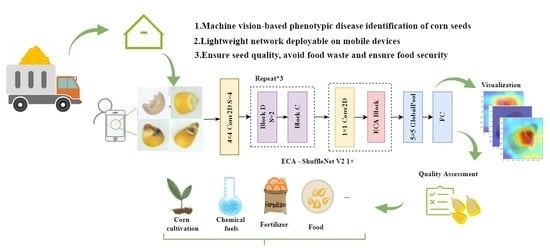
1. Introduction
Evaluating the quality agricultural products is an issue to which countries have always attached great importance. Recent years have seen the introduction of the concept of precision agriculture, with stricter requirements for the quality assessment of agricultural products, which is important to guarantee accurate identification and effective control of seed pests and diseases, in addition to playing a vital role in grain storage and distribution management, helping to ensure seed quality, avoid food waste, and ensure food security [
1,
2].
Fusarium graminearum, F.cepacia, F.proliferatum, and F.subglutinans are common causes of root, stalk, and cob rot in maize [
3]. Diseased seeds are an important source of primary infestation resulting in plant diseases, leading to the long-distance spread of plant diseases and reducing the germination rate of seeds [
4]. Infected seeds are not conducive to storage and can cause other seeds in the storage to become moldy, thus causing huge food losses and further leading to declines in seed quality or rendering these seeds altogether inedible [
5]. Traditional grain quality and safety assessments often use microbial experiments (e.g., spore counting, enzyme-linked immunosorbent assays). In spite of their excellent detection accuracy, these methods are time-consuming, labor-intensive, and destructive [
6]. As an important basis for evaluating the quality of seeds, the phenotypic detection of seeds is a non-destructive testing method. However, due to the influence of subjective factors in manual testing, the test results vary from person to person, and the detection efficiency is also low, which is easy to misjudge [
7,
8]. Therefore, quality inspectors urgently need a fast and objective method to detect diseases in corn seeds.
With deep learning’s ability to extract features efficiently and accurately, it has become widely used in agriculture, reducing the need for manual feature extraction and analysis and making great progress in crop disease detection. Using hyperspectral imaging technology and deep convolutional neural networks (DCNNs), Zhang et al. [
9] classified corn seeds with different degrees of freezing damage and reached a higher than 90% classification accuracy. Javanmardi et al. [
10] used deep convolutional neural networks to classify varieties of maize seeds with an accuracy of 98%. Wang et al. [
11] used hyperspectral imaging to identify aged maize seeds, which involved a full-spectrum classification model using the support vector machine (SVM) algorithm, principal component analysis (PCA), and ANOVA to reduce the data’s dimensionality and extract the feature wavelengths; they classified maize seeds harvested in different years with a prediction accuracy of 97.5%. Yang et al. [
12] used hyperspectral imaging (HSI) combined with sparse auto-encoders (SAEs) and convolutional neural network (CNN) algorithms to classify the mold grades of maize kernels. SAEs and a CNN were combined with an SVM classifier to construct the SAE-CNN-SVM model, and the results showed 99.47% and 98.94% correct recognition rates on the training and test sets, respectively.
A real-time method based on deep convolutional neural networks for the identification of maize leaf diseases was proposed by Mishra et al. [
13], and the model was deployed on a Raspberry Pi 3. It was used to identify maize leaf diseases, and their model achieved an accuracy of 88.46%. Meng et al. [
14] developed a spectral disease indices (SDIs) monitoring model based on in situ leaf reflection spectra to detect southern corn rust (SCR)-infected leaves and to classify the severity of SCR damage. The performance of the developed SCR-SDIs was evaluated by employing a support vector machine (SVM), and the model achieved an overall accuracy of 87% and 70% for SCR detection and severity classification, respectively. The authors also found that these spectral features were associated with the leaf pigments and water content. Albarrak et al. [
15] created a date fruit dataset containing eight categories and used the MobileNetV2 model for date fruit classification. The results showed that the classification accuracy was 99%. Padilla et al. [
16] used convolutional neural networks and OpenMP to detect leaf blight, leaf rust, and leaf spot in corn crops, and performed validation experiments on a Raspberry Pi, with measured accuracies of 93%, 89%, and 89%, respectively.
All of the above studies have provided positive results in agricultural product quality assessment and classification. However, hyperspectral imaging data acquired has high physical complexity, and analysis of hyperspectral data requires fast computers, sensitive detectors, and large data storage capacity [
17]. In addition, large convolutional neural networks or traditional machine learning models are difficult to deploy in agricultural production environments with limited computing resources.
To make deep learning models flexible for deployment on mobile platforms, scholars have proposed lightweight network structures such as GhostNet [
18], MobileNet [
19], ShuffleNet [
20], etc. Existing domestic and international methods for mobile crop disease identification include application realization on mobile devices such as cell phones, intelligent mobile monitoring robots, aerial monitoring drones, and other ground deployment methods [
21,
22]. In response to the above research, this paper proposes a lightweight corn seed disease identification method with an improved ShuffleNetV2. We started by using CycleGAN [
23] to solve the problem of the unbalanced corn seed disease dataset. Then, the ShuffleNetV2 and ECA [
24] modules are combined to improve network performance; the network structure is simplified to speed up network inference. Finally, an experimental evaluation was conducted on a corn seeds dataset [
25], and the results showed that the improved model was lighter and had better recognition accuracy compared with ShuffleNetV2, which shows the potential for crop pest recognition on mobile platforms with low computational power.
2. Dataset Preparation
The dataset in this paper is the public Corn Seeds Dataset [
25] provided by the laboratory in Hyderabad, India, which classifies corn seeds into four categories, pure, broken, discolored, and silkcut, for a total of 17,801 maps. The number of healthy seeds accounts for 40.8% of the original dataset, and the number of diseased seeds that are broken, discolored, and silkcut account for 32%, 17.4%, and 9.8% of the total, respectively. The number of corn seeds in each of the four categories is thus extremely unbalanced. Nagar S. et al. [
25] used the BigGAN [
26] method to generate 5000 pseudograms, but the dataset suffered from classification inaccuracies, making the recognition model network severely over-fitted.
Figure 1 shows a preview of the four categories of the Corn Seeds Dataset.
2.1. CycleGAN Data Augmentation
In this paper, each type of seed in the dataset is first manually corrected according to the original criteria:
- (1)
Pure: the seeds are full in appearance, with no visible breakage, mold, black rot, or cracks;
- (2)
Broken: the seeds are incomplete in appearance, with visible breaking, accompanied by a few mold infections and discoloration;
- (3)
Discolored: large areas of mold infection and black rot on the seed surface causing discoloration of the seeds, accompanied by a small amount of breakage;
- (4)
Silkcut: the basic type of seed is intact, with visible cracks on the surface, with a few accompanying discolorations or breakage at the cracks.
Next, we use the Cycle–Consistent Adversarial Networks (CycleGAN) to generate disease seeds in the dataset to solve the dataset’s imbalance problem. CycleGAN is an unsupervised generative adversarial network that does not require a one-to-one mapping relationship between training data for image-to-image translation.
Figure 2 illustrates the model structure of CycleGAN, where
A and
B represent two different styles of image domains;
a and
b represent the images in the
A and
B domains, respectively; and
G and
F represent the generators required for the mutual translation process of image domains
A and
B. The translation process from
A to
B can be described as follows:
a obtains a forged image
with the style of
B through generator
G, and the forged image
is input to generator
F to obtain the reconstructed image
. The translation of
B to
A follows the same process as above. The two discriminators,
and
, discriminate the forged image and calculate the probabilities
and
that
a and
belong to the
A domain and similarly obtain the probabilities
and
that
b and
belong to the
B domain. The computed probabilities between the discriminators
and
are used to define the adversarial loss of CycleGAN, which ensures that the generator and discriminator evolve with each other, thus allowing the generator to generate more realistic images; the
A and
B domains and the reconstructed image domains
and
are mapped using a cyclic consistent loss function to ensure an efficient mapping from domain
A to
B.
2.2. Loss Function
The loss function of CycleGAN consists of the adversarial loss
and the cycle consistent loss
, which ensures an efficient mapping of the two domains. The total adversarial loss is
with image domains
A and
B.
G is the mapping generator from image domain
A to
B with the adversarial loss function denoted as
;
F is the mapping generator from image domain
B to
A with the adversarial loss function denoted as
; and the total adversarial loss is
, as shown in the following Equations (1)–(4):
where
and
are the probability distributions of image domains
A and
B, respectively.
The cycle consistent loss function
is shown in Equation (
4). For each image
a from domain
A, the image translation cycle should be able to bring
a back to the original image, i.e.,
a ≈
. We call this forward cycle consistency. The cycle consistent loss of the
B to
A domain is similar:
b ≈
satisfies the backward cycle consistency, thus avoiding the situation of invalid adversarial loss. From the above Equations (1)–(4), the loss function Loss of CycleGAN can be obtained as in Equation (
5):
where
is weight of the cyclic consistency loss, controlling for the relative importance of adversarial and cyclic consistency losses.
2.3. Training Results
Table 1 below shows the distribution of pure, broken, discolored and silkcut seeds in this dataset. This paper uses CycleGAN to supplement the discolored and silkcut classes. The two training processes define image domain
A as pure and image domain
B as discolored or silkcut.
The CycleGAN image translation models for pure and discolored seeds are first trained. In its model training phase, for the input image domains A (pure) and B (discolored), the corresponding forged and reconstructed images are generated by the generative network; then, the gradient of the generative network is calculated and the weight parameters are updated; next, the gradient of the discriminative network is calculated and the weight parameters are updated; finally, the model parameters are saved. The training process for the pure to silkcut translation is the same as above.
The input image is processed by RandomHorizontalFlip, RandomCrop, and Normalize into the generator; the input and output image resolutions are unified at 128 × 128, the number of residual blocks is 6, the Adam optimizer is used, the batch size is 1, and a total of 200 iterations are trained from the beginning. The learning rate remains constant at 0.0002 for the first 100 iterations, and decreases linearly in the direction of 0 for the next 100 iterations. The experimental framework was PaddlePaddle 2.1.2 (Baidu; Beijing, China), Python 3.7 (Centrum Wiskunde & Informatica, Netherlands), and an NVIDIA Tesla V100 graphics card for model training.
The experimental procedure is shown in
Figure 3 and
Figure 4 The image translation process recordings for each 50 iterations of the two training sessions are shown separately. The first row of the figure are all real training images of image domains
A and
B, and the second row shows the corresponding generated images. The figure clearly shows that as the number of training epochs increases, the healthy maize seeds gradually possess the features of the diseased seeds, while the mold and cracks on the diseased seeds gradually disappear, representing that the CycleGAN network has learned the mapping relationship between the two image domains, completing the translation between pure and discolored (or pure and silkcut).
In the model testing phase, e.g., translating pure seeds to discolored, the latest saved model is first loaded; then, a batch of pure and discolored images are fed into CycleGAN; and after the test is completed, we will obtain a batch of fake images, such as the fake discolored seed images in
Figure 5. Finally, we save the resulting fake map and use it to solve the imbalance problem in the dataset.
Figure 5 shows the results of testing with the trained CycleGAN model, with pure translated into discolored on top and pure translated into silkcut on the bottom. The generated images, noted as fake discolored or fake silkcut, are similar to the discolored and silkcut corn seed features in the real dataset and can be used to balance the dataset.
3. Efficient Channel Attention
Attention mechanisms have been widely used in machine vision in recent years, and attention in neural networks allows the system to pay more attention to focused information and learn better attention weights. The ECA attention mechanism proposed by Wang et al. is an improvement on the SE (Squeeze-and-Excitation) [
27] module. The authors found that avoiding dimensionality reduction and a proper local cross-channel interaction strategy helped to improve the performance and efficiency of channel attention.
The ECA structure is shown in
Figure 6, with feature
passing through the Global Average Pooling (GAP) layer to obtain a
feature matrix, and the convolution operation is performed on this feature using a local weight sharing, one-dimensional convolution, followed by a sigmoid activation function to obtain the attention weights; then, the output feature matrix is derived by multiplying this weight with the input feature map. One-dimensional convolution involves an adaptive hyperparameter
k (convolution kernel size), which represents the coverage of local cross-channel interactions, as shown in Equation (
6):
where
denotes the oddest number closest to
x. ECA attention mechanisms have fewer parameters compared to SE, and appropriate cross-channel information interaction ensures that gains are brought to the network while introducing a small number of parameters.
4. ECA—ShuffleNetV2 Relevant Theories
The ultimate goal of deep learning development has always been to proceed to practical applications, which makes people more concerned about how to obtain the optimal results with limited resources. In this paper, we compare the Top-1 Accuracy of GhostNet, MobileNet V2, MobileNet V3, and ShuffleNetV2 series networks under CIFAR-10 [
28]. The following metrics were counted using the torchstate tool provided by PyTorch: FLOPs, Params, and Memory cost.
To meet the needs of mobile deployments, only lightweight versions of these networks are tested in this article. The detailed model versions are shown in
Table 2. We had to make a trade-off between accuracy and speed, and the table shows that MobileNet V2 1× achieved the highest Top-1 Accuracy on CIFAR-10, with ShuffleNetV2 1× following closely behind. However, the FLOPs, Params, and Memory costs of ShuffleNetV2 1× are much lower than those of MobileNet V2 1×. Without a doubt, ShuffleNet V2 1× won this race, and this was further confirmed in subsequent experiments. The subsequent experiments also focused on ShuffleNetV2 and MobileNet V2. To further refine the experiments, the rest of the networks are tested in the final inference speed comparison.
4.1. Depthwise Separable Convolution
Depthwise Separable Convolution (DSC) has been effective in making network structures more lightweight [
29]. It consisting of depthwise (DW) and pointwise (PW) convolution and has a relatively low number of parameters and computational cost in extracting features compared to ordinary convolution. One depthwise convolution is computed for only one channel of the input feature map. Pointwise convolution is similar to ordinary convolution with a size of
.
M is the number of channels in the input feature matrix. Thus, the number of output feature maps is equal to PW’s number of convolution kernels. Moreover, PW convolution solves the problem of DW convolution, which is the poor interaction between the feature information of different channels at the same spatial location, shown in
Figure 7.
4.2. Grouped Convolution and Channel Shuffle
Grouped convolution discretizes dense convolutional connections to build a sufficiently deep and wide neural network by replicating the grouped convolution. Compared with standard convolution, grouped convolution has less parameters, lower complexity, and helps to facilitate the parallelism in the model, but the lack of information exchange between different groups weakens the feature extraction ability of the network. ShuffleNet V1 uses the channel shuffle to remedy this deficiency [
30]. As shown in
Figure 8, Output Features 1 after group convolution are “shuffled” by the channel shuffle operation, which fully integrates the inter-group channel information without increasing the computational effort.
4.3. ShuffleNet V2
ShuffleNetV2 proposes that FLOPs are an indirect metric that cannot be equated with direct metrics such as speed and accuracy and proposes four guidelines for efficient network design:
- (1)
Maintain a constant number of convolutional input and output channel widths to minimize the memory access cost;
- (2)
The quantity of groups in group convolution is inversely proportional to the speed of network operation;
- (3)
Cautious fragmentation operations and reducing the count of network branches can improve operational efficiency;
- (4)
Reducing element-wise operations which have relatively small FLOPs but high memory access costs.
Starting from the four guidelines mentioned above, the authors present an improved ShuffleNetV2 network structure, illustrated in
Figure 9. The authors devised the channel split operation shown in
Figure 9a. In front of each unit, the input channel
C is divided into two equal branches. To avoid fragmentation, one branch is left unchanged. The other branch follows criterion (1) and consists of three convolutional layers with a constant number of channels and no longer uses
grouped convolution. Then, the two branches are concatenated in the depth direction, followed by a channel shuffling operation to enhance the information interaction between the channels.
Figure 9b shows the module with downsampling, and with channel splits removed, twice the number of channels of the output feature map as the input are obtained.
4.4. ECA—ShuffleNetV2
The basic component of a transformer is self-attention, which essentially performs Query-Key-Value operations at a global scale or within a larger window, which is the reason for the superior performance of transformers on downstream tasks [
31]. Recently, many scholars have applied large convolution kernels to CNNs. Liu et al. [
32] borrowed the large-scale window of the transformer in their paper and changed the size of the convolution kernels in CNNs from
to
. They experimentally concluded that
convolution kernels can achieve better detection on the ImageNet dataset with only a small increase in the number of parameters. Similarly Ding et al. [
33] state in their article that large convolution kernels are both more accurate and more efficient at this task.
In this paper, the two units of ShuffleNetV2 are improved by moving forward the depthwise convolution in the original branch 2, followed by two pointwise convolutions. The specific structure of branch 2 is shown in
Figure 10b, where a depthwise convolution of the size
is used instead of the
depthwise convolution in ShuffleNetV2, depicted in
Figure 10c.
In practical applications, the model needs to be reasonably designed according to the complexity of the task. The deep small kernel network has a large theoretical receptive field, but its effective receptive field is limited. We replace the CNN network with a deep small kernel with a shallow but large kernel and conclude in this paper that the method is effective and feasible.
This is a relatively simple four-classification task, and the model size should be appropriately reduced to improve detection efficiency. Therefore, in this paper, the number of repeats of Block C is reduced. Morevoer, the
DW convolution is connected after the channel shuffling of Block D with the downsampling function to further extract features. The modified unit of the network is shown in
Figure 11.
Figure 12 shows a comparison of the ShuffleNetV2
and the improved ECA-ShuffleNetV2
network structure, with the input image resolution changed to 160 × 160. Since the DW convolution is moved forward in the branch, the number of channels in the output feature layer is changed to [58, 116, 232, 464, 1024] here to guarantee that the number of channels of its output feature matrix is divisible by the number of channels of the input feature matrix. The
convolutional layer and the max pooling at the beginning of the ShuffleNetV2 are replaced by a
convolutional, a stride of 4, and the input image is downsampled
. We reduce the size of the network by changing the number of iterations of Block C in each stage from (3, 7, 3) to (1, 1, 1). Conv5 is then followed by the ECA attention module, and the appropriate cross-channel information interaction ensures that a small quantity of parameters are introduced while bringing gains to the network.
6. Conclusions and Future Work
This paper presents an improved ECA-ShuffleNetV2 network for corn seed disease identification. Production of the dataset requires only a low-cost digital camera. On a CPU device, this model has a single-threaded inference speed of about 9.71 ms and a classification accuracy of 96.28% on the validation set. It has 0.913 M parameters and 44.75 M FLOPs. The model is suitable for deployment on mobile devices, such as smart phones and portable laptops available to growers or quality assessment practitioners, as well as on offline mobile monitoring sites.
Compared with traditional machine learning, deep learning is more effective at pest and disease classification. In addition, the corn seed disease recognition model built in this paper has the advantages of high recognition accuracy, fast recognition speed, and relatively small model parameters. This model can quickly extract the characteristics of diseased seeds, which greatly reduces the workload of manual detection. It is possible for relevant departments to collect and produce different data sets of agricultural samples for quality evaluation model training of other cereals according to the actual situation, which has good application prospects.
Using machine vision, we can only obtain phenotypic information about seeds; however, that information does not describe their internal characteristics. Therefore, in the follow-up work, we will combine our method with hyperspectral technology in order to detect and classify seed diseases and enhance the model’s robustness so that it works well on similar tasks.
According to a comparison with related research at this stage, the limited number of seed samples chosen for this study is not representative of all diseases present in Chinese maize seeds today. Particularly, diseased seeds are much less common than healthy seeds, which is a huge test of our work. In the next study, a complete maize seed image acquisition system will be designed to address the problem. To build a comprehensive and accurate data set of maize seed diseases, we will collaborate with relevant departments. We will also ensure its classification quality in order to improve the practical utility of the model.
While the acquisition of datasets is critical, the success of supervised machine learning is not possible without high-quality data annotation. Manual annotation is labor-intensive and time-consuming, so relying only on manual annotation is not a wise decision. Our future research will use unsupervised machine learning to classify and label maize seed diseases. This will further reduce the workload and accelerate the deployment of the model in agricultural quality assessment.
In addition, this experiment did not consider the relationship between external factors, such as different corn growing regions, corn varieties, and climate in China, and corn seed diseases. These external factors are also crucial to the quality assessment of agricultural products and food waste reduction. As a result, follow-up work will include further experiments and investigations related to this topic.