Comparative Biochemical and Transcriptome Analyses Reveal Potential Candidate Genes Related to Pericarp Browning in Red Rice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
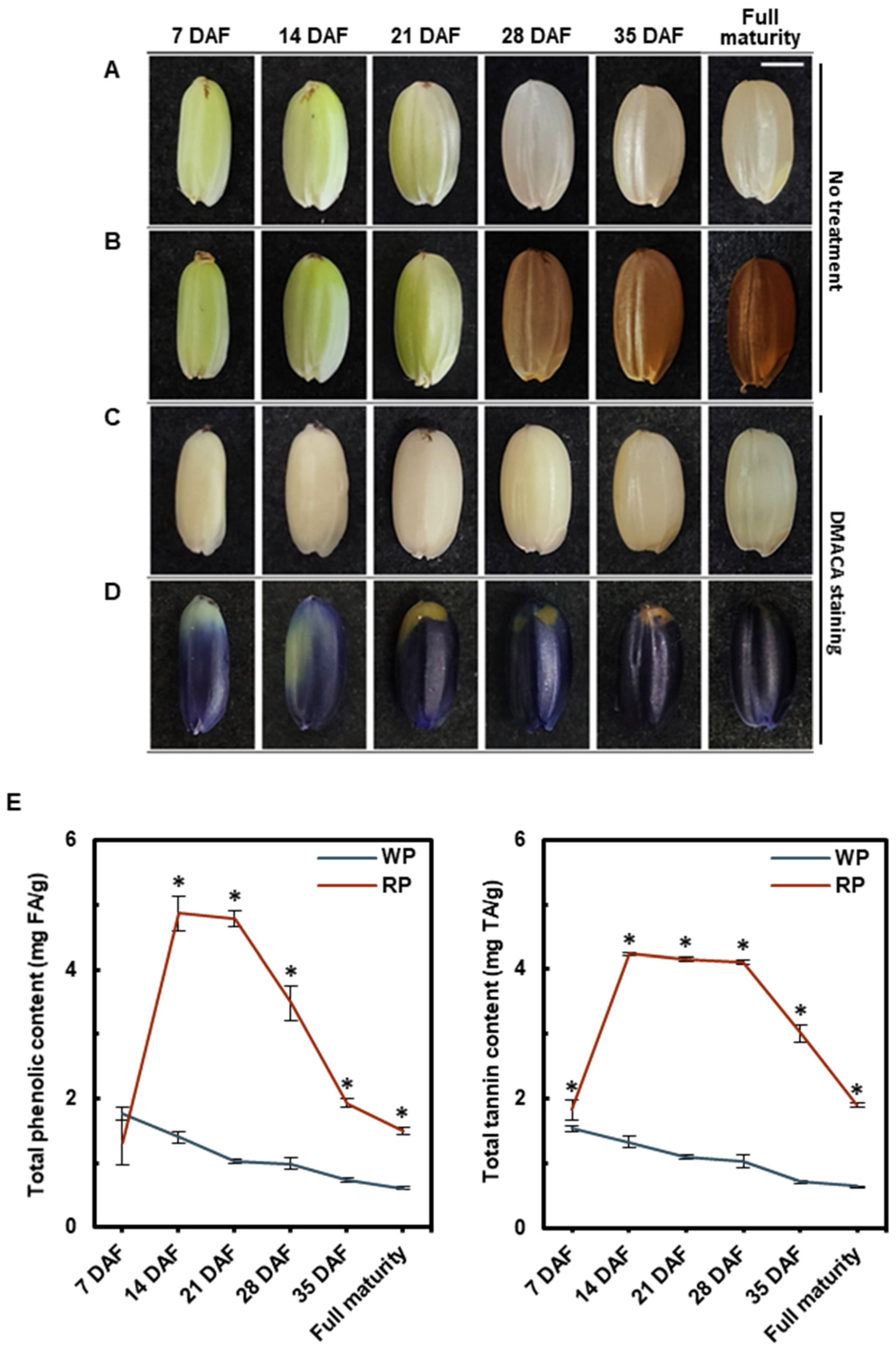
2.2. 4-Dimethylaminocinnamaldehyde Staining
2.3. Quantification of Total Phenolic and Tannin Content
2.4. RNA Extraction and Sequencing
2.5. Identification of Differentially Expressed Genes and Enrichment Analysis
2.6. Validation of Gene Expression via Quantitative Reverse Transcription Polymerase Chain Reaction
2.7. Statical Analysis of Experimental Data
3. Results
3.1. Pericarp Color Change and Proanthocyanidin Accumulation during Grain Development
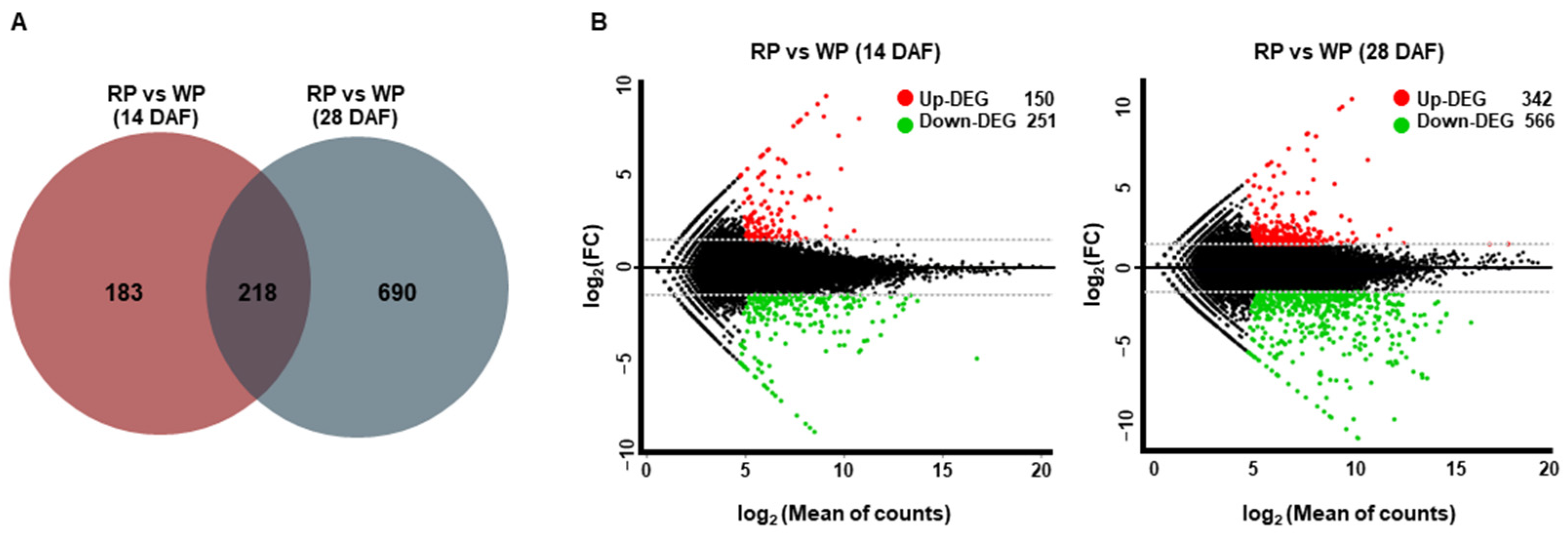
3.2. RNA Sequencing-Based Transcriptome Profiling and Detection of Differentially Expressed Genes
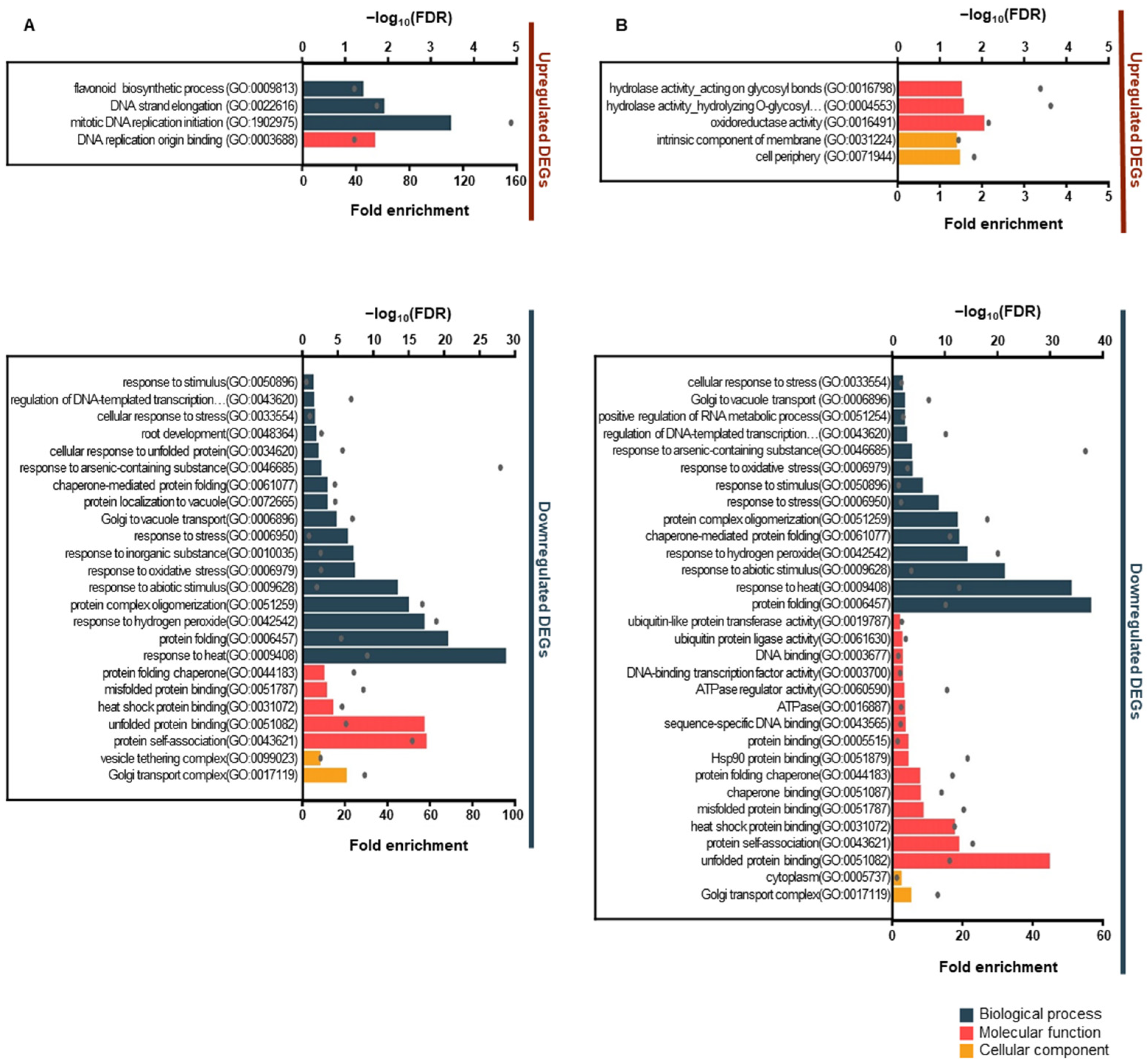
3.3. Gene Ontology and Kyoto Encyclopedia of Genes and Genome Pathway Enrichment Analysis
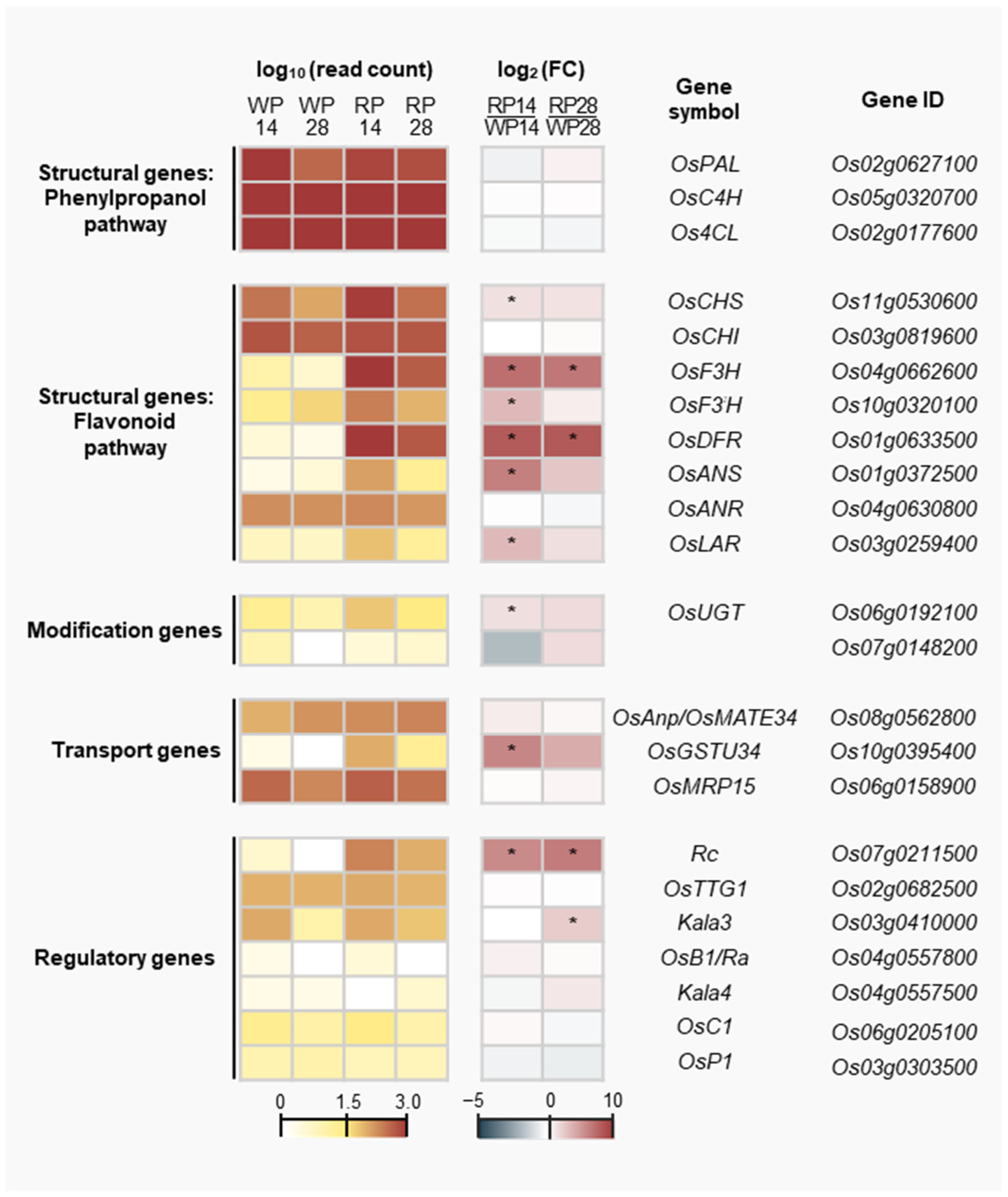
3.4. Expression Profile of Rice PA Biosynthesis-Related Genes
3.5. Potential Candidate Genes Related to Pericarp Browning
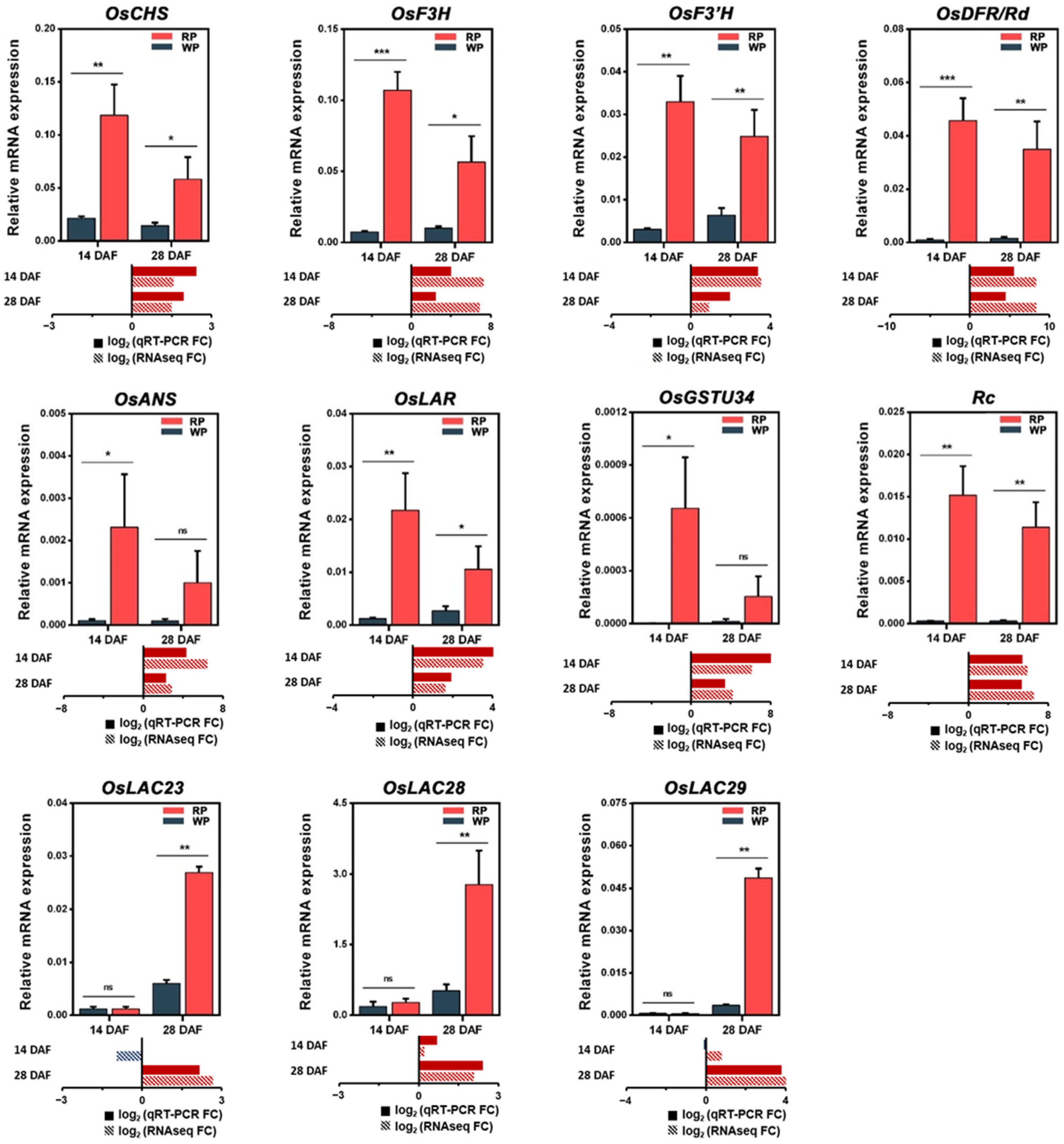
3.6. Quantitative Reverse Transcription-Polymerase Chain Reaction Validation of RNA Sequencing-Based Expression Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ahuja, U.; Ahuja, S.C.; Chaudhary, N.; Thakrar, R. Red Rices—Past, Present, and Future. Asian Agrihist. 2007, 11, 291–304. [Google Scholar]
- Mbanjo, E.G.N.; Kretzschmar, T.; Jones, H.; Ereful, N.; Blanchard, C.; Boyd, L.A.; Sreenivasulu, N. The Genetic Basis and Nutritional Benefits of Pigmented Rice Grain. Front. Genet. 2020, 11, 229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dixon, R.A.; Xie, D.Y.; Sharma, S.B. Proanthocyanidins—A Final Frontier in Flavonoid Research? New Phytol. 2005, 165, 9–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mannino, G.; Chinigò, G.; Serio, G.; Genova, T.; Gentile, C.; Munaron, L.; Bertea, C.M. Proanthocyanidins and Where to Find Them: A Meta-Analytic Approach to Investigate Their Chemistry, Biosynthesis, Distribution and Effect on Human Health. Antioxidants 2021, 10, 1229. [Google Scholar] [CrossRef] [PubMed]
- Marles, M.A.S.; Ray, H.; Gruber, M.Y. New Perspectives on Proanthocyanidin Biochemistry and Molecular Regulation. Phytochemistry 2003, 64, 367–383. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Pan, Q.H.; Shi, Y.; Duan, C.Q. Biosynthesis and Genetic Regulation of Proanthocyanidins in Plants. Molecules 2008, 13, 2674–2703. [Google Scholar] [CrossRef] [Green Version]
- Kitamura, S.; Shikazono, N.; Tanaka, A. TRANSPARENT TESTA 19 Is Involved in the Accumulation of Both Anthocyanins and Proanthocyanidins in Arabidopsis. Plant J. 2004, 37, 104–114. [Google Scholar] [CrossRef]
- Debeaujon, I.; Léon-Kloosterziel, K.M.; Koornneef, M. Influence of the Testa on Seed Dormancy, Germination, and Longevity in Arabidopsis. Plant Physiol. 2000, 122, 403–413. [Google Scholar] [CrossRef] [Green Version]
- Pourcel, L.; Routaboul, J.M.; Kerhoas, L.; Caboche, M.; Lepiniec, L.; Debeaujon, I. TRANSPARENT TESTA10 Encodes a Laccase-like Enzyme Involved in Oxidative Polymerization of Flavonoids in Arabidopsis Seed Coat. Plant Cell 2005, 17, 2966–2980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sweeney, M.T.; Thomson, M.J.; Pfeil, B.E.; McCouch, S. Caught Red-Handed: Rc Encodes a Basic Helix-Loop-Helix Protein Conditioning Red Pericarp in Rice. Plant Cell 2006, 18, 283–294. [Google Scholar] [CrossRef] [Green Version]
- Furukawa, T.; Maekawa, M.; Oki, T.; Suda, I.; Iida, S.; Shimada, H.; Takamure, I.; Kadowaki, K.I. The Rc and Rd Genes Are Involved in Proanthocyanidin Synthesis in Rice Pericarp. Plant J. 2007, 49, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [Green Version]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. Revigo summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-I: Intelligent Prioritization and Exploratory Visualization of Biological Functions for Gene Enrichment Analysis. Nucleic Acids Res. 2021, 49, 317–325. [Google Scholar] [CrossRef]
- Goufo, P.; Trindade, H. Rice Antioxidants: Phenolic Acids, Flavonoids, Anthocyanins, Proanthocyanidins, Tocopherols, Tocotrienols, c-Oryzanol, and Phytic Acid. Food Sci. Nutr. 2014, 2, 75–104. [Google Scholar] [CrossRef]
- Chen, M.H.; McClung, A.M.; Bergman, C.J. Concentrations of Oligomers and Polymers of Proanthocyanidins in Red and Purple Rice Bran and Their Relationships to Total Phenolics, Flavonoids, Antioxidant Capacity and Whole Grain Color. Food Chem. 2016, 208, 279–287. [Google Scholar] [CrossRef]
- Gu, X.Y.; Foley, M.E.; Horvath, D.P.; Anderson, J.V.; Feng, J.; Zhang, L.; Mowry, C.R.; Ye, H.; Suttle, J.C.; Kadowaki, K.I.; et al. Association between Seed Dormancy and Pericarp Color Is Controlled by a Pleiotropic Gene That Regulates Abscisic Acid and Flavonoid Synthesis in Weedy Red Rice. Genetics 2011, 189, 1515–1524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oikawa, T.; Maeda, H.; Oguchi, T.; Yamaguchi, T.; Tanabe, N.; Ebana, K.; Yano, M.; Ebitani, T.; Izawa, T. The Birth of a Black Rice Gene and Its Local Spread by Introgression. Plant Cell 2015, 27, 2401–2414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Butelli, E.; Martin, C. Engineering Anthocyanin Biosynthesis in Plants. Curr. Opin. Plant Biol. 2014, 19, 81–90. [Google Scholar] [CrossRef]
- Zhao, J.; Dixon, R.A. MATE Transporters Facilitate Vacuolar Uptake of Epicatechin 3′-O-Glucoside for Proanthocyanidin Biosynthesis in Medicago Truncatula and Arabidopsis. Plant Cell 2009, 21, 2323–2340. [Google Scholar] [CrossRef] [Green Version]
- Yoshimura, Y.; Zaima, N.; Moriyama, T.; Kawamura, Y. Different Localization Patterns of Anthocyanin Species in the Pericarp of Black Rice Revealed by Imaging Mass Spectrometry. PLoS ONE 2012, 7, e31285. [Google Scholar] [CrossRef]
- Zhao, J.; Pang, Y.; Dixon, R.A. The Mysteries of Proanthocyanidin Transport and Polymerization. Plant Physiol. 2010, 153, 437–443. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J. Flavonoid Transport Mechanisms: How to Go, and with Whom. Trends Plant Sci. 2015, 20, 576–585. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Sharma, N.; Kapoor, P.; Chunduri, V.; Pandey, A.K.; Garg, M. Spotlight on the Overlapping Routes and Partners for Anthocyanin Transport in Plants. Physiol. Plant. 2021, 171, 868–881. [Google Scholar] [CrossRef]
- Xu, W.; Grain, D.; Bobet, S.; Le Gourrierec, J.; Thévenin, J.; Kelemen, Z.; Lepiniec, L.; Dubos, C. Complexity and Robustness of the Flavonoid Transcriptional Regulatory Network Revealed by Comprehensive Analyses of MYB-BHLH-WDR Complexes and Their Targets in Arabidopsis Seed. New Phytol. 2014, 202, 132–144. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, M.T.; Thomson, M.J.; Yong, G.C.; Yong, J.P.; Williamson, S.H.; Bustamante, C.D.; McCouch, S.R. Global Dissemination of a Single Mutation Conferring White Pericarp in Rice. PLoS Genet. 2007, 3, 1418–1424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Lin, Y.; Chen, S.; Liu, H.; Chen, Z.; Fan, M.; Hu, T.; Mei, F.; Chen, J.; Chen, L.; et al. CRISPR/Cas9-Mediated Functional Recovery of the Recessive Rc Allele to Develop Red Rice. Plant Biotechnol. J. 2019, 17, 2096–2105. [Google Scholar] [CrossRef] [Green Version]
- Queiroz, C.; Mendes Lopes, M.L.; Fialho, E.; Valente-Mesquita, V.L. Polyphenol Oxidase: Characteristics and Mechanisms of Browning Control. Food Rev. Int. 2008, 24, 361–375. [Google Scholar] [CrossRef]
- Mayer, A.M.; Staples, R.C. Laccase: New Functions for an Old Enzyme. Phytochemistry 2002, 60, 551–565. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Lu, K.; Qu, C.; Liang, Y.; Wang, R.; Chai, Y.; Li, J. Gene Silencing of BnTT10 Family Genes Causes Retarded Pigmentation and Lignin Reduction in the Seed Coat of Brassica Napus. PLoS ONE 2013, 8, e61247. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Zhang, X.; Zhong, R.; Liu, B.; Zhang, X.; Fang, F.; Zhang, Z.; Pang, X. Laccase-Mediated Flavonoid Polymerization Leads to the Pericarp Browning of Litchi Fruit. J. Agric. Food Chem. 2021, 69, 15218–15230. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zhu, Q.; Zhou, X.; Zhang, X.; Dang, Z.; Liang, S.; Li, G.; Zhang, Z.; Fang, F.; Pang, X. Characterization of a Pericarp Browning Related LACCASE 14-4 from Longan Fruit with a Focus on (Epi)Catechin Oxidative Polymerization. Postharvest Biol. Technol. 2022, 185, 111802. [Google Scholar] [CrossRef]
- Liu, M.; Dong, H.; Wang, M.; Liu, Q. Evolutionary Divergence of Function and Expression of Laccase Genes in Plants. J. Genet. 2020, 99, 23. [Google Scholar] [CrossRef]
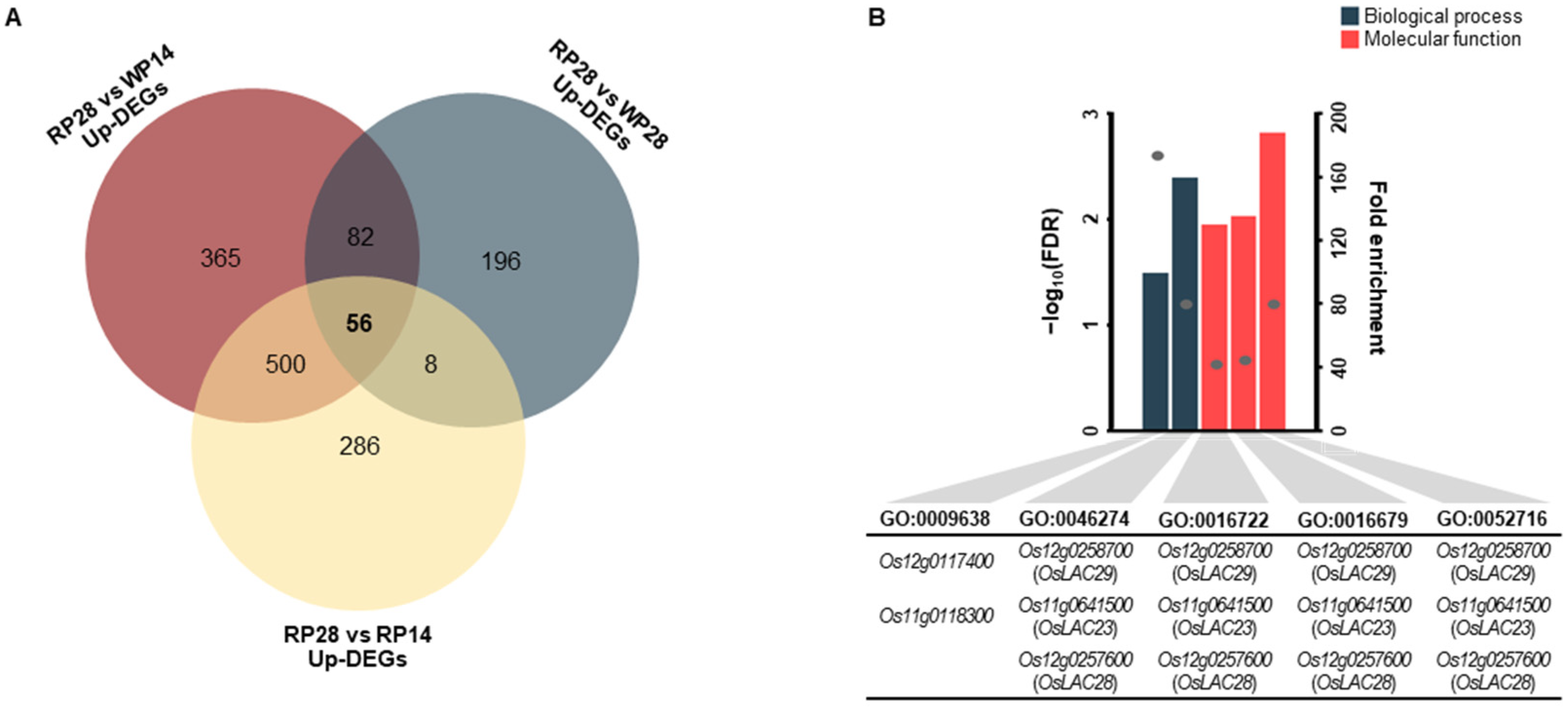
| Stage | DEG Set | KEGG ID | Pathway Name | Count | Expected Count | FDR |
|---|---|---|---|---|---|---|
| 14 DAF | Upregulated | dosa00941 | Flavonoid biosynthesis | 6 | 41 | 7.12 × 10−7 |
| Upregulated | dosa03030 | DNA replication | 4 | 57 | 1.09 × 10−3 | |
| Downregulated | dosa04141 | Protein processing in endoplasmic reticulum | 20 | 210 | 3.91 × 10−15 | |
| 28 DAF | Upregulated | dosa00904 | Diterpenoid biosynthesis | 4 | 41 | 4.25 × 10−2 |
| Downregulated | dosa04141 | Protein processing in endoplasmic reticulum | 27 | 210 | 2.95 × 10−14 | |
| Downregulated | dosa04626 | Plant-pathogen interaction | 10 | 192 | 2.75 × 10−2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, G.; Park, J.W.; Park, J.; Han, A.-R.; Hong, M.J.; Jo, Y.D.; Kim, J.-B.; Kim, S.H.; Choi, H.-I. Comparative Biochemical and Transcriptome Analyses Reveal Potential Candidate Genes Related to Pericarp Browning in Red Rice. Agriculture 2023, 13, 183. https://doi.org/10.3390/agriculture13010183
Lee G, Park JW, Park J, Han A-R, Hong MJ, Jo YD, Kim J-B, Kim SH, Choi H-I. Comparative Biochemical and Transcriptome Analyses Reveal Potential Candidate Genes Related to Pericarp Browning in Red Rice. Agriculture. 2023; 13(1):183. https://doi.org/10.3390/agriculture13010183
Chicago/Turabian StyleLee, Gileung, Jae Wan Park, Jisu Park, Ah-Reum Han, Min Jeong Hong, Yeong Deuk Jo, Jin-Baek Kim, Sang Hoon Kim, and Hong-Il Choi. 2023. "Comparative Biochemical and Transcriptome Analyses Reveal Potential Candidate Genes Related to Pericarp Browning in Red Rice" Agriculture 13, no. 1: 183. https://doi.org/10.3390/agriculture13010183
APA StyleLee, G., Park, J. W., Park, J., Han, A.-R., Hong, M. J., Jo, Y. D., Kim, J.-B., Kim, S. H., & Choi, H.-I. (2023). Comparative Biochemical and Transcriptome Analyses Reveal Potential Candidate Genes Related to Pericarp Browning in Red Rice. Agriculture, 13(1), 183. https://doi.org/10.3390/agriculture13010183








