Helicobacter pylori Is Present at Quantifiable Levels in Raw Vegetables in the Mediterranean Area of Spain
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples and Isolation of Helicobacter pylori
2.2. DNA Extraction and Quantification by qPCR
2.3. Sequencing of Presumptive Positive H. pylori Samples
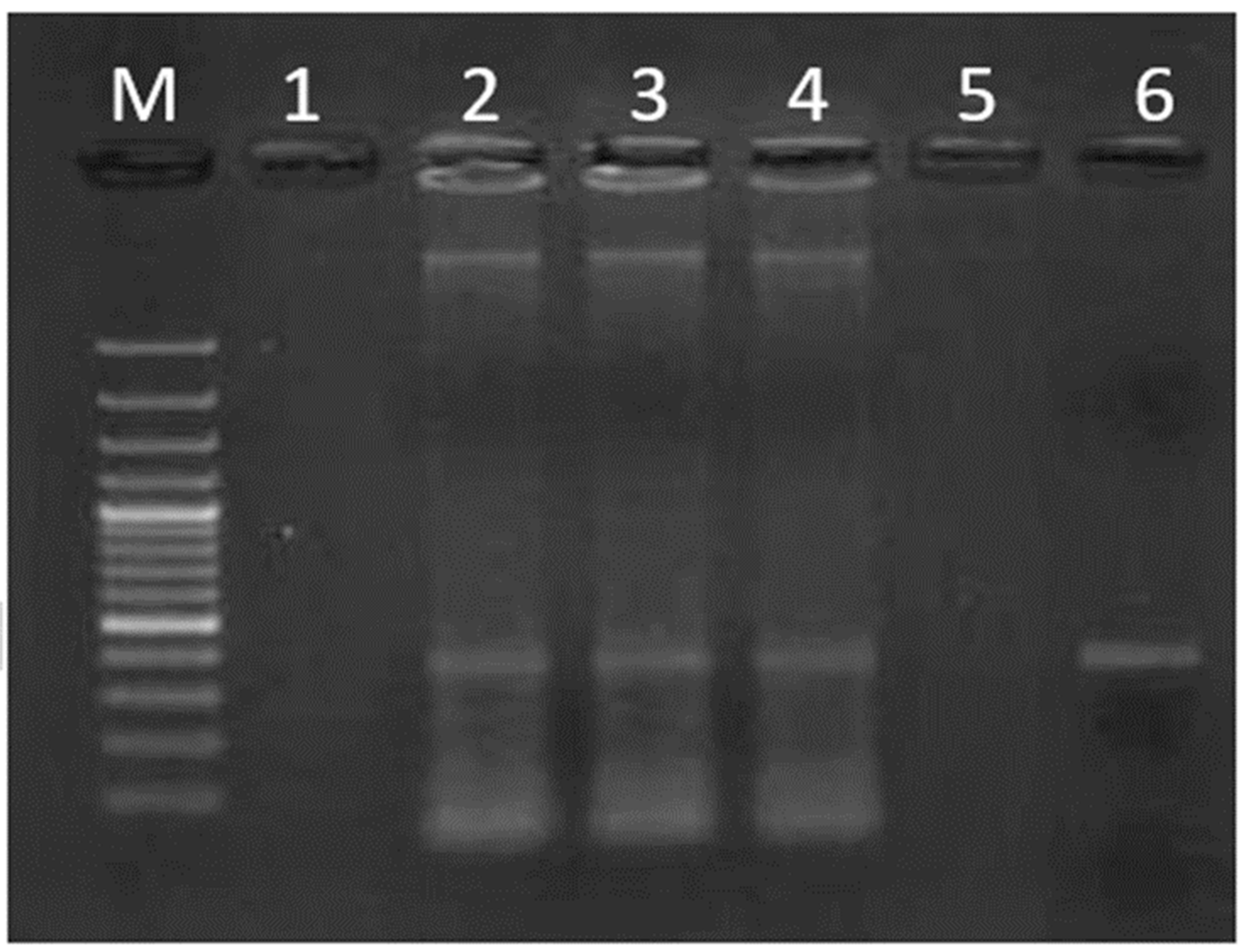
3. Results
Prevalence of Helicobacter pylori in Fresh Vegetables
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- EU Statistics. Fruits and Vegetables Consumption in EU. 2018. Available online: https://ec.europa.eu/eurostat/statistics-explained/index.php/Fruit_and_vegetable_consumption_statistics (accessed on 20 January 2022).
- Harvey, R.R.; Zakhour, C.M.; Gould, L.H. Foodborne disease outbreaks associated with organic foods in the United States. J. Food Prot. 2016, 79, 1953–1958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blau, K.; Bettermann, A.; Jechalke, S.; Fornefeld, E.; Vanrobaeys, Y.; Stalder, T.; Top, E.M.; Smalla, K. The transferable resistome of produce. Am. Soc. Microbiol. 2018, 9, e01300–e01318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venneman, K.; Huybrechts, I.; Gunter, M.J.; Vandendaele, L.; Herrero, R.; Van Herck, K. The epidemiology of Helicobacter pylori infection in Europe and the impact of lifestyle on its natural evolution toward stomach cancer after infection: A systematic review. Helicobacter 2018, 23, e12483. [Google Scholar] [CrossRef] [PubMed]
- Hooi, J.K.Y.; Lai, W.Y.; Ng, W.K.; Suen Massess, M.Y.; Underwood, F.E.; Tanyingoh, D.; Malfertheiner, P.; Graham, D.Y.; Wong, V.W.S.; Wu, J.C.Y.; et al. Global prevalence of Helicobacter pylori infection: Systematic review and meta-analysis. Gastroenterology 2017, 53, 420–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization (WHO). WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. 2017. Available online: https://www.who.int/news-room/detail/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 20 January 2022).
- Gisbert, J.P. “Rescue” regimens after Helicobacter pylori treatment failure. World J. Gastroenterol. 2008, 14, 5385–5402. [Google Scholar] [CrossRef] [PubMed]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Biological Agents. V100 B. A review of human carcinogens. IARC Monogr. Eval. Carcinog. Risks Hum. 2012, 100, 1–441. [Google Scholar]
- Azevedo, N.F.; Almeida, C.; Fernandes, I.; Cerqueira, L.; Dias, S.; Keevil, C.W.; Vieira, M.J. Survival of gastric and enterohepatic Helicobacter spp. in water: Implications for transmission. Appl. Environ. Microbiol. 2008, 74, 1805–1811. [Google Scholar] [CrossRef] [Green Version]
- Konishi, K.; Saito, N.; Shoji, E.; Takeda, H.; Kato, M.; Asaka, M.; Ooi, H.K. Helicobacter pylori: Longer survival in deep ground water and sea water than in a nutrient-rich environment. Apmis 2007, 115, 1285–1291. [Google Scholar] [CrossRef]
- Bai, X.; Xi, C.; Wu, J. Survival of Helicobacter pylori in the wastewater treatment process and the receiving river in Michigan, USA. J. Water Health 2016, 14, 692–698. [Google Scholar] [CrossRef] [Green Version]
- Zamani, M.; Vahedi, A.; Maghdouri, Z.; Shokri-Shirvani, J. Role of food in environmental transmission of Helicobacter pylori. Caspian J. Intern. Med. 2017, 8, 146–152. [Google Scholar]
- Quaglia, N.C.; Dambrosio, A. Helicobacter pylori: A foodborne pathogen? World J. Gastroenterol. 2018, 24, 3472–3487. [Google Scholar] [CrossRef]
- Leja, M.; Grinberga-Derica, I.; Bilgilier, C.; Steininger, C. Review: Epidemiology of Helicobacter pylori infection. Helicobacter 2019, 24 (Suppl. 1), e12635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atapoor, S.; Safarpoor Dehkordi, F.; Rahimi, E. Detection of Helicobacter pylori in various types of vegetables and salads. Jundishapur J. Microbiol. 2014, 7, e10013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamada, M.; Elbehiry, A.; Marzouk, E.; Moussa, I.M.; Hessain, A.M.; Alhaji, J.H.; Heme, H.A.; Zahran, R.; Abdeen, E. Helicobacter pylori in a poultry slaughterhouse: Prevalence, genotyping and antibiotic resistance pattern. Saudi J. Biol. Sci. 2018, 25, 1072–1078. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Mesonero, L.; Moreno, Y.; Alonso, J.L.; Ferrús, M.A. Detection of viable Helicobacter pylori inside free-living amoebae in wastewater and drinking water samples from Eastern Spain. Environ. Microbiol. 2018, 19, 4103–4112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdel-Moein, K.A.; Saeed, H.; Samir, A. Novel detection of Helicobacter pylori in fish: A possible public health concern. Acta Trop. 2015, 152, 141–144. [Google Scholar] [CrossRef] [PubMed]
- Vesga, F.J.; Moreno, Y.; Ferrús, M.A.; Campos, C.; Trespalacios, C.C. Detection of Helicobacter pylori in drinking water treatment plants in Bogotá, Colombia, using cultural and molecular techniques. Int. J. Hyg. Environ. Health 2018, 221, 595–601. [Google Scholar] [CrossRef]
- Allende, A.; Monaghan, J. Irrigation water quality for leafy crops: A perspective of risks and potential solutions. Int. J. Environ. Res. Public Health 2015, 12, 7457–7477. [Google Scholar] [CrossRef] [Green Version]
- Rahman, S.M.E.; Mele, M.A.; Lee, Y.T.; Islam, M.Z. Consumer preference, quality, and safety of organic and conventional fresh fruits, vegetables, and cereals. Foods 2021, 10, 105. [Google Scholar] [CrossRef]
- Yahaghi, E.; Khamesipour, F.; Mashayekhi, F.; Safarpoor Dehkordi, F.; Hossein Sakhaei, M.; Masoudimanesh, M.; Khayyat Khameneie, M. Helicobacter pylori in vegetables and salads: Genotyping and antimicrobial resistance properties. Biomed. Res. Int. 2014, 2014, 757941. [Google Scholar] [CrossRef] [Green Version]
- Pina-Pérez, M.C.; González, A.; Moreno, Y.; Ferrús, M.A. Helicobacter pylori growth pattern in reference media and extracts from selected minimally processed vegetables. Food Control 2018, 86, 389–396. [Google Scholar] [CrossRef]
- Buck, A.; Oliver, J.D. Survival of spinach-associated Helicobacter pylori in the viable but nonculturable state. Food Control 2010, 21, 1150–1154. [Google Scholar] [CrossRef]
- Aktas, D.; Bagirova, M.; Allahverdiyev, A.M.; Abamor, E.S.; Safarov, T.; Kocazeybek, B.S. Resuscitation of the Helicobacter pylori coccoid forms by resuscitation promoter factor obtained from Micrococcus luteus. Curr. Microbiol. 2020, 77, 2093–2103. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.G.; Loke, M.F.; Goh, K.L.; Vadivelu, J.; Ho, B. Biofilm formation enhances Helicobacter pylori survivability in vegetables. Food Microbiol. 2017, 62, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Huang, L. Optimization of a new mathematical model for bacterial growth. Food Control 2013, 32, 283–288. [Google Scholar] [CrossRef]
- Baranyi, J.; Csernus, C.; Beczner, J. Error analysis in predictive modelling demonstrated on mould data. Int. J. Food Microbiol. 2014, 170, 78–82. [Google Scholar] [CrossRef]
- Nilsson, H.O.; Blom, J.; Abu-Al-Soud, W.; Ljungh, A.A.; Andersen, L.; Wadström, T. Effect of cold starvation, acid stress, and nutrients on metabolic activity of Helicobacter pylori. Appl. Environ. Microbiol. 2002, 68, 11–19. [Google Scholar] [CrossRef] [Green Version]
- Santiago, P.; Moreno, Y.; Ferrús, M.A. Identification of viable Helicobacter pylori in drinking water supplies by cultural and molecular techniques. Helicobacter 2015, 20, 252–259. [Google Scholar] [CrossRef]
- Pina-Pérez, M.C.; González, A.; Moreno, Y.; Ferrús, M.A. Helicobacter pylori detection in shellfish: A real-time quantitative polymerase chain reaction approach. Foodborne Path. Dis. 2018, 16, 137–143. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [Green Version]
- Solnick, J.V.; Hansen, L.M.; Canfield, D.R.; Parsonnet, J. Determination of the infectious dose of Helicobacter pylori during primary and secondary infection in rhesus monkeys (Macaca mulatta). Infect. Immun. 2001, 69, 6887–6892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrero, R.L.; Thiberge, J.M.; Kansau, I.; Wuscher, N.; Huerre, M.; Labigne, A. The GroES homolog of Helicobacter pylori confers protective immunity against mucosal infection in mice. Proc. Natl. Acad. Sci. USA 1995, 92, 6499–6503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pagliai, G.; Sofi, F.; Vannetti, F.; Caiani, S.; Pasquini, G.; Molino Lova, R.; Cecchi, F.; Sorbi, S.; Macchi, C. Mediterranean diet, food consumption and risk of late-life depression: The mugello study. J. Nutr. Health Aging 2018, 22, 569–574. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Ferrús, M.; González, A.; Pina-Pérez, M.C.; Ferrús, M.A. Helicobacter pylori Is Present at Quantifiable Levels in Raw Vegetables in the Mediterranean Area of Spain. Agriculture 2022, 12, 339. https://doi.org/10.3390/agriculture12030339
García-Ferrús M, González A, Pina-Pérez MC, Ferrús MA. Helicobacter pylori Is Present at Quantifiable Levels in Raw Vegetables in the Mediterranean Area of Spain. Agriculture. 2022; 12(3):339. https://doi.org/10.3390/agriculture12030339
Chicago/Turabian StyleGarcía-Ferrús, Miguel, Ana González, María Consuelo Pina-Pérez, and Maria Antonia Ferrús. 2022. "Helicobacter pylori Is Present at Quantifiable Levels in Raw Vegetables in the Mediterranean Area of Spain" Agriculture 12, no. 3: 339. https://doi.org/10.3390/agriculture12030339
APA StyleGarcía-Ferrús, M., González, A., Pina-Pérez, M. C., & Ferrús, M. A. (2022). Helicobacter pylori Is Present at Quantifiable Levels in Raw Vegetables in the Mediterranean Area of Spain. Agriculture, 12(3), 339. https://doi.org/10.3390/agriculture12030339





