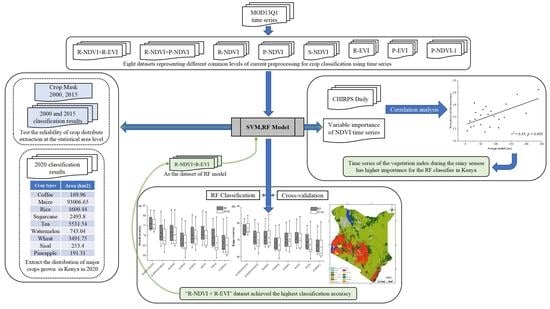
Effectiveness of Common Preprocessing Methods of Time Series for Monitoring Crop Distribution in Kenya
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Data
- R-NDVI: raw NDVI time series (23 bands, 23 NDVI image sequences per year).
- R-EVI: raw EVI time series (23 bands, 23 EVI image sequences per year).
- S-NDVI: smoothed NDVI time series (23 bands, 23 NDVI image sequences by smoothing).
- P-NDVI: phenological parameters obtained from the original NDVI time series (26 bands, i.e., 26 vegetation phenometrics extracted from two growing seasons).
- P-EVI: phenological parameters obtained from the original EVI time series (26 bands, vegetation phenometrics extracted from two growing seasons).
- P-NDVI-1: phenological parameters obtained from the original NDVI time series (13 bands, i.e., 13 vegetation phenometrics extracted from the first growing season).
- R-NDVI + R-EVI: a combination of original NDVI and original EVI time series (46 bands, 23 NDVI + 23 EVI).
- R-NDVI + P-NDVI: a combination of original NDVI time series and NDVI-derived phenological parameters (49 bands, 23 NDVI + 26 vegetation phenometrics).
2.3. Methods
2.3.1. Vegetation Index Time Series Smoothing and Phenometrics’ Extraction
- Beginning of the season: The date from the minimum value at the left edge to a user-defined value (usually a proportion of the seasonal amplitude).
- End of the season: The date from the minimum value at the right edge to the user-defined value.
- Length of the season: Days from the beginning to the end of the growing season.
- Base level: The average of the minimum values around the complete growing season.
- Time for the mid of the season: The average of the dates corresponds to the increase to 80% of the peak and the decrease to 80% of the peak.
- Largest data value for the fitted function during the season: The peak of the fitted growing season curve.
- Seasonal amplitude: The difference between the growing season peak and the base value.
- Left derivative: The ratio of the difference between 20% and 80% of the left peak to the corresponding time difference.
- Right derivative: The absolute value of the ratio of the difference between 20% and 80% of the peak on the right side and the corresponding time difference.
- Large seasonal integral: The integral value of the fitted curve from the beginning to the end of the growing season.
- Small seasonal integral: The integral value of the difference between the fitted curve and the base value from the beginning to the end of the growing season.
- Value for the beginning of the season: The value of the curve fit corresponding to the beginning of the growing season.
- Value for the end of the season: The curve-fit value corresponding to the end of the growing season.
2.3.2. Classification Strategy
2.3.3. Statistical Learning Algorithm
2.3.4. Accuracy Assessment
2.3.5. Variable Importance Analysis
3. Results
3.1. Raw Vegetation Index Time Series, Smoothing Effect, and Phenological Images
3.2. Classification Accuracy
3.3. Validation of Classification Strategy
3.4. Planting Area Extraction
3.5. Variable Importance Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A


References
- Luciani, R.; Laneve, G.; Jahjah, M. Agricultural Monitoring, an Automatic Procedure for Crop Mapping and Yield Estimation: The Great Rift Valley of Kenya Case. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2019, 12, 2196–2208. [Google Scholar] [CrossRef]
- FAO. Guidelines on the Use of Remote Sensing Products to Improve Agricultural Crop Production Forecast Statistics in Sub-Saharan African Countries; FAO: Rome, Italy, 2018. [Google Scholar] [CrossRef]
- Lowder, S.K.; Skoet, J.; Raney, T. The Number, Size, and Distribution of Farms, Smallholder Farms, and Family Farms Worldwide. World Dev. 2016, 87, 16–29. [Google Scholar] [CrossRef] [Green Version]
- Samberg, L.H.; Gerber, J.; Ramankutty, N.; Herrero, M.; West, P. Subnational distribution of average farm size and smallholder contributions to global food production. Environ. Res. Lett. 2016, 11, 124010. [Google Scholar] [CrossRef]
- Smith, J.H.; Stehman, S.V.; Wickham, J.D.; Yang, L. Effects of landscape characteristics on land-cover class accuracy. Remote Sens. Environ. 2003, 84, 342–349. [Google Scholar] [CrossRef]
- Piiroinen, R.; Heiskanen, J.; Mõttus, M.; Pellikka, P. Classification of crops across heterogeneous agricultural landscape in Kenya using AisaEAGLE imaging spectroscopy data. Int. J. Appl. Earth Obs. Geoinform. 2015, 39, 1–8. [Google Scholar] [CrossRef]
- Jin, Z.; Azzari, G.; Burke, M.; Aston, S.; Lobell, D.B. Mapping Smallholder Yield Heterogeneity at Multiple Scales in Eastern Africa. Remote Sens. 2017, 9, 931. [Google Scholar] [CrossRef] [Green Version]
- Mosomtai, G.; Odindi, J.; Abdel-Rahman, E.M.; Babin, R.; Fabrice, P.; Mutanga, O.; Tonnang, H.E.Z.; David, G.; Landmann, T. Landscape fragmentation in coffee agroecological subzones in central Kenya: A multiscale remote sensing approach. J. Appl. Remote Sens. 2020, 14, 044513. [Google Scholar] [CrossRef]
- Richard, K.; Abdel-Rahman, E.M.; Subramanian, S.; Nyasani, J.O.; Thiel, M.; Jozani, H.; Borgemeister, C.; Landmann, T. Maize Cropping Systems Mapping Using RapidEye Observations in Agro-Ecological Landscapes in Kenya. Sensors 2017, 17, 2537. [Google Scholar] [CrossRef] [Green Version]
- Maingi, J.K.; Marsh, S.E. Assessment of environmental impacts of river basin development on the riverine forests of eastern Kenya using multi-temporal satellite data. Int. J. Remote Sens. 2001, 22, 2701–2729. [Google Scholar] [CrossRef]
- Tottrup, C. Sensing, Improving tropical forest mapping using multi-date Landsat TM data and pre-classification image smoothing. Int. J. Remote Sens. 2004, 25, 717–730. [Google Scholar] [CrossRef]
- Hashim, M.; Pour, A.B.; Onn, C.H. Optimizing cloud removal from satellite remotely sensed data for monitoring vegetation dynamics in humid tropical climate. IOP Conf. Ser. Earth Environ. Sci. 2014, 18, 12010. [Google Scholar] [CrossRef] [Green Version]
- Vithanage, J.; Miller, S.N.; Driese, K. Land cover characterization for a watershed in Kenya using MODIS data and Fourier algorithms. J. Appl. Remote Sens. 2016, 10, 045015. [Google Scholar] [CrossRef]
- Baldyga, T.J.; Miller, S.N.; Driese, K.L.; Gichaba, C.M. Assessing land cover change in Kenya’s Mau Forest region using remotely sensed data. Afric. J. Ecol. 2008, 46, 46–54. [Google Scholar] [CrossRef]
- Moody, A.; Johnson, D.M. Land-Surface Phenologies from AVHRR Using the Discrete Fourier Transform. Remote Sens. Environ. 2001, 75, 305–323. [Google Scholar] [CrossRef]
- Mwaniki, W.M.; Möller, S.M. Knowledge based multi-source, time series classification: A case study of central region of Kenya. Appl. Geogr. 2015, 60, 58–68. [Google Scholar] [CrossRef]
- Luciani, R.; Laneve, G.; Jahjah, M. Developing a classification method for periodically updating agricultural maps in Kenya. In 2016 IEEE International Geoscience and Remote Sensing Symposium (IGARSS); IEEE: Beijing, China, 2016; pp. 3543–3546. [Google Scholar] [CrossRef]
- Luciani, R.; Laneve, G.; Jahjah, M.; Collins, M. Crop species classification: A phenology based approach. In 2017 IEEE International Geoscience and Remote Sensing Symposium (IGARSS); IEEE: Fort Worth, TX, USA, 2017; pp. 4390–4393. [Google Scholar] [CrossRef]
- Gachoki, S.M. Estimating Vegetation Phenology at 30m Resolution with Multi-Temporal Optical Imagery for a Rangeland Site in Kenya. Master’s Thesis, University of Twente, Enschede, The Netherlands, 2018. [Google Scholar]
- Jin, Z.; Azzari, G.; You, C.; Di Tommaso, S.; Aston, S.; Burke, M.; Lobell, D.B. Smallholder maize area and yield mapping at national scales with Google Earth Engine. Remote Sens. Environ. 2019, 228, 115–128. [Google Scholar] [CrossRef]
- Sun, R.; Chen, S.; Su, H.; Mi, C.; Jin, N. The Effect of NDVI Time Series Density Derived from Spatiotemporal Fusion of Multisource Remote Sensing Data on Crop Classification Accuracy. ISPRS Int. J. Geo-Inf. 2019, 8, 502. [Google Scholar] [CrossRef] [Green Version]
- Shao, Y.; Lunetta, R.S.; Wheeler, B.; Iiames, J.S.; Campbell, J.B. An evaluation of time-series smoothing algorithms for land-cover classifications using MODIS-NDVI multi-temporal data. Remote Sens. Environ. 2016, 174, 258–265. [Google Scholar] [CrossRef]
- Brown, J.; Kastens, J.H.; Coutinho, A.C.; Victoria, D.D.C.; Bishop, C.R. Classifying multiyear agricultural land use data from Mato Grosso using time-series MODIS vegetation index data. Remote Sens. Environ. 2013, 130, 39–50. [Google Scholar] [CrossRef] [Green Version]
- Atkinson, P.; Jeganathan, C.; Dash, J.; Atzberger, C. Inter-comparison of four models for smoothing satellite sensor time-series data to estimate vegetation phenology. Remote Sens. Environ. 2012, 123, 400–417. [Google Scholar] [CrossRef]
- Zhong, L.; Hu, L.; Yu, L.; Gong, P.; Biging, G.S. Automated mapping of soybean and corn using phenology. ISPRS J. Photogramm. Remote Sens. 2016, 119, 151–164. [Google Scholar] [CrossRef] [Green Version]
- Valero, S.; Morin, D.; Inglada, J.; Sepulcre, G.; Arias, M.; Hagolle, O.; Dedieu, G.; Bontemps, S.; Defourny, P.; Koetz, B. Production of a Dynamic Cropland Mask by Processing Remote Sensing Image Series at High Temporal and Spatial Resolutions. Remote Sens. 2016, 8, 55. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Lu, D.; Moran, E.; Batistella, M.; Dutra, L.V.; Sanches, I.D.A.; da Silva, R.F.B.; Huang, J.; Luiz, A.J.B.; de Oliveira, M.A.F. Mapping croplands, cropping patterns, and crop types using MODIS time-series data. Int. J. Appl. Earth Obs. Geoinf. 2018, 69, 133–147. [Google Scholar] [CrossRef]
- Pelletier, C.; Valero, S.; Inglada, J.; Champion, N.; Dedieu, G. Assessing the robustness of Random Forests to map land cover with high resolution satellite image time series over large areas. Remote Sens. Environ. 2016, 187, 156–168. [Google Scholar] [CrossRef]
- Kuchler, P.C.; Bégué, A.; Simões, M.; Gaetano, R.; Arvor, D.; Ferraz, R.P. Assessing the optimal preprocessing steps of MODIS time series to map cropping systems in Mato Grosso, Brazil. Int. J. Appl. Earth Obs. Geoinf. 2020, 92, 102150. [Google Scholar] [CrossRef]
- Picoli, M.C.A.; Camara, G.; Sanches, I.; Simões, R.; Carvalho, A.; Maciel, A.; Coutinho, A.; Esquerdo, J.; Antunes, J.; Begotti, R.; et al. Big earth observation time series analysis for monitoring Brazilian agriculture. ISPRS J. Photogramm. Remote Sens. 2018, 145, 328–339. [Google Scholar] [CrossRef]
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. An Introduction to Statistical Learning; Springer: New York, NY, USA, 2013; Volume 103. [Google Scholar]
- Muthoni, F.K.; Odongo, V.O.; Ochieng, J.; Mugalavai, E.M.; Mourice, S.K.; Hoesche-Zeledon, I.; Mwila, M.; Bekunda, M. Long-term spatial-temporal trends and variability of rainfall over Eastern and Southern Africa. Theor. Appl. Climatol. 2018, 137, 1869–1882. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Chen, S.; Aluoch, S.O.; Mosongo, P.S.; Cao, J.; Hu, C. Maize production status and yield limiting factors of Kenya. Chin. J. Eco-Agric. 2018, 26, 567–573. [Google Scholar]
- Zhang, Y.; Song, C.; Band, L.E.; Sun, G.; Li, J. Reanalysis of global terrestrial vegetation trends from MODIS products: Browning or greening? Remote Sens. Environ. 2017, 191, 145–155. [Google Scholar] [CrossRef] [Green Version]
- Funk, C.; Peterson, P.; Landsfeld, M.; Pedreros, D.; Verdin, J.; Shukla, S.; Husak, G.; Rowland, J.; Harrison, L.; Hoell, A.; et al. The climate hazards infrared precipitation with stations—A new environmental record for monitoring extremes. Sci. Data 2015, 2, 150066. [Google Scholar] [CrossRef] [Green Version]
- Jönsson, P.; Eklundh, L. TIMESAT—A program for analyzing time-series of satellite sensor data. Comput. Geosci. 2004, 30, 833–845. [Google Scholar] [CrossRef] [Green Version]
- De Castro, A.I.; Six, J.; Plant, R.E.; Peña, J.M. Mapping Crop Calendar Events and Phenology-Related Metrics at the Parcel Level by Object-Based Image Analysis (OBIA) of MODIS-NDVI Time-Series: A Case Study in Central California. Remote Sens. 2018, 10, 1745. [Google Scholar] [CrossRef] [Green Version]
- Richard, K.; Abdel-Rahman, E.M.; Mohamed, S.A.; Ekesi, S.; Borgemeister, C.; Landmann, T. Importance of Remotely-Sensed Vegetation Variables for Predicting the Spatial Distribution of African Citrus Triozid (Trioza erytreae) in Kenya. ISPRS Int. J. Geo-Inf. 2018, 7, 429. [Google Scholar] [CrossRef] [Green Version]
- Makori, D.M.; Fombong, A.T.; Abdel-Rahman, E.M.; Nkoba, K.; Ongus, J.; Irungu, J.; Mosomtai, G.; Makau, S.; Mutanga, O.; Odindi, J.; et al. Predicting Spatial Distribution of Key Honeybee Pests in Kenya Using Remotely Sensed and Bioclimatic Variables: Key Honeybee Pests Distribution Models. ISPRS Int. J. Geo-Inf. 2017, 6, 66. [Google Scholar] [CrossRef] [Green Version]
- Vigani, M.; Dudu, H.; Solano-Hermosilla, G. Estimation of Food Demand Parameters in Ethiopia: A Quadratic Almost Ideal Demand System (QUAIDS) Approach; Joint Research Centre: Ispra, Italy, 2019. [Google Scholar]
- Mabiso, A.; Pauw, K.; Benin, S. Agricultural growth and poverty reduction in Kenya: Technical analysis for the Agricultural Sectoral Development Strategy (ASDS)—Medium Term Investment Plan (MTIP). Reg. Strateg. Anal. Knowl. Support Syst. (ReSAKSS) Work. Pap. 2015, 35. Available online: https://ebrary.ifpri.org/utils/getfile/collection/p15738coll2/id/127063/filename/127274.pdf (accessed on 10 December 2021).
- Forkuor, G.; Conrad, C.; Thiel, M.; Landmann, T.; Barry, B. Evaluating the sequential masking classification approach for improving crop discrimination in the Sudanian Savanna of West Africa. Comput. Electron. Agric. 2015, 118, 380–389. [Google Scholar] [CrossRef]
- Forkuor, G.; Conrad, C.; Thiel, M.; Ullmann, T.; Zoungrana, E. Integration of Optical and Synthetic Aperture Radar Imagery for Improving Crop Mapping in Northwestern Benin, West Africa. Remote Sens. 2014, 6, 6472–6499. [Google Scholar] [CrossRef] [Green Version]
- Inglada, J.; Arias, M.; Tardy, B.; Hagolle, O.; Valero, S.; Morin, D.; Dedieu, G.; Sepulcre, G.; Bontemps, S.; Defourny, P.; et al. Assessment of an Operational System for Crop Type Map Production Using High Temporal and Spatial Resolution Satellite Optical Imagery. Remote Sens. 2015, 7, 12356–12379. [Google Scholar] [CrossRef] [Green Version]
- Onojeghuo, A.O.; Blackburn, G.A.; Wang, Q.; Atkinson, P.M.; Kindred, D.; Miao, Y. Mapping paddy rice fields by applying machine learning algorithms to multi-temporal Sentinel-1A and Landsat data. Int. J. Remote Sens. 2017, 39, 1042–1067. [Google Scholar] [CrossRef] [Green Version]
- Mudereri, B.T.; Dube, T.; Niassy, S.; Kimathi, E.; Landmann, T.; Khan, Z.; Abdel-Rahman, E.M. Is it possible to discern Striga weed (Striga hermonthica) infestation levels in maize agro-ecological systems using in-situ spectroscopy? Int. J. Appl. Earth Obs. Geoinf. 2020, 85, 102008. [Google Scholar] [CrossRef]
- Shao, Y.; Lunetta, R.S. Comparison of support vector machine, neural network, and CART algorithms for the land-cover classification using limited training data points. ISPRS J. Photogramm. Remote Sens. 2012, 70, 78–87. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Wiens, T.S.; Dale, B.C.; Boyce, M.S.; Kershaw, G.P. Three way k-fold cross-validation of resource selection functions. Ecol. Model. 2008, 212, 244–255. [Google Scholar] [CrossRef]
- Gillieson, D.; Lawson, T.; Searle, L. Applications of High Resolution Remote Sensing in Rainforest Ecology and Management. Living A Dyn. Trop. For. Landsc. 2008, 334–348. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer Science & Business Media: New York, NY, USA, 2009. [Google Scholar]
- Alexandridis, T.K.; Gitas, I.; Silleos, N.G. An estimation of the optimum temporal resolution for monitoring vegetation condition on a nationwide scale using MODIS/Terra data. Int. J. Remote Sens. 2008, 29, 3589–3607. [Google Scholar] [CrossRef]









| Parameters | Settings |
|---|---|
| Curve-fitting model | Savitzky–Golay filtering |
| Seasonality parameter | 0 (0 will attempt to fit two seasons) |
| Spike method | 3 (STL original) |
| No. of envelope iterations | 1 |
| Adaptation strength | 3 |
| Window size Season start/end values | 4 (only for Savitzky–Golay filtering) 0.2 |
| R-NDVI + R-EVI | R-NDVI + P-NDVI | R-NDVI | P-NDVI | S-NDVI | R-EVI | P-EVI | P-NDVI-1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OA | Kappa | OA | Kappa | OA | Kappa | OA | Kappa | OA | Kappa | OA | Kappa | OA | Kappa | OA | Kappa | |
| Arid North | 73.78 | 62.27 | 69.85 | 56.88 | 70.7 | 58.03 | 66.35 | 52.22 | 69.72 | 56.65 | 68.09 | 54.13 | 66.64 | 52.6 | 65.48 | 50.88 |
| Arid South | 78.14 | 56.96 | 77.11 | 55.23 | 77.27 | 55.49 | 75.35 | 51.9 | 76.87 | 54.72 | 78.17 | 56.83 | 78.38 | 51.92 | 74.92 | 51.24 |
| Cities | 82.86 | 75.1 | 82.14 | 73.92 | 82.86 | 75.06 | 78.57 | 68.64 | 80.48 | 71.42 | 82.38 | 74.35 | 78.57 | 68.84 | 78.33 | 68.57 |
| Coast | 73.31 | 61.75 | 73.63 | 62.33 | 73.23 | 61.72 | 67.36 | 53.47 | 71.64 | 59.56 | 71.17 | 58.65 | 68.39 | 55 | 63.22 | 47.66 |
| High Rainfall | 73.24 | 57.17 | 68.57 | 49.79 | 68.74 | 49.53 | 65.66 | 46.4 | 67.09 | 48.14 | 70.05 | 51.76 | 65.86 | 46.56 | 63.82 | 44.26 |
| Semi-Arid North | 72.63 | 57.45 | 67.78 | 49.97 | 66.9 | 48.47 | 63.8 | 43.57 | 65.51 | 47.35 | 68.13 | 50.57 | 64.37 | 44.19 | 63.2 | 42.91 |
| Semi-Arid South | 72.99 | 63.17 | 67.5 | 55.76 | 68.02 | 56.19 | 64.79 | 52.35 | 67.01 | 55.38 | 65.02 | 51.9 | 65.23 | 52.86 | 61.62 | 48.22 |
| Turkana | 80.16 | 68.74 | 64.62 | 44.6 | 65.88 | 49.94 | 64.23 | 43.93 | 63.84 | 43.37 | 70.76 | 54.2 | 64.49 | 44.44 | 62.79 | 41.85 |
| R-NDVI + R-EVI | R-NDVI + P-NDVI | R-NDVI | P-NDVI | S-NDVI | R-EVI | P-EVI | P-NDVI-1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OA | Kappa | OA | Kappa | OA | Kappa | OA | Kappa | OA | Kappa | OA | Kappa | OA | Kappa | OA | Kappa | |
| Arid North | 74.49 | 63.9 | 66.4 | 52.95 | 67.75 | 54.68 | 64.9 | 50.31 | 69.12 | 56.29 | 64.8 | 50.44 | 64.27 | 50.0 | 64.53 | 49.61 |
| Arid South | 77.41 | 57.62 | 75.85 | 54.5 | 75.58 | 53.93 | 75.65 | 53.16 | 73.59 | 50.97 | 73.95 | 51.49 | 74.62 | 52.0 | 73.36 | 49.22 |
| Cities | 82.62 | 74.72 | 77.86 | 67.77 | 83.1 | 75.38 | 77.38 | 66.91 | 79.52 | 70.12 | 81.9 | 73.81 | 79.29 | 69.64 | 79.05 | 69.4 |
| Coast | 72.99 | 61.69 | 70.85 | 58.72 | 72.76 | 61.02 | 63.86 | 48.51 | 71.8 | 59.9 | 69.9 | 57.03 | 64.65 | 49.66 | 60.92 | 44.39 |
| High Rainfall | 71.76 | 58.4 | 61.59 | 45.52 | 64.79 | 47.3 | 58.89 | 41.44 | 62.55 | 46.11 | 65.37 | 48.95 | 58.82 | 41.71 | 60.37 | 41.06 |
| Semi-Arid North | 73.26 | 59.96 | 68.73 | 52.53 | 66.52 | 49.36 | 63.04 | 44.13 | 67.63 | 50.25 | 67.53 | 50.66 | 62.15 | 42.75 | 61.58 | 41.31 |
| Semi-Arid South | 73.33 | 64.82 | 65.41 | 54.44 | 65.31 | 54.11 | 63.4 | 51.54 | 66.03 | 54.96 | 64.17 | 52.62 | 63.4 | 51.54 | 59.84 | 46.8 |
| Turkana | 78.85 | 66.7 | 65.8 | 46.31 | 67.62 | 49.37 | 63.84 | 43.35 | 65.27 | 45.57 | 71.28 | 54.95 | 63.58 | 42.96 | 61.88 | 40.41 |
| Coffee | Maize | Rice | Sugarcane | Tea | Watermelon | Wheat | Sisal | Pineapple | |
|---|---|---|---|---|---|---|---|---|---|
| Area (km2) | 169.96 | 93006.65 | 1600.44 | 2493.80 | 5531.54 | 743.04 | 3491.75 | 253.40 | 191.31 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ni, R.; Zhu, X.; Lei, Y.; Li, X.; Dong, W.; Zhang, C.; Chen, T.; Mburu, D.M.; Hu, C. Effectiveness of Common Preprocessing Methods of Time Series for Monitoring Crop Distribution in Kenya. Agriculture 2022, 12, 79. https://doi.org/10.3390/agriculture12010079
Ni R, Zhu X, Lei Y, Li X, Dong W, Zhang C, Chen T, Mburu DM, Hu C. Effectiveness of Common Preprocessing Methods of Time Series for Monitoring Crop Distribution in Kenya. Agriculture. 2022; 12(1):79. https://doi.org/10.3390/agriculture12010079
Chicago/Turabian StyleNi, Rui, Xiaohui Zhu, Yuping Lei, Xiaoxin Li, Wenxu Dong, Chuang Zhang, Tuo Chen, David M. Mburu, and Chunsheng Hu. 2022. "Effectiveness of Common Preprocessing Methods of Time Series for Monitoring Crop Distribution in Kenya" Agriculture 12, no. 1: 79. https://doi.org/10.3390/agriculture12010079
APA StyleNi, R., Zhu, X., Lei, Y., Li, X., Dong, W., Zhang, C., Chen, T., Mburu, D. M., & Hu, C. (2022). Effectiveness of Common Preprocessing Methods of Time Series for Monitoring Crop Distribution in Kenya. Agriculture, 12(1), 79. https://doi.org/10.3390/agriculture12010079