Analysis of Metabolomic Changes in Lettuce Leaves under Low Nitrogen and Phosphorus Deficiencies Stresses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Metabolite Extraction
2.3. Non-Targeted LC–MS Analysis
2.4. Data Processing, Statistical Analysis, and Metabolic Pathway Analysis
3. Results
3.1. Metabolic Biomarkers Screening
3.2. Cluster Analysis of Metabolic Biomarkers
3.3. KEGG Metabolic Pathway Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Yoneyama, K.; Xie, X.; Kim, H.I.; Kisugi, T.; Nomura, T.; Sekimoto, H.; Yokota, T.; Yoneyama, K. How do nitrogen and phosphorus deficiencies affect strigolactone production and exudation? Planta 2012, 235, 1197–1207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larbat, R.; Olsen, K.M.; Slimestad, R.; Lovdal, T.; Benard, C.; Verheul, M.; Bourgaud, F.; Robin, C.; Lillo, C. Influence of repeated short-term nitrogen limitations on leaf phenolics metabolism in tomato. Phytochemistry 2012, 77, 119–128. [Google Scholar] [CrossRef]
- Cao, Y.W.; Qu, R.J.; Tang, X.Q.; Sun, L.Q.; Chen, Q.Q.; Miao, Y.J. UPLC-Triple TOF-MS/MS based metabolomics approach to reveal the influence of nitrogen levels on Isatis indigotica seedling leaf. Sci. Hortic. 2020, 266, 109280. [Google Scholar] [CrossRef]
- Carstensen, A.; Herdean, A.; Schmidt, S.B.; Sharma, A.; Spetea, C.; Pribil, M.; Husted, S. The impacts of phosphorus deficiency on the photosynthetic electron transport chain. Plant Physiol. 2018, 177, 271–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdel-Rahman, E.M.; Ahmed, F.B.; van den Berg, M. Estimation of sugarcane leaf nitrogen concentration using in situ spectroscopy. Int. J. Appl. Earth Obs. Geoinf. 2010, 12 (Suppl. S1), S52–S57. [Google Scholar] [CrossRef]
- Story, D.; Kacira, M.; Kubota, C.; Akoglu, A. Morphological and Textural Plant Feature Detection Using Machine Vision for Intelligent Plant Health, Growth and Quality Monitoring. Acta Hortic. 2011, 893, 299–306. [Google Scholar] [CrossRef]
- Chen, L.S.; Huang, S.H.; Sun, Y.Y.; Zhu, E.Y.; Wang, K. Rapid Identification of Potassium Nutrition Stress in Rice Based on Machine Vision and Object-Oriented Segmentation. J. Spectrosc. 2019, 2019, 4623545. [Google Scholar] [CrossRef]
- Mao, H.P.; Gao, H.Y.; Zhang, X.D.; Kumi, F. Nondestructive measurement of total nitrogen in lettuce by integrating spectroscopy and computer vision. Sci. Hortic. 2015, 184, 1–7. [Google Scholar] [CrossRef]
- Kalaji, H.M.; Jajoo, A.; Oukarroum, A.; Brestic, M.; Zivcak, M.; Samborska, I.A.; Cetner, M.D.; Lukasik, I.; Goltsev, V.; Ladle, R.J. Chlorophyll a fluorescence as a tool to monitor physiological status of plants under abiotic stress conditions. Acta Physiol. Plant. 2016, 38, 102. [Google Scholar] [CrossRef] [Green Version]
- Gao, H.Y.; Mao, H.P.; Zhang, X.D. Determination of lettuce nitrogen content using spectroscopy with efficient wavelength selection and extreme learning machine. Zemdirb. Agric. 2015, 102, 51–58. [Google Scholar] [CrossRef] [Green Version]
- Mayerle, J.; Kalthoff, H.; Reszka, R.; Kamlage, B.; Peter, E.; Schniewind, B.; Maldonado, S.G.; Pilarsky, C.; Heidecke, C.-D.; Schatz, P.; et al. Metabolic biomarker signature to differentiate pancreatic ductal adenocarcinoma from chronic pancreatitis. Gut 2018, 67, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Wani, S.H.; Kumar, V.; Shriram, V.; Sah, S.K. Phytohormones and their metabolic engineering for abiotic stress tolerance in crop plants. Crop J. 2016, 4, 162–176. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Ma, X.M.; Wang, X.C.; Liu, J.H.; Huang, B.Y.; Guo, X.Y.; Xiong, S.P.; La, G.X. UPLC-QTOF analysis reveals metabolomic changes in the flag leaf of wheat (Triticum aestivum L.) under low-nitrogen stress. Plant Physiol. Biochem. 2017, 111, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Chen, H.; Jiang, Y.; Wang, J.; Khan, A.; Li, P.; Cao, C. Metabolomic analysis reveals metabolites and pathways involved in grain quality traits of high-quality rice cultivars under a dry cultivation system. Food Chem. 2020, 326, 126845. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef]
- Meena, K.K.; Sorty, A.M.; Bitla, U.M.; Choudhary, K.; Gupta, P.; Pareek, A.; Singh, D.P.; Prabha, R.; Sahu, P.K.; Gupta, V.K.; et al. Abiotic Stress Responses and Microbe-Mediated Mitigation in Plants: The Omics Strategies. Front. Plant Sci. 2017, 8, 172. [Google Scholar] [CrossRef]
- Sung, J.; Lee, S.; Lee, Y.; Ha, S.; Song, B.; Kim, T.; Waters, B.M.; Krishnan, H.B. Metabolomic profiling from leaves and roots of tomato (Solanum lycopersicum L.) plants grown under nitrogen, phosphorus or potassium-deficient condition. Plant Sci. 2015, 241, 55–64. [Google Scholar] [CrossRef]
- Cevallos-Cevallos, J.M.; Garcia-Torres, R.; Etxeberria, E.; Reyes-De-Corcuera, J.I. GC-MS analysis of headspace and liquid extracts for metabolomic differentiation of citrus Huanglongbing and zinc deficiency in leaves of ‘Valencia’ sweet orange from commercial groves. Phytochem. Anal. 2011, 22, 236–246. [Google Scholar] [CrossRef]
- Shen, T.H.; Xiong, Q.Q.; Zhong, L.; Shi, X.; Cao, C.H.; He, H.H.; Chen, X.R. Analysis of main metabolisms during nitrogen deficiency and compensation in rice. Acta Physiol. Plant. 2019, 41, 68. [Google Scholar] [CrossRef]
- Watanabe, M.; Walther, D.; Ueda, Y.; Kondo, K.; Ishikawa, S.; Tohge, T.; Burgos, A.; Brotman, Y.; Fernie, A.R.; Hoefgen, R.; et al. Metabolomic markers and physiological adaptations for high phosphate utilization efficiency in rice. Plant Cell Environ. 2020, 43, 2066–2079. [Google Scholar] [CrossRef]
- Kitazaki, K.; Fukushima, A.; Nakabayashi, R.; Okazaki, Y.; Kobayashi, M.; Mori, T.; Nishizawa, T.; Reyes-Chin-Wo, S.; Michelmore, R.W.; Saito, K.; et al. Metabolic Reprogramming in Leaf Lettuce Grown under Different Light Quality and Intensity Conditions Using Narrow-Band LEDs. Sci. Rep. 2018, 8, 7914. [Google Scholar] [CrossRef]
- Mbong, V.B.M.; Ampofo-Asiama, J.; Hertog, M.L.; Geeraerd, A.H.; Nicolai, B.M. The effect of temperature on the metabolic response of lamb’s lettuce (Valerianella locusta, (L), Laterr.) cells to sugar starvation. Postharvest Biol. Technol. 2017, 125, 1–12. [Google Scholar] [CrossRef]
- Lucini, L.; Rouphael, Y.; Cardarelli, M.; Canaguier, R.; Kumar, P.; Colla, G. The effect of a plant-derived biostimulant on metabolic profiling and crop performance of lettuce grown under saline conditions. Sci. Hortic. 2015, 182, 124–133. [Google Scholar] [CrossRef]
- Tamura, Y.; Mori, T.; Nakabayashi, R.; Kobayashi, M.; Saito, K.; Okazaki, S.; Wang, N.; Kusano, M. Metabolomic Evaluation of the Quality of Leaf Lettuce Grown in Practical Plant Factory to Capture Metabolite Signature. Front. Plant Sci. 2018, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wei, S.; Liu, B.; Guo, D.; Zheng, B.; Feng, L.; Liu, Y.; Tomas-Barberan, F.A.; Luo, L.; Huang, D. A novel integrated non-targeted metabolomic analysis reveals significant metabolite variations between different lettuce (Lactuca sativa. L) varieties. Hortic. Res. 2018, 5, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, H.P.; Hang, T.; Zhang, X.D.; Lu, N. Both Multi-Segment Light Intensity and Extended Photoperiod Lighting Strategies, with the Same Daily Light Integral, Promoted Lactuca sativa L. Growth and Photosynthesis. Agronomy 2019, 9, 857. [Google Scholar] [CrossRef] [Green Version]
- Luo, P.; Yin, P.; Zhang, W.; Zhou, L.; Lu, X.; Lin, X.; Xu, G. Optimization of large-scale pseudotargeted metabolomics method based on liquid chromatography-mass spectrometry. J. Chromatogr. A 2016, 1437, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Trygg, J.; Holmes, E.; Lundstedt, T. Chemometrics in metabonomics. J. Proteome Res. 2007, 6, 469–479. [Google Scholar] [CrossRef]
- Shi, M.Y.; Wang, S.S.; Zhang, Y.; Wang, S.; Zhao, J.; Feng, H.; Sun, P.P.; Fang, C.B.; Xie, X.B. Genome-wide characterization and expression analysis of ATP-binding cassette (ABC) transporters in strawberry reveal the role of FvABCC11 in cadmium tolerance. Sci. Hortic. 2020, 271, 1–16. [Google Scholar] [CrossRef]
- Patra, B.; Schluttenhofer, C.; Wu, Y.M.; Pattanaik, S.; Yuan, L. Transcriptional regulation of secondary metabolite biosynthesis in plants. Biochim. Biophys. Acta Gene Regul. Mech. 2013, 1829, 1236–1247. [Google Scholar] [CrossRef]
- Ozyigit, Y.; Bilgen, M. Use of Spectral Reflectance Values for Determining Nitrogen, Phosphorus, and Potassium Contents of Rangeland Plants. J. Agric. Sci. Technol. 2013, 15, 1537–1545. [Google Scholar]
- Romualdo, L.M.; Luz, P.H.C.; Devechio, F.F.S.; Marin, M.A.; Zuniga, A.M.G.; Bruno, O.M.; Herling, V.R. Use of artificial vision techniques for diagnostic of nitrogen nutritional status in maize plants. Comput. Electron. Agric. 2014, 104, 63–70. [Google Scholar] [CrossRef]
- Story, D.; Kacira, M.; Kubota, C.; Akoglu, A.; An, L.L. Lettuce calcium deficiency detection with machine vision computed plant features in controlled environments. Comput. Electron. Agric. 2010, 74, 238–243. [Google Scholar] [CrossRef]
- Rubio-Wilhelmi, M.M.; Sanchez-Rodriguez, E.; Rosales, M.A.; Begona, B.; Rios, J.J.; Romero, L.; Blumwald, E.; Ruiz, J.M. Effect of cytokinins on oxidative stress in tobacco plants under nitrogen deficiency. Environ. Exp. Bot. 2011, 72, 167–173. [Google Scholar] [CrossRef]
- Wittenmayer, L.; Merbach, W. Plant responses to drought and phosphorus deficiency: Contribution of phytohormones in root-related processes. J. Plant Nutr. Soil Sci. 2010, 168, 531–540. [Google Scholar] [CrossRef]
- Jiang, J.; Wang, Y.P.; Yang, Y.H.; Yu, M.X.; Wang, C.; Yan, J.H. Interactive effects of nitrogen and phosphorus additions on plant growth vary with ecosystem type. Plant Soil 2019, 440, 523–537. [Google Scholar] [CrossRef]
- Mirande-Ney, C.; Tcherkez, G.; Balliau, T.; Zivy, M.; Gilard, F.; Cui, J.; Ghashghaie, J.; Lamade, E. Metabolic leaf responses to potassium availability in oil palm (Elaeis guineensis Jacq.) trees grown in the field. Environ. Exp. Bot. 2020, 175, 104062. [Google Scholar] [CrossRef]
- Sun, Y.L.; Mu, C.H.; Chen, Y.; Kong, X.P.; Xu, Y.C.; Zheng, H.X.; Zhang, H.; Wang, Q.C.; Xue, Y.F.; Li, Z.X.; et al. Comparative transcript profiling of maize inbreds in response to long-term phosphorus deficiency stress. Plant Physiol. Biochem. 2016, 109, 467–481. [Google Scholar] [CrossRef] [Green Version]






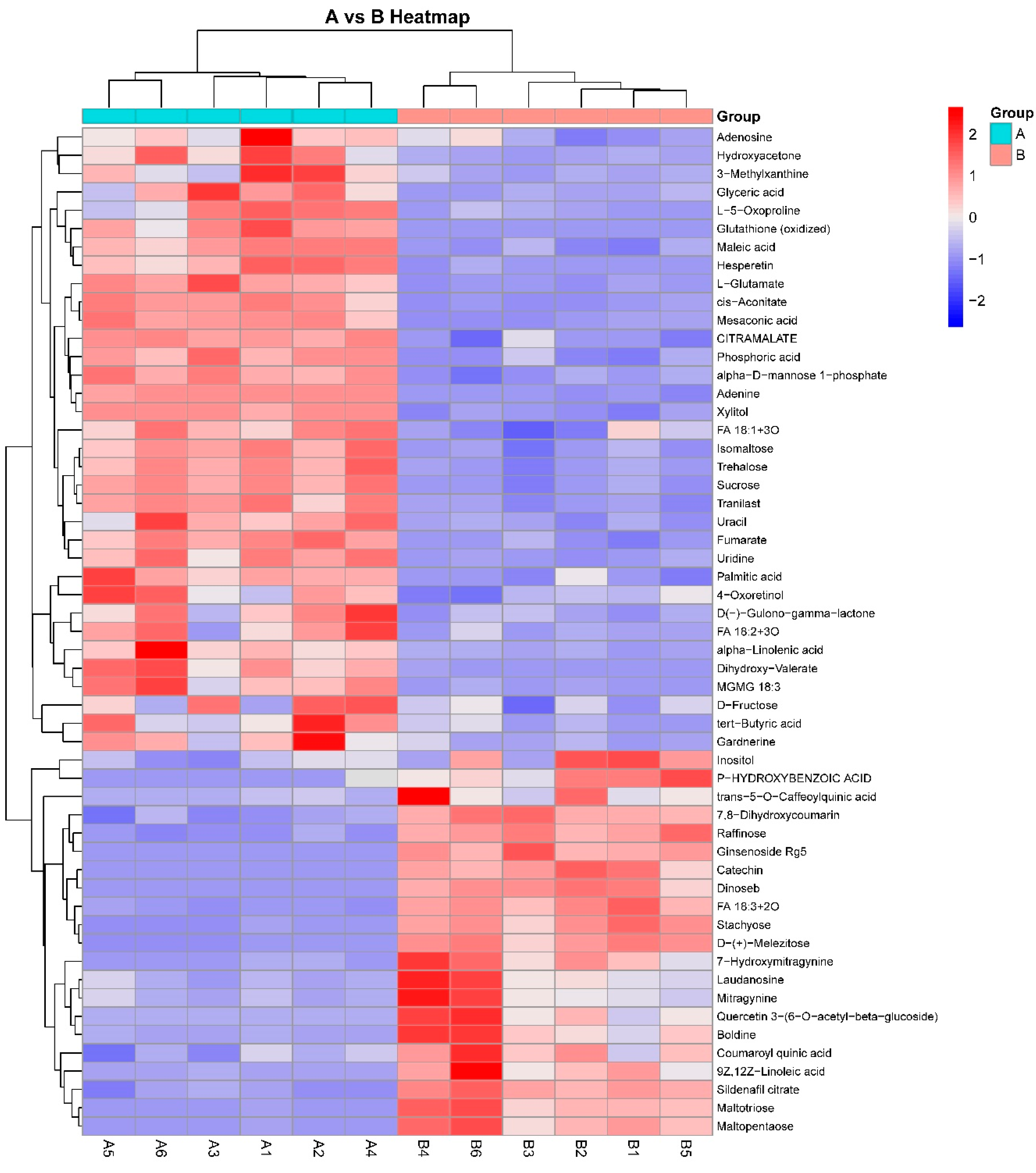
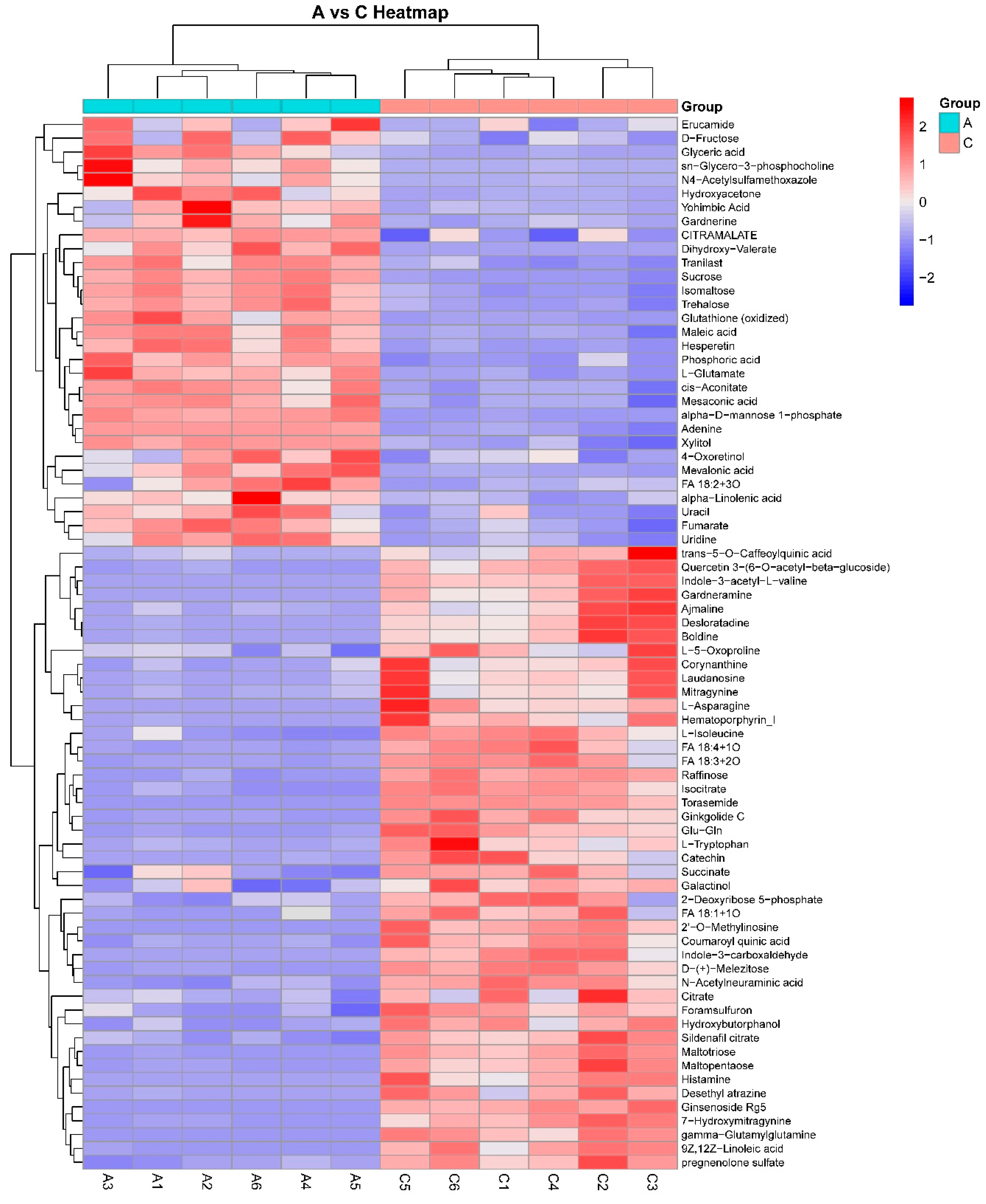

| Name | ESI Mode | m/z | RT (min) | |
|---|---|---|---|---|
| Upregulation | Inositol | Negative | 179.0586 | 9.03 |
| P-hydroxybenzoic acid | Negative | 137.0247 | 0.72 | |
| Stachyose | Negative | 725.2340 | 12.05 | |
| Dinoseb | Negative | 239.0704 | 0.99 | |
| 7,8-Dihydroxycoumarin | Negative | 177.0211 | 4.42 | |
| Downregulation | Adenosine | Positive | 268.1028 | 3.96 |
| Palmitic acid | Negative | 255.2326 | 1.13 | |
| L-5-Oxoproline | Positive | 130.0497 | 9.68 | |
| 3-Methylxanthine | Negative | 165.0412 | 8.65 | |
| tert-Butyric acid | Negative | 115.0787 | 1.82 | |
| D(-)-Gulono-gamma-lactone | Negative | 177.0418 | 2.55 | |
| FA 18:2 + 3O | Negative | 329.2314 | 3.22 | |
| MGMG 18:3 | Negative | 559.3070 | 2.56 |
| Name | ESI Mode | m/z | RT (min) | |
|---|---|---|---|---|
| Upregulation | Succinate | Negative | 117.0217 | 9.20 |
| L-Tryptophan | Negative | 203.0822 | 6.44 | |
| L-Asparagine | Negative | 131.0461 | 9.57 | |
| Citrate | Negative | 191.0211 | 11.99 | |
| Indole-3-carboxaldehyde | Negative | 144.0468 | 0.87 | |
| Isocitrate | Negative | 191.0193 | 11.30 | |
| Histamine | Positive | 112.0865 | 6.75 | |
| L-Isoleucine | Negative | 130.0877 | 6.64 | |
| 2-Deoxyribose 5-phosphate | Negative | 213.0164 | 3.09 | |
| Galactinol | Negative | 341.1062 | 11.85 | |
| L-5-Oxoproline | Positive | 130.0497 | 9.68 | |
| gamma-Glutamylglutamine | Positive | 276.1186 | 10.66 | |
| Ajmaline | Positive | 327.2030 | 2.13 | |
| Desethyl atrazine | Positive | 188.0702 | 6.43 | |
| Ginkgolide C | Negative | 439.1220 | 8.51 | |
| Corynanthine | Positive | 355.2086 | 6.11 | |
| pregnenolone sulfate | Negative | 395.1898 | 3.78 | |
| N-Acetylneuraminic acid | Negative | 308.0955 | 6.96 | |
| Glu-Gln | Negative | 274.1040 | 10.66 | |
| 2’-O-Methylinosine | Negative | 281.0865 | 8.92 | |
| FA 18:4 + 1O | Negative | 291.1943 | 1.09 | |
| FA 18:1 + 1O | Negative | 297.2427 | 1.06 | |
| Torasemide | Negative | 347.1167 | 11.04 | |
| Foramsulfuron | Negative | 451.1034 | 0.52 | |
| Indole-3-acetyl-L-valine | Positive | 275.1346 | 10.49 | |
| Desloratadine | Positive | 311.1336 | 8.51 | |
| Hydroxybutorphanol | Positive | 344.2264 | 1.69 | |
| Gardneramine | Positive | 413.2110 | 8.46 | |
| Hematoporphyrin_I | Positive | 599.2871 | 5.69 | |
| Downregulation | Mevalonic acid | Negative | 295.1385 | 2.49 |
| sn-Glycero-3-phosphocholine | Positive | 258.1099 | 9.75 | |
| N4-Acetylsulfamethoxazole | Positive | 296.0658 | 9.75 | |
| Yohimbic Acid | Negative | 339.1631 | 2.54 | |
| Erucamide | Positive | 338.3422 | 0.80 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, H.; Mao, H.; Ullah, I. Analysis of Metabolomic Changes in Lettuce Leaves under Low Nitrogen and Phosphorus Deficiencies Stresses. Agriculture 2020, 10, 406. https://doi.org/10.3390/agriculture10090406
Gao H, Mao H, Ullah I. Analysis of Metabolomic Changes in Lettuce Leaves under Low Nitrogen and Phosphorus Deficiencies Stresses. Agriculture. 2020; 10(9):406. https://doi.org/10.3390/agriculture10090406
Chicago/Turabian StyleGao, Hongyan, Hanping Mao, and Ikram Ullah. 2020. "Analysis of Metabolomic Changes in Lettuce Leaves under Low Nitrogen and Phosphorus Deficiencies Stresses" Agriculture 10, no. 9: 406. https://doi.org/10.3390/agriculture10090406





