Kato, T.; Kawaguchi, Y.; Kunita, A.; et al. KRAS variant allele frequency, but not mutation positivity, associates with survival of patients with pancreatic cancer. Cancer Sci. 2022, 113, 3097–3109. [Google Scholar] [CrossRef] [PubMed]
- Nauheim, D.; Moskal, D.; Renslo, B.; Chadwick, M.; Jiang, W.; Yeo, C.J.; Nevler, A.; Bowne, W.; Lavu, H. KRAS mutation allele frequency threshold alters prognosis in right-sided resected pancreatic cancer. J. Surg. Oncol. 2022, 126, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, E.S.; Balaji, U.; Mannakee, B.; Vail, P.; Eslinger, C.; Moxom, C.; Mansour, J.; Witkiewicz, A.K. Pancreatic cancer cell lines as patient-derived avatars: Genetic characterisation and functional utility. Gut 2018, 67, 508–520. [Google Scholar] [CrossRef]
- Biankin, A.V.; Waddell, N.; Kassahn, K.S.; Gingras, M.-C.; Muthuswamy, L.B.; Johns, A.L.; Miller, D.K.; Wilson, P.J.; Patch, A.-M.; Wu, J.; et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012, 491, 399–405. [Google Scholar] [CrossRef]
- Kessler, D.; Gmachl, M.; Mantoulidis, A.; Martin, L.J.; Zoephel, A.; Mayer, M.; Gollner, A.; Covini, D.; Fischer, S.; Gerstberger, T.; et al. Drugging an undruggable pocket on KRAS. Proc. Natl. Acad. Sci. USA 2019, 116, 15823–15829. [Google Scholar] [CrossRef]
- Punekar, S.R.; Velcheti, V.; Neel, B.G.; Wong, K.K. The current state of the art and future trends in RAS-targeted cancer therapies. Nat. Rev. Clin. Oncol. 2022, 19, 637–655. [Google Scholar] [CrossRef] [PubMed]
- ClinicalTrials.Gov. NCT03608631: iExosomes in Treating Participants with Metastatic Pancreas Cancer with KrasG12D Mutation. Available online: https://clinicaltrials.gov/study/NCT03608631 (accessed on 14 February 2024).
- Choi, J.K.; Cho, H.; Moon, B.S. Small Molecule Destabilizer of beta-Catenin and Ras Proteins Antagonizes Growth of K-Ras Mutation-Driven Colorectal Cancers Resistant to EGFR Inhibitors. Target Oncol. 2020, 15, 645–657. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-K.; Cho, Y.-H.; Cha, P.-H.; Yoon, J.-S.; Ro, E.J.; Jeong, W.-J.; Park, J.; Kim, H.; Kim, T.I.; Min, D.S.; et al. A small molecule approach to degrade RAS with EGFR repression is a potential therapy for KRAS mutation-driven colorectal cancer resistance to cetuximab. Exp. Mol. Med. 2018, 50, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, E.C.; Drezner, N.; Li, X.; Mishra-Kalyani, P.S.; Liu, Y.; Zhao, H.; Bi, Y.; Liu, J.; Rahman, A.; Wearne, E.; et al. FDA Approval Summary: Sotorasib for KRAS G12C-Mutated Metastatic NSCLC. Clin. Cancer Res. 2022, 28, 1482–1486. [Google Scholar] [CrossRef]
- Strickler, J.H.; Satake, H.; George, T.J.; Yaeger, R.; Hollebecque, A.; Garrido-Laguna, I.; Schuler, M.; Burns, T.F.; Coveler, A.L.; Falchook, G.S.; et al. Sotorasib in KRAS p.G12C-Mutated Advanced Pancreatic Cancer. N. Engl. J. Med. 2023, 388, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Bekaii-Saab, T.S.; Yaeger, R.; Spira, A.I.; Pelster, M.S.; Sabari, J.K.; Hafez, N.; Barve, M.; Velastegui, K.; Yan, X.; Shetty, A.; et al. Adagrasib in Advanced Solid Tumors Harboring a KRAS(G12C) Mutation. J. Clin. Oncol. 2023, 41, 4097–4106. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Allen, S.; Blake, J.F.; Bowcut, V.; Briere, D.M.; Calinisan, A.; Dahlke, J.R.; Fell, J.B.; Fischer, J.P.; Gunn, R.J.; et al. Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS(G12D) Inhibitor. J. Med. Chem. 2022, 65, 3123–3133. [Google Scholar] [CrossRef]
- ClinicalTrials.Gov. NCT05379985: Study of RMC-6236 in Patients with Advanced Solid Tumors Harboring Specific Mutations in RAS. Available online: https://www.clinicaltrials.gov/study/NCT05379985. (accessed on 14 February 2024).
- Revolution-Medicines. Revolution Medicines Presents Encouraging Clinical Data for RMC-6236 and RMC-6291 at 2023 Triple Meeting. Available online: https://ir.revmed.com/news-releases/news-release-details/revolution-medicines-presents-encouraging-clinical-data-rmc-6236 (accessed on 14 February 2024).
- Drugging RAS: Moving Beyond KRASG12C. Cancer Discov. 2023, 13, OF7. [CrossRef]
- ClinicalTrials.Gov. NCT03592888: DC Vaccine in Pancreatic Cancer. Available online: https://clinicaltrials.gov/study/NCT03592888 (accessed on 14 February 2024).
- ClinicalTrials.Gov. NCT04117087: Pooled Mutant KRAS-Targeted Long Peptide Vaccine Combined with Nivolumab and Ipilimumab for Patients with Resected MMR-p Colorectal and Pancreatic Cancer. Available online: https://clinicaltrials.gov/study/NCT04117087 (accessed on 14 February 2024).
- ClinicalTrials.Gov. NCT04853017: A Study of ELI-002 in Subjects with KRAS Mutated Pancreatic Ductal Adenocarcinoma (PDAC) and Other Solid Tumors (AMPLIFY-201). Available online: https://clinicaltrials.gov/study/NCT04853017 (accessed on 14 February 2024).
- ClinicalTrials.Gov. NCT03745326: Administering Peripheral Blood Lymphocytes Transduced with a Murine T-Cell Receptor Recognizing the G12D Variant of Mutated RAS in HLA-A*11:01 Patients. Available online: https://clinicaltrials.gov/study/NCT03745326 (accessed on 14 February 2024).
- ClinicalTrials.Gov. NCT04146298: Mutant KRAS G12V-specific TCR Transduced T Cell Therapy for Advanced Pancreatic Cancer. Available online: https://clinicaltrials.gov/study/NCT04146298 (accessed on 14 February 2024).
- Network NCC. Pancreatic Adenocarcinoma (Version 1.2024). Available online: https://www.nccn.org/professionals/physician_gls/pdf/pancreatic.pdf (accessed on 25 March 2024).
- Nicolle, R.; Gayet, O.; Duconseil, P.; Vanbrugghe, C.; Roques, J.; Bigonnet, M.; Blum, Y.; Elarouci, N.; Armenoult, L.; Ayadi, M.; et al. A transcriptomic signature to predict adjuvant gemcitabine sensitivity in pancreatic adenocarcinoma. Ann. Oncol. 2021, 32, 250–260. [Google Scholar] [CrossRef]
- Nicolle, R.; Bachet, J.B.; Harlé, A.; Iovanna, J.; Hammel, P.; Rebours, V.; Turpin, A.; Ben Abdelghani, M.; Wei, A.; Mitry, E.; et al. Prediction of Adjuvant Gemcitabine Sensitivity in Resectable Pancreatic Adenocarcinoma Using the GemPred RNA Signature: An Ancillary Study of the PRODIGE-24/CCTG PA6 Clinical Trial. J. Clin. Oncol. 2024, 42, 1067–1076. [Google Scholar] [CrossRef]
- Ecker, B.L.; Tao, A.J.; Janssen, Q.P.; Walch, H.S.; Court, C.M.; Balachandran, V.P.; Crane, C.H.; D’Angelica, M.I.; Drebin, J.A.; Kingham, T.P.; et al. Genomic Biomarkers Associated with Response to Induction Chemotherapy in Patients with Localized Pancreatic Ductal Adenocarcinoma. Clin. Cancer Res. 2023, 29, 1368–1374. [Google Scholar] [CrossRef] [PubMed]
- Brar, G.; Blais, E.M.; Bender, R.J.; Brody, J.R.; Sohal, D.; Madhavan, S.; Picozzi, V.J.; Hendifar, A.E.; Chung, V.M.; Halverson, D.; et al. Multi-omic molecular comparison of primary versus metastatic pancreatic tumours. Br. J. Cancer 2019, 121, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Chao, T.; Wang, Z.-X.; Bowne, W.B.; Yudkoff, C.J.; Torjani, A.; Swaminathan, V.; Kavanagh, T.R.; Roadarmel, A.; Sholevar, C.J.; Cannaday, S.; et al. Association of Mutant KRAS Alleles with Morphology and Clinical Outcomes in Pancreatic Ductal Adenocarcinoma. Arch. Pathol. Lab. Med. 2024. [Google Scholar] [CrossRef] [PubMed]
| Pancreatic Cancer Stage | Degree of Spread | 5-Year Survival Rate (%) |
|---|---|---|
| Stage I | Localized | 44 |
| Stage II/III | Regional | 15 |
| Stage IV | Distant | 3 |
| Study | Cohort | Findings |
|---|---|---|
| Hendifar et al. [19] | Advanced disease | KRAS G12R and KRAS G12V have greater overall survival than KRAS G12D in advanced-stage PDAC. |
| Chen et al. [12] | Advanced disease | ctDNA analysis revealed mutant KRAS was associated with shorter OS than patients with wild-type KRAS. |
| Diehl et al. [18] | Advanced Disease | KRAS G12R was associated with improved OS and PFS compared with non-G12R. |
| Ogura et al. [13] | Advanced disease | KRAS G12D and G12R mutations had shorter overall survival than G12V, which had shorter survival than wild-type KRAS in unresectable pancreatic cancer patients. |
| Dai et al. [9] | Resectable Disease | KRAS G12D was associated with worse survival compared with wild-type KRAS. |
| Kawesha et al. [20] | Resectable Disease | KRAS G12V and KRAS wild-type had longer overall survival than KRAS G12D in patients who had undergone resection for PDAC. |
| Schultz et al. [24] | Resectable Disease | Mutant KRAS was not associated with RFS and OS compared with wild-type KRAS. |
| McIntyre et al. [14] | Resectable Disease | Mutant KRAS correlated with worse survival compared with wild-type KRAS. |
| Shen et al. [21] | Resectable Disease | KRAS G12D in resectable PDAC patients had shorter overall survival than non-G12D patients. |
| Guo et al. [23] | Resectable disease | ctDNA analysis revealed mutant KRAS detection to be strongly associated with worse OS and RFS. G12D mutations were strongly, correlated with poor OS and RFS. |
| Bournet et al. [17] | All stages | Mutant KRAS and wild-type KRAS had comparable survival. G12D had significantly worse survival. |
| Yousef et al. [15] | All stages | KRAS G12D and Q61H mutations had shorter overall survival, while G12R and wild-type KRAS had improved and comparable survival. |
| Windon et al. [10] | All stages | Mutant KRAS correlated with worse survival compared with wild-type KRAS. |
| Zhou et al. [16] | All stages | KRAS mutation status did not correlate with survival. |
| Shoucair et al. [22] | All stages | KRAS G12D co-occurrence with mutant TP53 was associated with improved survival. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nusrat, F.; Khanna, A.; Jain, A.; Jiang, W.; Lavu, H.; Yeo, C.J.; Bowne, W.; Nevler, A. The Clinical Implications of KRAS Mutations and Variant Allele Frequencies in Pancreatic Ductal Adenocarcinoma. J. Clin. Med. 2024, 13, 2103. https://doi.org/10.3390/jcm13072103
Nusrat F, Khanna A, Jain A, Jiang W, Lavu H, Yeo CJ, Bowne W, Nevler A. The Clinical Implications of KRAS Mutations and Variant Allele Frequencies in Pancreatic Ductal Adenocarcinoma. Journal of Clinical Medicine. 2024; 13(7):2103. https://doi.org/10.3390/jcm13072103
Chicago/Turabian StyleNusrat, Faria, Akshay Khanna, Aditi Jain, Wei Jiang, Harish Lavu, Charles J. Yeo, Wilbur Bowne, and Avinoam Nevler. 2024. "The Clinical Implications of KRAS Mutations and Variant Allele Frequencies in Pancreatic Ductal Adenocarcinoma" Journal of Clinical Medicine 13, no. 7: 2103. https://doi.org/10.3390/jcm13072103
APA StyleNusrat, F., Khanna, A., Jain, A., Jiang, W., Lavu, H., Yeo, C. J., Bowne, W., & Nevler, A. (2024). The Clinical Implications of KRAS Mutations and Variant Allele Frequencies in Pancreatic Ductal Adenocarcinoma. Journal of Clinical Medicine, 13(7), 2103. https://doi.org/10.3390/jcm13072103





