AgroGenome: Interactive Genomic-Based Web Server Developed Based on Data Collected for Accessions Stored in Polish Genebank
Abstract
:1. Introduction
2. Materials and Methods
2.1. AgroGenome Portal Application Architecture
2.2. Collecting Data to Develop AgroGenome portal
2.2.1. Plant Material
2.2.2. Passport Data
2.2.3. Phenotypic Data
- Descriptors used for barley:
- Descriptors used for wheat: https://old.vurv.cz/Ewdb/asp/IPGRI_descr_1985.pdf (accessed on 29 October 2021);
- Descriptors used for soybean: https://www.bioversityinternational.org/fileadmin/_migrated/uploads/tx_news/Descriptors_for_soyabean_252.pdf (accessed on 29 October 2021).
2.2.4. Molecular Data Using DArTseq and GWAS Analysis
- A.
- DNA extraction and quantification
- B.
- Genotyping using DArTseq and GWAS analysis
- B.1.
- Data Filtering ProcessGenotypes were genotyped by Diversity Arrays Technology Pty Ltd, Building 3, Level D, University of Canberra, Monana Street, Bruce, ACT, 2617, Australia, using DArTseq [30]. SNP calls were made against: Hordeum vulgare Morex v2, T. aestivum Chinese Spring (CS) IWGSC RefSeq v1.0 (https://wheat-urgi.versailles.inra.fr/Seq-Repository/Assemblies - accessed on 8 September 2021) and soybean available in phytozome (https://phytozome-next.jgi.doe.gov/- acessed on 10 October 2021) [47].DArT data were handled in the same manner for all crops. That is, we used the DArTR v1.1.11 package [47] in the R programming language. SNPs and genotypes were removed if SNP markers contained > 5% missing data and genotypes contained > 10% missing data. SNPs with a reproducibility score of (RepAvg) < 100% were removed. Where SNPs originated from the same fragment, a random SNP was retained while the others were discarded. Noninformative monomorphic SNPs were removed, as were rare SNPs with a minor allele frequency of <1%.
- B.2.
- Genome-wide association studies (GWAS)GWAS analysis was conducted using the GAPIT v2018.08.18 R package [48,49]. We used the recently developed Bayesian-information and Linkage-disequilibrium Iteratively Nested Keyway (BLINK) model, which has been shown to produce fewer false positives, identify more true positives and scale to very large data sets [50]. Physical genome positions of markers were derived from the DArTseq SNP genotype file. Since GAPIT can only handle complete data, only markers with a physical position on one of the chromosomes and zero missing data were used as input to the GWAS analysis. Bonferroni and FDR thresholds were used. DArTseq markers with FDR and Bonferroni p = 0.01 thresholds were taken as significantly associated with the evaluated trait. In order to show the distribution of SNPs over the chromosome, Manhattan plots were also generated. The significance levels for GWAS analysis on the Manhattan plots were as follows: solid line represented the Bonferroni FDR multiple test threshold (p = 0.01), and dashed green line represented the FDR threshold (FDR adjusted ≤ 0.05). In order to show the distribution of SNPs over the chromosome, Manhattan plots were also generated.
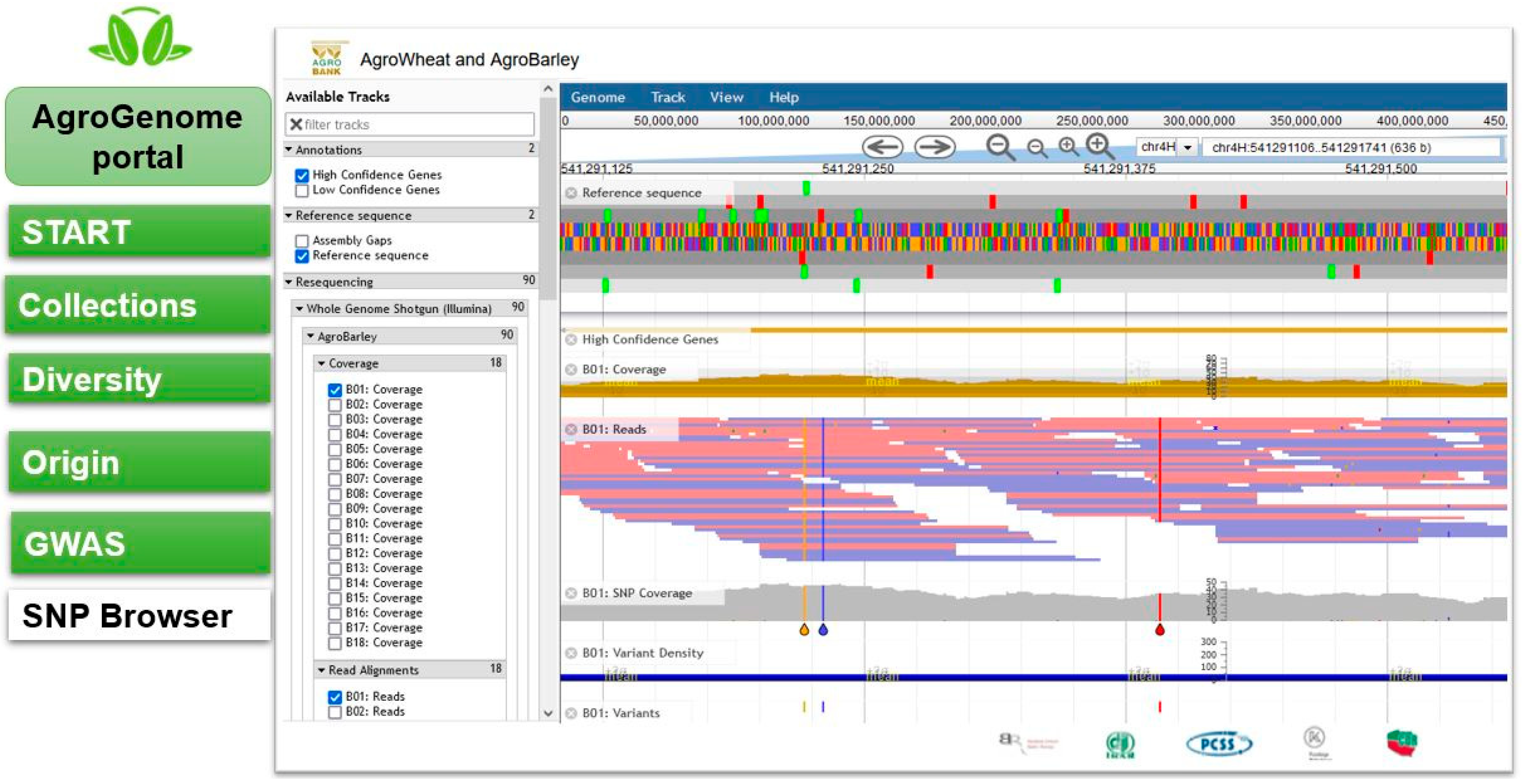
2.3. SNP Browser
2.3.1. Barley SNP Browser
- A.
- Barley Genotype Selection for WGSDistance matrices, provided by Diversity Arrays, were partitioned (clustered) into k clusters around medoids using the pam () method available in the cluster package [51,52] and performed in the R statistical programming language [53]. The pam-algorithm searchers for k representative genotypes/clusters were used such that the sum of dissimilarities between genotypes in a cluster and its representative genotype was minimized. Therefore, the number of clusters, k, was set to 18, the number of genotypes to be selected for whole-genome sequencing (WGS). Some selections were made to ensure a preference for Polish genotypes if one was close to the medoid genotype.
- B.
- Sequencing and read processingSequencing of full genomes of 18 spring barley accessions using the NGS method was carried out using the newest NovaSeq600 platform (Illumina), generating 2 × 150 PE reads. Raw reads were preprocessed through trimmomatic v0.39 (http://www.usadellab.org/cms/?page=trimmomatica, accessed on 10 October 2021) to remove adapters, low-quality bases and short reads. Specifically, the following command line argument was used: ILLUMINACLIP:TruSeq3-PE.fa:2:30:10:3:true LEADING:2 TRAILING:2 SLIDINGWINDOW:4:15 MINLEN:36.Reads were processed using the approach described in Watson-Haigh et al. [43]. Briefly, QC reads were aligned to the Barley Morex v2 genome assembly [54] using Minimap v2.17 [55], and variants were called using a SAMtools v1.9 [56] and BCFtool v1.9 calling pipeline, which required a minimum mapping quality of 20 and minimum base call quality of 30. Processing was parallelized per chromosome to facilitate timely completion of the analysis. Read alignment coverage and variant density (variants per 10 kbp) files were generated in BigWig format.All data have been made available as visualization tracks within a JBrowse [57] instance (http://62.3.171.115/jbrowse/?data=data%2Fbarley_morex_v2, accessed on 10 October 2021).
2.3.2. Wheat SNP Browser
- A.
- Genotype Selection for WESDistance matrices, provided by Diversity Arrays, were partitioned (clustered) into k clusters around medoids using the pam () method available in the cluster package [51,52] and performed in the R statistical programming language [55]. The pam-algorithm searchers for k representative genotypes/clusters were used such that the sum of dissimilarities between genotypes in a cluster and its representative genotype was minimized. Therefore, the number of clusters, k, was set to 48, the number of genotypes to be selected for whole-exome sequencing (WES). Some selections were made to ensure a preference for Polish genotypes if one was close to the medoid genotype.
- B.
- Sequencing and read processingThe selected genotypes based on the DArTseq data accessions were sequenced on an Illumina NovaSeq, generating 2 × 150 PE reads. Raw reads were preprocessed through trimmomatic v0.39 to remove adapters, low-quality bases and short reads. Specifically, the following command line argument was used: ILLUMINACLIP:TruSeq3-PE.fa:2:30:10:3:true LEADING:2 TRAILING:2 SLIDINGWINDOW:4:15 MINLEN:36.Reads were processed using the approach described in Watson-Haigh et al. [43]. Briefly, QC reads were aligned to the IWGSC RefSeq v1.0 genome assembly [58] using Minimap v2.17 [55], and variants were called using a SAMtools v1.9 [58] and BCFtool v1.9 calling pipeline, which required a minimum mapping quality of 20 and minimum base call quality of 30. Processing was parallelized per chromosome to facilitate timely completion of the analysis. Read alignment coverage and variant density (variants per 10 kbp) files were generated in BigWig format.All data have been made available as visualization tracks within a JBrowse [57] instance (http://62.3.171.115/jbrowse/?data=data%2Fwheat_CS_v1.0, accessed on 10 October 2021).
2.4. Collecting DNA Samples for Genebank
2.5. Collecting Reference Materials for Herbarium and Photo Documentation
3. Results
3.1. AgroGenome Portal Summary Presentations
3.2. AgroGenome Passport Data Presentation
3.3. AgroGenome GWAS Results Presentation
3.4. AgroGenome SNP Browser Presentation
4. Discussion
5. Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weise, S.; Lohwasser, U.; Oppermann, M. Document or Lose It—On the Importance of Information Management for Genetic Resources Conservation in Genebanks. Plants 2020, 9, 1050. [Google Scholar] [CrossRef] [PubMed]
- Commission on Genetic Resources for Food and Agriculture Food and Agriculture Organization of the United Nations. The Second Report on the State of the World’s Animal Genetic Resources for Food and Agriculture; FAO: Rome, Italy, 2007; ISBN 978-92-5-108820-3. [Google Scholar]
- Jia, J.; Li, H.; Zhang, X.; Li, Z.; Qiu, L. Genomics-based plant germplasm research (GPGR). Crop. J. 2017, 5, 166–174. [Google Scholar] [CrossRef]
- Diez, M.J.; De La Rosa, L.; Martín, I.; Guasch, L.; Cartea, M.E.; Mallor, C.; Casals, J.; Simó, J.; Rivera, A.; Anastasio, G.; et al. Plant Genebanks: Present Situation and Proposals for Their Improvement. the Case of the Spanish Network. Front. Plant Sci. 2018, 9, 1794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milner, S.G.; Jost, M.; Taketa, S.; Mazón, E.R.; Himmelbach, A.; Oppermann, M.; Weise, S.; Knüpffer, H.; Basterrechea, M.; König, P.; et al. Genebank genomics highlights the diversity of a global barley collection. Nat. Genet. 2018, 51, 319–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czembor, J.H.; Gryziak, G.; Zaczyński, M.; Wlodarczyk, S.; Podyma, W. Gromadzenie i zachowanie zasobów genowych roślin użytkowych w Polsce 2015–2017. Biul. IHAR 2018, 15–16. [Google Scholar]
- Czembor, J.H.; Gryziak, G.; Zaczyński, M.; Puchta, M.; Czembor, E. Gromadzenie i zachowanie zasobów genowych roślin użytkowych w Polsce—Artykuł przeglądowy Część 1. Gromadzenie zasobów genowych roślin użytkowych w trakcie ekspedycji krajowych i zagranicznych. Agron. Sci. 2018, 72, 135–146. [Google Scholar] [CrossRef]
- Czembor, J.H.; Gryziak, G.; Zaczyński, M.; Puchta, M.; Czembor, E. Gromadzenie i zachowanie zasobów genowych roślin użytkowych w Polsce—Artykuł przeglądowy Część 2. Przechowywanie zasobów genowych w formie nasion, prowadzenie herbarium, baz danych i udostępnianie zasobów genowych. Agron. Sci. 2018, 72, 147–154. [Google Scholar] [CrossRef]
- Purugganan, M.D.; Fuller, D.Q. The nature of selection during plant domestication. Nature 2009, 457, 843–848. [Google Scholar] [CrossRef]
- Ingvordsen, C.H. Climate Change Effects on Plant Ecosystems—Genetic Resources for Future Barley Breeding. Ph.D. Thesis, Technical University of Denmark (DTU), Lyngby, Denmark, April 2014. [Google Scholar]
- Nguyen, G.N.; Norton, S.L. Genebank Phenomics: A Strategic Approach to Enhance Value and Utilization of Crop Germplasm. Plants 2020, 9, 817. [Google Scholar] [CrossRef]
- Marone, D.; Russo, M.; Mores, A.; Ficco, D.; Laidò, G.; Mastrangelo, A.; Borrelli, G. Importance of Landraces in Cereal Breeding for Stress Tolerance. Plants 2021, 10, 1267. [Google Scholar] [CrossRef]
- Ansaldi, B.H.; Franks, S.J.; Weber, J.J. The influence of environmental factors on breeding system allocation at large spatial scales. AoB PLANTS 2018, 10, ply069. [Google Scholar] [CrossRef]
- Singh, D.; Ziems, L.A.; Dracatos, P.M.; Pourkheirandish, M.; Tshewang, S.; Czembor, P.; German, S.; Fowler, R.A.; Snyman, L.; Platz, G.J.; et al. Genome-wide association studies provide insights on genetic architecture of resistance to leaf rust in a worldwide barley collection. Mol. Breed. 2018, 38, 43. [Google Scholar] [CrossRef]
- Hickey, L.T.; Hafeez, A.N.; Robinson, H.; Jackson, S.A.; Leal-Bertioli, S.C.M.; Tester, M.; Gao, C.; Godwin, I.D.; Hayes, B.J.; Wulff, B.B.H. Breeding crops to feed 10 billion. Nat. Biotechnol. 2019, 37, 744–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piechota, U.; Czembor, P.C.; Słowacki, P.; Czembor, J.H. Identifying a novel powdery mildew resistance gene in a barley landrace from Morocco. J. Appl. Genet. 2019, 60, 243–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rebetzke, G.; Jimenez-Berni, J.A.; Fischer, R.; Deery, D.; Smith, D. Review: High-throughput phenotyping to enhance the use of crop genetic resources. Plant Sci. 2019, 282, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Volk, G.M.; Byrne, P.F.; Coyne, C.J.; Flint-Garcia, S.; Reeves, P.A.; Richards, C. Integrating Genomic and Phenomic Approaches to Support Plant Genetic Resources Conservation and Use. Plants 2021, 10, 2260. [Google Scholar] [CrossRef]
- Czembor, E.; Czembor, J.H.; Suchecki, R.; Watson-Haigh, N.S. DArT-based evaluation of soybean germplasm from Polish Gene Bank. BMC Res. Notes 2021, 14, 343. [Google Scholar] [CrossRef]
- Czembor, J.H.; Czembor, E.; Suchecki, R.; Watson-Haigh, N.S. Genome-Wide Association Study for Powdery Mildew and Rusts Adult Plant Resistance in European Spring Barley from Polish Gene Bank. Agronomy 2021, 12, 7. [Google Scholar] [CrossRef]
- Czembor, J.H.; Czembor, E. Genome-Wide Association Study of Agronomic Traits in European Spring Barley from Polish Gene Bank. Agronomy 2022, 12, 2135. [Google Scholar] [CrossRef]
- Smykal, P.; Aubert, G.; Burstin, J.; Coyne, C.J.; Ellis, N.T.H.; Flavell, A.J.; Ford, R.; Hýbl, M.; Macas, J.; Neumann, P.; et al. Pea (Pisum sativum L.) in the Genomic Era. Agronomy 2012, 2, 74–115. [Google Scholar] [CrossRef]
- Gilliham, M.; Able, J.A.; Roy, S.J. Translating knowledge about abiotic stress tolerance to breeding programmes. Plant J. 2017, 90, 898–917. [Google Scholar] [CrossRef] [PubMed]
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.D.; Schroeder, J.I. Genetic strategies for improving crop yields. Nature 2019, 575, 109–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raggi, L.; Caproni, L.; Negri, V. Landrace added value and accessibility in Europe: What a collection of case studies tells us. Biodivers. Conserv. 2021, 30, 1031–1048. [Google Scholar] [CrossRef]
- Cobb, J.N.; Biswas, P.S.; Platten, J.D. Back to the future: Revisiting MAS as a tool for modern plant breeding. Theor. Appl. Genet. 2018, 132, 647–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, A.; Verma, R.P.S.; Singh, A.; Sharma, H.K.; Devi, G. Barley landraces: Ecological heritage for edaphic stress adaptations and sustainable production. Environ. Sustain. Indic. 2020, 6, 100035. [Google Scholar] [CrossRef]
- Varshney, R.K.; Singh, V.K.; Hickey, J.M.; Xun, X.; Marshall, D.F.; Wang, J.; Edwards, D.; Ribau, J.-M. Analytical and Decision Support Tools for Genomics-Assisted Breeding. Trends Plant Sci. 2016, 4, 354–363. [Google Scholar] [CrossRef] [Green Version]
- van Bemmelen van der Plaat, A.; van Treuren, R.; van Hintum, T.J.L. Reliable genomic strategies for species classification of plant genetic resources. BMC Bioinform. 2021, 22, 173. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A Robust, Simple Genotyping-by-Sequencing (GBS) Approach for High Diversity Species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Zhang, X.; Li, H.; Zheng, H.; Zhang, J.; Olsen, M.S.; Varshney, R.K.; Prasanna, B.M.; Qian, Q. Smart breeding driven by big data, artificial intelligence, and integrated genomic-enviromic prediction. Mol. Plant 2022, 15, 1664–1695. [Google Scholar] [CrossRef]
- Schaid, D.J.; Chen, W.; Larson, N.B. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat. Rev. Genet. 2018, 19, 491–504. [Google Scholar] [CrossRef]
- Varshney, R.K.; Roorkiwal, M.; Sorrells, M.E.; Molecular, N.; Strategies, B. Genomic Selection for Crop Improvement; Springer: Cham, Switzerland, 2017; ISBN 9783319631684. [Google Scholar]
- Wenzl, P.; Raman, H.; Wang, J.; Zhou, M.; Huttner, E.; Kilian, A. A DArT platform for quantitative bulked segregant analysis. BMC Genom. 2007, 8, 196. [Google Scholar] [CrossRef] [PubMed]
- Uffelmann, E.; Huang, Q.Q.; Munung, N.S.; de Vries, J.; Okada, Y.; Martin, A.R.; Martin, H.C.; Lappalainen, T.; Posthuma, D. Genome-wide association studies. Nat. Rev. Methods Prim. 2021, 1, 59. [Google Scholar] [CrossRef]
- Desgroux, A.; L’Anthoëne, V.; Roux-Duparque, M.; Rivière, J.-P.; Aubert, G.; Tayeh, N.; Moussart, A.; Mangin, P.; Vetel, P.; Piriou, C.; et al. Genome-wide association mapping of partial resistance to Aphanomyces euteiches in pea. BMC Genom. 2016, 17, 124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gali, K.K.; Sackville, A.; Tafesse, E.G.; Lachagari, V.R.; McPhee, K.; Hybl, M.; Mikić, A.; Smýkal, P.; McGee, R.; Burstin, J.; et al. Genome-Wide Association Mapping for Agronomic and Seed Quality Traits of Field Pea (Pisum sativum L.). Front. Plant Sci. 2019, 10, 1538. [Google Scholar] [CrossRef]
- Tsai, H.-Y.; Janss, L.L.; Andersen, J.R.; Orabi, J.; Jensen, J.D.; Jahoor, A.; Jensen, J. Genomic prediction and GWAS of yield, quality and disease-related traits in spring barley and winter wheat. Sci. Rep. 2020, 10, 3347. [Google Scholar] [CrossRef] [Green Version]
- Kilian, A.; Huttner, E.; Wenzl, P.; Jaccoud, D.; Carling, J.; Caig, V.; Evers, M.; Heller-Uszynska, K.; Uszynski, G.; Cayla, C.; et al. The fast and the cheap: SNP and DArT-based whole genome profiling for crop improvement. In The Wake of the Double Helix: From the Green Revolution to the Gene Revolution, Proceedings of the International Congress, Bologna, Italy, 27–31 May 2003; Tuberosa, R., Phillips, R.L., Gale, M., Eds.; Avenue Media: Bologna, Italy, 2005; pp. 443–461. [Google Scholar]
- Brown, H.E.; Huth, N.I.; Holzworth, D.P.; Teixeira, E.I.; Zyskowski, R.F.; Hargreaves, J.N.; Moot, D.J. Plant Modelling Framework: Software for building and running crop models on the APSIM platform. Environ. Model. Softw. 2014, 62, 385–398. [Google Scholar] [CrossRef] [Green Version]
- König, P.; Beier, S.; Basterrechea, M.; Schüler, D.; Arend, D.; Mascher, M.; Stein, N.; Scholz, U.; Lange, M. BRIDGE—A Visual Analytics Web Tool for Barley Genebank Genomics. Front. Plant Sci. 2020, 11, 701. [Google Scholar] [CrossRef]
- Wang, W.; Wang, Z.; Li, X.; Ni, Z.; Hu, Z.; Xin, M.; Peng, H.; Yao, Y.; Sun, Q.; Guo, W. SnpHub: An easy-to-set-up web server framework for exploring large-scale genomic variation data in the post-genomic era with applications in wheat. Gigascience 2020, 9, giaa060. [Google Scholar] [CrossRef]
- Watson-haigh, N.S.; Suchecki, R.; Kalashyan, E.; Garcia, M.; Baumann, U. DAWN: A resource for yielding in-sights into the diversity among wheat genomes. BMC Genom. 2018, 19, 941. [Google Scholar] [CrossRef] [Green Version]
- Qiu, L.-J.; Xing, L.-L.; Guo, Y.; Wang, J.; Jackson, S.A.; Chang, R.-Z. A platform for soybean molecular breeding: The utilization of core collections for food security. Plant Mol. Biol. 2013, 83, 41–50. [Google Scholar] [CrossRef] [Green Version]
- Grant, D.; Nelson, R.T.; Cannon, S.B.; Shoemaker, R.C. SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res. 2009, 38, D843–D846. [Google Scholar] [CrossRef] [PubMed]
- Doyle, A.; Doyle, J.L. Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zhang, Z. GAPIT Version 3: Boosting Power and Accuracy for Genomic Association and Prediction. Genom. Proteom. Bioinform. 2021, 19, 629–640. [Google Scholar] [CrossRef]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.; Liu, X.; Zhou, Y.; Summers, R.M.; Zhang, Z. BLINK: A package for the next level of genome-wide association studies with both individuals and markers in the millions. Gigascience 2018, 8, giy154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reynolds, A.P.; Richards, G.; de la Iglesia, B.; Rayward-Smith, V.J. Clustering Rules: A Comparison of Partitioning and Hierarchical Clustering Algorithms. J. Math. Model. Algorithms 2006, 5, 475–504. [Google Scholar] [CrossRef]
- Schubert, E.; Rousseeuw, P.J. Faster k-Medoids Clustering: Improving the PAM, CLARA, and CLARANS Algorithms. In Similarity Search and Applications, Proceedings of the 12th International Conference, SISAP 2019, Newark, NJ, USA, 2–4 October 2019; Springer: Cham, Switzerland, 2019; pp. 171–187. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. A Languanguage and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: www.r-project.org/index.html (accessed on 15 November 2022).
- Monat, C.; Padmarasu, S.; Lux, T.; Wicker, T.; Gundlach, H.; Himmelbach, A.; Ens, J.; Li, C.; Muehlbauer, G.J.; Schulman, A.H.; et al. TRITEX: Chromosome-scale sequence assembly of Triticeae genomes with open-source tools. Genome Biol. 2019, 20, 284. [Google Scholar] [CrossRef] [Green Version]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Buels, R.; Yao, E.; Diesh, C.M.; Hayes, R.D.; Munoz-Torres, M.; Helt, G.; Goodstein, D.M.; Elsik, C.G.; Lewis, S.E.; Stein, L.; et al. JBrowse: A dynamic web platform for genome visualization and analysis. Genome Biol. 2016, 17, 66. [Google Scholar] [CrossRef]
- The International Wheat Genome Sequencing Consortium (IWGSC); Appels, R.; Eversole, K.; Feuillet, C.; Keller, B.; Rogers, J.; Stein, N.; Pozniak, C.J.; Stein, N.; Choulet, F.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Jiang, Y.; Wang, Z.; Gou, Z.; Lyu, J.; Li, W.; Yu, Y.; Shu, L.; Zhao, Y.; Ma, Y.; et al. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 2015, 33, 408–414. [Google Scholar] [CrossRef] [Green Version]
- Chapman, J.A.; Mascher, M.; Buluç, A.; Barry, K.; Georganas, E.; Session, A.; Strnadova, V.; Jenkins, J.; Sehgal, S.; Oliker, L.; et al. A whole-genome shotgun approach for assembling and anchoring the hexaploid bread wheat genome. Genome Biol. 2015, 16, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, F.; Pasam, R.; Shi, F.; Kant, S.; Keeble-Gagnere, G.; Kay, P.; Forrest, K.; Fritz, A.; Hucl, P.; Wiebe, K.; et al. Exome sequencing highlights the role of wild-relative introgression in shaping the adaptive landscape of the wheat genome. Nat. Genet. 2019, 51, 896–904. [Google Scholar] [CrossRef]
- Cheng, H.; Liu, J.; Wen, J.; Nie, X.; Xu, L.; Chen, N.; Li, Z.; Wang, Q.; Zheng, Z.; Li, M.; et al. Frequent intra- and inter-species introgression shapes the landscape of genetic variation in bread wheat. Genome Biol. 2019, 20, 136. [Google Scholar] [CrossRef] [PubMed]




| Common Name | Species | Accessions Number | |
|---|---|---|---|
| Total | Polish Origin | ||
| Barley | Hordeum vulgare L. | 461 | 146 |
| Common wheat | Triticum aestivum L. | 428 | 118 |
| Durum and dicoccum wheat | T. dicoccum (Schrank) Schuebl., T. durum Desf. | 75 | 11 |
| Soybean | Glycine max. | 196 | 80 |
| Pea | Pisum sativum L. | 184 | 115 |
| Total | 1344 | 470 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Czembor, J.H.; Czembor, E.; Krystek, M.; Pukacki, J. AgroGenome: Interactive Genomic-Based Web Server Developed Based on Data Collected for Accessions Stored in Polish Genebank. Agriculture 2023, 13, 193. https://doi.org/10.3390/agriculture13010193
Czembor JH, Czembor E, Krystek M, Pukacki J. AgroGenome: Interactive Genomic-Based Web Server Developed Based on Data Collected for Accessions Stored in Polish Genebank. Agriculture. 2023; 13(1):193. https://doi.org/10.3390/agriculture13010193
Chicago/Turabian StyleCzembor, Jerzy H., Elzbieta Czembor, Marcin Krystek, and Juliusz Pukacki. 2023. "AgroGenome: Interactive Genomic-Based Web Server Developed Based on Data Collected for Accessions Stored in Polish Genebank" Agriculture 13, no. 1: 193. https://doi.org/10.3390/agriculture13010193
APA StyleCzembor, J. H., Czembor, E., Krystek, M., & Pukacki, J. (2023). AgroGenome: Interactive Genomic-Based Web Server Developed Based on Data Collected for Accessions Stored in Polish Genebank. Agriculture, 13(1), 193. https://doi.org/10.3390/agriculture13010193









