Genome-Wide Association Screens for Anterior Cruciate Ligament Tears
Abstract
:1. Introduction
- 1.
- Genes encoding for collagen
- 2.
- Genes encoding for proteoglycans
- 3.
- Genes encoding for matrix metalloproteinases
- 4.
- Genes encoding for angiogenesis-associated signaling cascades and growth differentiation hormone factors
- 5.
- Genes encoding for elastin and fibrillin
- 6.
- Genes encoding for interleukins
- 7.
- Genes encoding for b-fibrinogen
2. Materials and Methods
3. Results
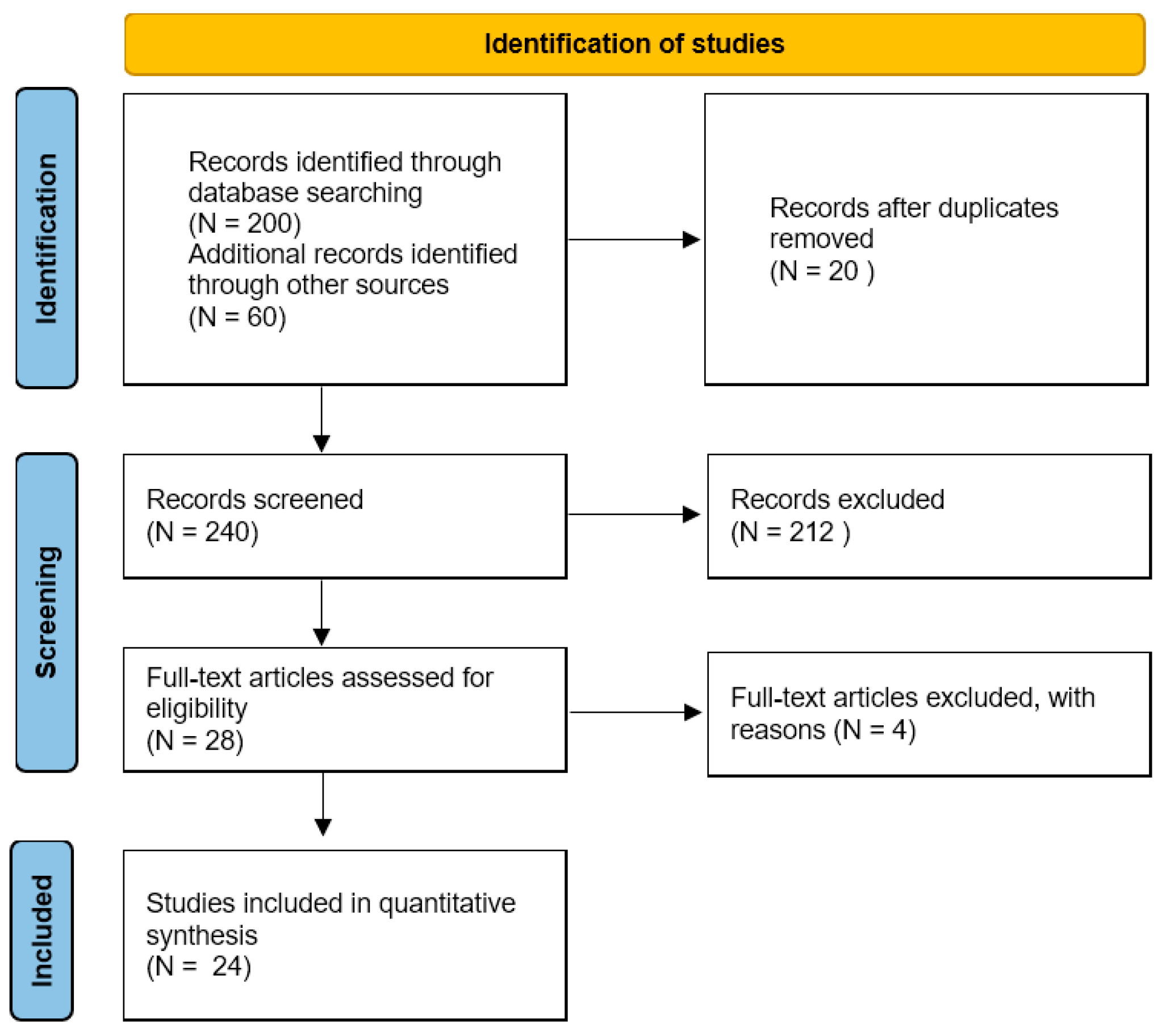
3.1. Study Selection
3.2. Study Characteristics
- a.
- Collagen
- b.
- Proteoglycans
- c.
- Matrix Metalloproteinases
- d.
- Angiogenesis-Associated Signaling Cascades and Growth Differentiation Hormone Factors
- e.
- Elastin and Fibrillin
- f.
- Interleukins
- g.
- B-fibrinogen
| Study | Year | Design | Patients with ACL Ruptures (n) | Controls (n) | Gene | Product | Variant | Association with ACL Ruptures (Differences in Patients with ACL Ruptures vs. Control) |
|---|---|---|---|---|---|---|---|---|
| Ficek et al. [25] | 2013 | Case– control | 91 | 143 | COL1A1 | Collagen | rs1800012 and rs1107946 | No significant differences |
| Ficek et al. [26] | 2014 | Case– control | 91 | 143 | COL12A1 | Collagen | rs970547 | No significant differences |
| Khoschnau et a [27] | 2008 | Case– control | 233 | 325 | COL1A1 | Collagen | rs1800012 | No significant differences |
| Khoury et al. [18] | 2015 | Case– control | 141 | 219 | ELN | Elastin | rs2071307 | No significant differences |
| - | - | - | - | - | FBN2 | Fibrillin-2 | rs331079 | No significant differences |
| Malila et al. [28] | 2011 | Case– control | 86 | 100 | MMP3 | Matrix metalloproteinase | –1612 | No significant differences |
| Mannion et al. [11] | 2014 | Case– control | 227 | 234 | ACAN | Proteoglycans | rs2351491, rs1042631, and rs1516797 | rs2351491 and rs1042631: no significant differences ACAN rs1516797: was significantly under-represented in the controls group (p = 0.024) |
| - | - | - | - | BGN | rs1126499 and rs1042103 | No significant differences | ||
| - | - | - | - | DCN | rs13312816 and rs516115 | rs13312816: no significant differences; rs516115: significant association | ||
| - | - | - | - | FMOD | rs7543148 and rs10800912 | No significant differences | ||
| - | - | - | - | LUM | rs2268578 | No significant differences | ||
| O’Connell et al. [29] | 2015 | Case– control | 242 (South African population) 91 (Polish population) | 235 (South African population) 91 (Polish population) | COL3A1 and COL6A1 | Collagen | rs1800255 and rs35796750 | rs1800255; significant association; rs35796750: no significant differences |
| Posthumus et al. [30] | 2009 | Case– control | 117 | 130 | COL1A1 | Collagen | rs1800012 | rs1800012: significant association |
| Posthumus et al. [31] | 2009 | Case– control | 129 | 216 | COL5A1 | Collagen | rs12722 and rs13946 | No significant differences |
| Posthumus et al. [32] | 2010 | Case– control | 129 | 216 | COL12A1 | Collagen | rs970547 and rs240736 | No significant differences |
| Posthumus et al. [14] | 2012 | Case– control | 129 | 216 | MMP1, MMP3, MMP10, and MMP12 | Matrix metalloproteinase | rs1799750, rs679620, rs486055, and rs2276109 | No significant differences |
| Rahim et al. [16] | 2014 | Case– control | 227 | 227 | VEGFA, KDR, NGFB, and HIF1A | Angiogenesis-associated signaling cascade genes | rs699947, rs1570360, rs2010963, rs1870377, rs2071559, rs6678788, and rs11549465 | No significant differences |
| Raleigh et al. [17] | 2013 | Case– control | 126 | 216 | GDF5 | Growth differentiation factor | rs143383 | No significant differences |
| Stepien-Słodkowska et al. [33] | 2013 | Case– control | 138 | 183 | COL1A1 | Collagen | rs1800012 | Significant association |
| Stepien-Słodkowska et al. [34] | 2015 | Case– control | 138 | 183 | COL3A1 | Collagen | rs1800255 | Significant association |
| Stepien-Słodkowska et al. [35] | 2015 | Case– control | 138 | 183 | COL3A1 | Collagen | rs13946 and rs12722 | No significant differences |
| Cięszczyk et al. [10] | 2017 | Case– control | 143 | 229 | ACAN | Proteoglycans | rs1516797 | Significant association T/T (p > 0.041) |
| BGN | rs1042103 and rs1126499 | BGN rs1042103 A allele: significant association in the male | ||||||
| DCN | rs516115 | No significant differences | ||||||
| VEGFA | Angiogenesis-associated signaling cascade genes | rs699947 | No significant differences | |||||
| Lulinska-Kuklik et al. [23] | 2018 | Case– control | 229 | 192 | MMP3 | Matrix metalloproteinases | rs591058C/T and rs679620 G/A | Significant association: MMP3 rs591058C and rs679620 G alleles were significantly over-represented in cases compared to controls (p = 0.021) |
| MMP8 | Matrix metalloproteinases | rs11225395C/T | No significant differences | |||||
| TIMP2 | Tissue inhibitors of metalloproteases | rs4789932 G/A | No significant differences | |||||
| Lulinska et al. [13] | 2020 | Case– control | 228 | 202 | MMP1 | Matrix metalloproteinases | rs1799750, →G | No significant differences |
| MMP10 | Matrix metalloproteinases | rs486055 C > T | No significant differences | |||||
| MMP12 | Matrix metalloproteinases | rs2276109 T > C | No significant differences | |||||
| Lulinska-Kuklik et al. [19] | 2019 | Case– control | 229 | 194 | IL1B | Interleukins | rs16944 and rs1143627 | No significant differences |
| IL6 | Interleukins | rs1800795 | Significant association | |||||
| IL6R | Interleukins | rs2228145 | No significant differences | |||||
| Shukla et al. [24] | 2020 | Case– control | 90 | 76 | VEGFA | Angiogenesis-associated signaling cascade genes | rs699947 and rs35569394 | Significant association: the A allele (rs699947) and I allele (rs35569394) were significantly over-represented in the ACL group |
| Shukla et al. [36] | 2020 | Case– control | 90 | 76 | COLIA1 | Angiogenesis-associated signaling cascade genes | COLIA1 Sp1 + 1245 G > T | No significant differences |
| Willard et al. [22] | 2017 | Case– control | 227 | 234 | BGN | Proteoglycans | rs1126499 and rs1042103 | Significant association |
| COL5A1 | Collagen | rs12722 | Significant association | |||||
| DCN | Proteoglycans | rs516115 | Significant association | |||||
| Zhao et al. [20] | 2020 | Case– control | 101 | 110 | COL1A1 | Collagen | rs1800012 | No significant differences |
| COL5A1 | Collagen | rs12722 and rs13946 | No significant differences | |||||
| COL12A1 | Collagen | rs970547 and rs240736 | Significant association | |||||
| FGB | B-fibrinogen | rs1800787, rs1800788, rs1800789, rs1800790, rs1800791, and rs2227389 | rs1800789 and rs1800791: no significant differences; rs1800787, rs1800788, rs1800790, and rs2227389: significant association; rs1800787, rs1800788, rs1800790, and rs2227389 genotypes in the b-fib promoter region: association with ACL injuries |
3.3. Risk of Bias
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Giuliani, J.R.; Kilcoyne, K.G.; Rue, J.P. Anterior cruciate ligament anatomy: A review of the anteromedial and posterolateral bundles. J. Knee Surg. 2009, 22, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Adhitya, I.P.G.S.; Kurniawati, I.; Sawa, R.; Wijaya, T.F.; Dewi, N.P.A.C. The Risk Factors and Preventive Strategies of Poor Knee Functions and Osteoarthritis after Anterior Cruciate Ligament Reconstruction: A Narrative Review. Phys. Ther. Res. 2023, 26, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Longo, U.G.; Nagai, K.; Salvatore, G.; Cella, E.; Candela, V.; Cappelli, F.; Ciccozzi, M.; Denaro, V. Epidemiology of Anterior Cruciate Ligament Reconstruction Surgery in Italy: A 15-Year Nationwide Registry Study. J. Clin. Med. 2021, 10, 223. [Google Scholar] [CrossRef] [PubMed]
- Longo, U.G.; Buchmann, S.; Franceschetti, E.; Maffulli, N.; Denaro, V. A systematic review of single-bundle versus double-bundle anterior cruciate ligament reconstruction. Br. Med. Bull. 2012, 103, 147–168. [Google Scholar] [CrossRef] [PubMed]
- Flynn, R.K.; Pedersen, C.L.; Birmingham, T.B.; Kirkley, A.; Jackowski, D.; Fowler, P.J. The familial predisposition toward tearing the anterior cruciate ligament: A case control study. Am. J. Sports Med. 2005, 33, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Longo, U.G.; King, J.B.; Denaro, V.; Maffulli, N. Double-bundle arthroscopic reconstruction of the anterior cruciate ligament: Does the evidence add up? J. Bone Jt. Surg. 2008, 90, 995–999. [Google Scholar] [CrossRef] [PubMed]
- Longo, U.G.; Salvatore, G.; Ruzzini, L.; Risi Ambrogioni, L.; de Girolamo, L.; Vigano, M.; Facchini, F.; Cella, E.; Candela, V.; Ciccozzi, M.; et al. Trends of anterior cruciate ligament reconstruction in children and young adolescents in Italy show a constant increase in the last 15 years. Knee Surg. Sports Traumatol. Arthrosc. Off. J. ESSKA 2021, 29, 1728–1733. [Google Scholar] [CrossRef] [PubMed]
- September, A.V.; Schwellnus, M.P.; Collins, M. Tendon and ligament injuries: The genetic component. Br. J. Sports Med. 2007, 41, 241–246; discussion 246. [Google Scholar] [CrossRef]
- Kaynak, M.; Nijman, F.; van Meurs, J.; Reijman, M.; Meuffels, D.E. Genetic Variants and Anterior Cruciate Ligament Rupture: A Systematic Review. Sports Med. 2017, 47, 1637–1650. [Google Scholar] [CrossRef]
- Cieszczyk, P.; Willard, K.; Gronek, P.; Zmijewski, P.; Trybek, G.; Gronek, J.; Weber-Rajek, M.; Stastny, P.; Petr, M.; Lulinska-Kuklik, E.; et al. Are genes encoding proteoglycans really associated with the risk of anterior cruciate ligament rupture? Biol. Sport 2017, 34, 97–103. [Google Scholar] [CrossRef]
- Mannion, S.; Mtintsilana, A.; Posthumus, M.; van der Merwe, W.; Hobbs, H.; Collins, M.; September, A.V. Genes encoding proteoglycans are associated with the risk of anterior cruciate ligament ruptures. Br. J. Sports Med. 2014, 48, 1640–1646. [Google Scholar] [CrossRef] [PubMed]
- Kumar, L.; Bisen, M.; Khan, A.; Kumar, P.; Patel, S.K.S. Role of Matrix Metalloproteinases in Musculoskeletal Diseases. Biomedicines 2022, 10, 2477. [Google Scholar] [CrossRef]
- Lulinska, E.; Gibbon, A.; Kaczmarczyk, M.; Maciejewska-Skrendo, A.; Ficek, K.; Leonska-Duniec, A.; Wilk, M.; Leznicka, K.; Michalowska-Sawczyn, M.; Huminska-Lisowska, K.; et al. Matrix Metalloproteinase Genes (MMP1, MMP10, MMP12) on Chromosome 11q22 and the Risk of Non-Contact Anterior Cruciate Ligament Ruptures. Genes. 2020, 11, 766. [Google Scholar] [CrossRef] [PubMed]
- Posthumus, M.; Collins, M.; van der Merwe, L.; O’Cuinneagain, D.; van der Merwe, W.; Ribbans, W.J.; Schwellnus, M.P.; Raleigh, S.M. Matrix metalloproteinase genes on chromosome 11q22 and the risk of anterior cruciate ligament (ACL) rupture. Scand. J. Med. Sci. Sports 2012, 22, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Halper, J. Advances in the use of growth factors for treatment of disorders of soft tissues. Adv. Exp. Med. Biol. 2014, 802, 59–76. [Google Scholar] [CrossRef]
- Rahim, M.; Gibbon, A.; Hobbs, H.; van der Merwe, W.; Posthumus, M.; Collins, M.; September, A.V. The association of genes involved in the angiogenesis-associated signaling pathway with risk of anterior cruciate ligament rupture. J. Orthop. Res. Off. Publ. Orthop. Res. Soc. 2014, 32, 1612–1618. [Google Scholar] [CrossRef]
- Raleigh, S.M.; Posthumus, M.; O’Cuinneagain, D.; van der Merwe, W.; Collins, M. The GDF5 gene and anterior cruciate ligament rupture. Int. J. Sports Med. 2013, 34, 364–367. [Google Scholar] [CrossRef]
- Khoury, L.E.; Posthumus, M.; Collins, M.; van der Merwe, W.; Handley, C.; Cook, J.; Raleigh, S.M. ELN and FBN2 gene variants as risk factors for two sports-related musculoskeletal injuries. Int. J. Sports Med. 2015, 36, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Lulinska-Kuklik, E.; Maculewicz, E.; Moska, W.; Ficek, K.; Kaczmarczyk, M.; Michalowska-Sawczyn, M.; Huminska-Lisowska, K.; Buryta, M.; Chycki, J.; Cieszczyk, P.; et al. Are IL1B, IL6 and IL6R Gene Variants Associated with Anterior Cruciate Ligament Rupture Susceptibility? J. Sports Sci. Med. 2019, 18, 137–145. [Google Scholar]
- Zhao, D.; Zhang, Q.; Lu, Q.; Hong, C.; Luo, T.; Duan, Q.; Shu, S.; Lv, J.; Zhao, W. Correlations Between the Genetic Variations in the COL1A1, COL5A1, COL12A1, and beta-fibrinogen Genes and Anterior Cruciate Ligament Injury in Chinese Patients(a). J. Athl. Train. 2020, 55, 515–521. [Google Scholar] [CrossRef]
- Altman, D.G.; Schulz, K.F.; Moher, D.; Egger, M.; Davidoff, F.; Elbourne, D.; Gøtzsche, P.C.; Lang, T.; Trials, C.G.C.S.o.R. The revised CONSORT statement for reporting randomized trials: Explanation and elaboration. Ann. Intern. Med. 2001, 134, 663–694. [Google Scholar] [CrossRef] [PubMed]
- Willard, K.; Mannion, S.; Saunders, C.J.; Collins, M.; September, A.V. The interaction of polymorphisms in extracellular matrix genes and underlying miRNA motifs that modulate susceptibility to anterior cruciate ligament rupture. J. Sci. Med. Sport 2018, 21, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Lulinska-Kuklik, E.; Rahim, M.; Moska, W.; Maculewicz, E.; Kaczmarczyk, M.; Maciejewska-Skrendo, A.; Ficek, K.; Cieszczyk, P.; September, A.V.; Sawczuk, M. Are MMP3, MMP8 and TIMP2 gene variants associated with anterior cruciate ligament rupture susceptibility? J. Sci. Med. Sport 2019, 22, 753–757. [Google Scholar] [CrossRef] [PubMed]
- Shukla, M.; Gupta, R.; Pandey, V.; Rochette, J.; Dhandapany, P.S.; Tiwari, P.K.; Amrathlal, R.S. VEGFA Promoter Polymorphisms rs699947 and rs35569394 Are Associated With the Risk of Anterior Cruciate Ligament Ruptures Among Indian Athletes: A Cross-sectional Study. Orthop. J. Sports Med. 2020, 8, 2325967120964472. [Google Scholar] [CrossRef] [PubMed]
- Ficek, K.; Cieszczyk, P.; Kaczmarczyk, M.; Maciejewska-Karlowska, A.; Sawczuk, M.; Cholewinski, J.; Leonska-Duniec, A.; Stepien-Slodkowska, M.; Zarebska, A.; Stepto, N.K.; et al. Gene variants within the COL1A1 gene are associated with reduced anterior cruciate ligament injury in professional soccer players. J. Sci. Med. Sport 2013, 16, 396–400. [Google Scholar] [CrossRef] [PubMed]
- Ficek, K.; Stepien-Slodkowska, M.; Kaczmarczyk, M.; Maciejewska-Karlowska, A.; Sawczuk, M.; Cholewinski, J.; Leonska-Duniec, A.; Zarebska, A.; Cieszczyk, P.; Zmijewski, P. Does the A9285G Polymorphism in Collagen Type XII alpha1 Gene Associate with the Risk of Anterior Cruciate Ligament Ruptures? Balk. J. Med. Genet. BJMG 2014, 17, 41–46. [Google Scholar] [CrossRef]
- Khoschnau, S.; Melhus, H.; Jacobson, A.; Rahme, H.; Bengtsson, H.; Ribom, E.; Grundberg, E.; Mallmin, H.; Michaelsson, K. Type I collagen alpha1 Sp1 polymorphism and the risk of cruciate ligament ruptures or shoulder dislocations. Am. J. Sports Med. 2008, 36, 2432–2436. [Google Scholar] [CrossRef] [PubMed]
- Malila, S.; Yuktanandana, P.; Saowaprut, S.; Jiamjarasrangsi, W.; Honsawek, S. Association between matrix metalloproteinase-3 polymorphism and anterior cruciate ligament ruptures. Genet. Mol. Res. GMR 2011, 10, 4158–4165. [Google Scholar] [CrossRef]
- O’Connell, K.; Knight, H.; Ficek, K.; Leonska-Duniec, A.; Maciejewska-Karlowska, A.; Sawczuk, M.; Stepien-Slodkowska, M.; O’Cuinneagain, D.; van der Merwe, W.; Posthumus, M.; et al. Interactions between collagen gene variants and risk of anterior cruciate ligament rupture. Eur. J. Sport Sci. 2015, 15, 341–350. [Google Scholar] [CrossRef]
- Posthumus, M.; September, A.V.; Keegan, M.; O’Cuinneagain, D.; Van der Merwe, W.; Schwellnus, M.P.; Collins, M. Genetic risk factors for anterior cruciate ligament ruptures: COL1A1 gene variant. Br. J. Sports Med. 2009, 43, 352–356. [Google Scholar] [CrossRef]
- Posthumus, M.; September, A.V.; O’Cuinneagain, D.; van der Merwe, W.; Schwellnus, M.P.; Collins, M. The COL5A1 gene is associated with increased risk of anterior cruciate ligament ruptures in female participants. Am. J. Sports Med. 2009, 37, 2234–2240. [Google Scholar] [CrossRef] [PubMed]
- Posthumus, M.; September, A.V.; O’Cuinneagain, D.; van der Merwe, W.; Schwellnus, M.P.; Collins, M. The association between the COL12A1 gene and anterior cruciate ligament ruptures. Br. J. Sports Med. 2010, 44, 1160–1165. [Google Scholar] [CrossRef] [PubMed]
- Stepien-Slodkowska, M.; Ficek, K.; Eider, J.; Leonska-Duniec, A.; Maciejewska-Karlowska, A.; Sawczuk, M.; Zarebska, A.; Jastrzebski, Z.; Grenda, A.; Kotarska, K.; et al. The +1245g/t polymorphisms in the collagen type I alpha 1 (col1a1) gene in polish skiers with anterior cruciate ligament injury. Biol. Sport 2013, 30, 57–60. [Google Scholar] [CrossRef] [PubMed]
- Stepien-Slodkowska, M.; Ficek, K.; Maciejewska-Karlowska, A.; Sawczuk, M.; Zietek, P.; Krol, P.; Zmijewski, P.; Pokrywka, A.; Cieszczyk, P. Overrepresentation of the COL3A1 AA genotype in Polish skiers with anterior cruciate ligament injury. Biol. Sport 2015, 32, 143–147. [Google Scholar] [CrossRef]
- Stepien-Slodkowska, M.; Ficek, K.; Kaczmarczyk, M.; Maciejewska-Karlowska, A.; Sawczuk, M.; Leonska-Duniec, A.; Stepinski, M.; Zietek, P.; Krol, P.; Chudecka, M.; et al. The Variants Within the COL5A1 Gene are Associated with Reduced Risk of Anterior Cruciate Ligament Injury in Skiers. J. Hum. Kinet. 2015, 45, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Shukla, M.; Gupta, R.; Pandey, V.; Tiwari, P.K.; Amrathlal, R.S. COLIA1 + 1245 G > T Sp1 Binding Site Polymorphism is Not Associated with ACL Injury Risks Among Indian Athletes. Indian J. Orthop. 2020, 54, 647–654. [Google Scholar] [CrossRef] [PubMed]
- de Sire, A.; Demeco, A.; Marotta, N.; Moggio, L.; Palumbo, A.; Iona, T.; Ammendolia, A. Anterior Cruciate Ligament Injury Prevention Exercises: Could a Neuromuscular Warm-Up Improve Muscle Pre-Activation before a Soccer Game? A Proof-of-Principle Study on Professional Football Players. Appl. Sci. 2021, 11, 4958. [Google Scholar] [CrossRef]
- de Sire, A.; Marotta, N.; Demeco, A.; Moggio, L.; Paola, P.; Marotta, M.; Iona, T.; Invernizzi, M.; Leigheb, M.; Ammendolia, A. Electromyographic Assessment of Anterior Cruciate Ligament Injury Risk in Male Tennis Players: Which Role for Visual Input? A Proof-of-Concept Study. Diagnostics 2021, 11, 997. [Google Scholar] [CrossRef]
- Marotta, N.; de Sire, A.; Gimigliano, A.; Demeco, A.; Moggio, L.; Vescio, A.; Iona, T.; Ammendolia, A. Impact of COVID-19 lockdown on the epidemiology of soccer muscle injuries in Italian Serie A professional football players. J. Sports Med. Phys. Fit. 2022, 62, 356–360. [Google Scholar] [CrossRef]
- Demeco, A.; de Sire, A.; Marotta, N.; Spanò, R.; Lippi, L.; Palumbo, A.; Iona, T.; Gramigna, V.; Palermi, S.; Leigheb, M.; et al. Match Analysis, Physical Training, Risk of Injury and Rehabilitation in Padel: Overview of the Literature. Int. J. Environ. Res. Public. Health 2022, 19, 4153. [Google Scholar] [CrossRef]
- Bernad, M.; Martinez, M.E.; Escalona, M.; Gonzalez, M.L.; Gonzalez, C.; Garces, M.V.; Del Campo, M.T.; Martin Mola, E.; Madero, R.; Carreno, L. Polymorphism in the type I collagen (COLIA1) gene and risk of fractures in postmenopausal women. Bone 2002, 30, 223–228. [Google Scholar] [CrossRef] [PubMed]
| Criterion | Question |
|---|---|
| Case | Have the cases been clearly and sufficiently defined? |
| Control | Have the controls been clearly and adequately defined? |
| Selection bias | Has selection bias been adequately addressed and excluded? |
| Defined exposure | Has the exposure been clearly defined, and is the method employed to assess this exposure deemed appropriate? |
| Determination | Was blinding to exposure status maintained prior to determining the presence of the disease? |
| Confounding | Have the primary confounding factors been identified and adequately accounted for in both the study design and analysis? |
| Case | Control | Selection Bias | Defined Eposure | Determination Exposure | Confouding | Overall | |
|---|---|---|---|---|---|---|---|
| Ficek et al. [25] | + | + | ? | + | ? | + | ? |
| Ficek et al. [26] | + | + | ? | + | ? | + | ? |
| Khoschnau et al. [27] | + | ? | + | + | + | - | - |
| Khoury et al. [18] | + | + | ? | + | ? | - | - |
| Malila et al. [28] | + | + | ? | + | ? | - | - |
| Mannion et al. [11] | + | + | ? | + | ? | - | - |
| O’Connell et al. [29] | + | ? | ? | + | + | - | - |
| Posthumus et al. [30] | + | + | ? | + | ? | - | - |
| Posthumus et al. [31] | + | + | ? | + | ? | + | ? |
| Posthumus et al. [32] | + | + | ? | + | ? | + | ? |
| Posthumus et al. [14] | + | + | ? | + | ? | + | ? |
| Rahim et al. [16] | + | ? | ? | + | ? | - | - |
| Raleigh et al. [17] | + | + | ? | + | ? | + | ? |
| Stepien-Słodkowska et al. [33] | + | + | ? | + | ? | ? | ? |
| Stepien-Słodkowska et al. [34] | + | + | ? | + | ? | ? | ? |
| Stepien-Słodkowska et al. [35] | + | + | ? | + | ? | ? | ? |
| Cięszczyk et al. [10] | + | + | ? | + | ? | ? | ? |
| Lulinska-Kuklik et al. [23] | + | + | ? | + | ? | + | ? |
| Lulinska et al. [13] | + | + | ? | + | ? | + | ? |
| Lulinska-Kuklik et al. [19] | + | + | ? | + | ? | + | ? |
| Shukla et al. [24] | + | + | ? | + | ? | - | - |
| Shukla et al. [36] | + | + | ? | + | ? | ? | ? |
| Willard et al. [22] | + | + | ? | + | ? | + | ? |
| Zhao et al. [20] | + | + | ? | + | ? | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Candela, V.; Longo, U.G.; Berton, A.; Salvatore, G.; Forriol, F.; de Sire, A.; Denaro, V. Genome-Wide Association Screens for Anterior Cruciate Ligament Tears. J. Clin. Med. 2024, 13, 2330. https://doi.org/10.3390/jcm13082330
Candela V, Longo UG, Berton A, Salvatore G, Forriol F, de Sire A, Denaro V. Genome-Wide Association Screens for Anterior Cruciate Ligament Tears. Journal of Clinical Medicine. 2024; 13(8):2330. https://doi.org/10.3390/jcm13082330
Chicago/Turabian StyleCandela, Vincenzo, Umile Giuseppe Longo, Alessandra Berton, Giuseppe Salvatore, Francisco Forriol, Alessandro de Sire, and Vincenzo Denaro. 2024. "Genome-Wide Association Screens for Anterior Cruciate Ligament Tears" Journal of Clinical Medicine 13, no. 8: 2330. https://doi.org/10.3390/jcm13082330







