Molecular Characterization of Three Chemosensory Proteins from Carposina sasakii
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insect Rearing and Sample Collection
2.2. Total RNA Extraction and cDNA Synthesis
2.3. cDNA Cloning and Sequence Analysis
2.4. Expression Levels of Three CSPs mRNA from C. sasakii
2.5. Expression and Purification of Recombinant CsasCSPs Proteins
2.6. Fluorescence Competitive Binding Assay
3. Results
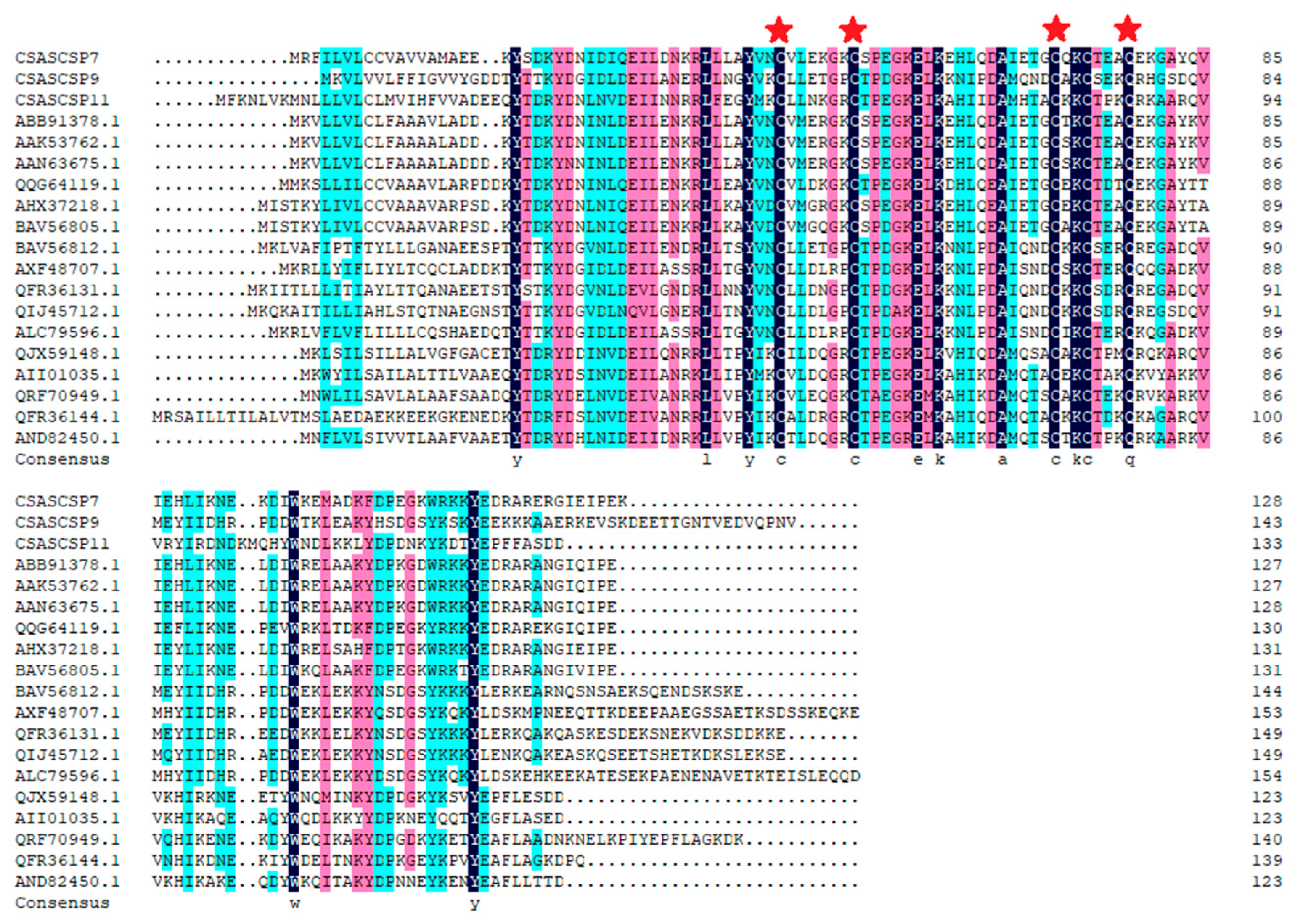
3.1. Sequence Analysis of CsasCSPs
3.2. Tissue Expression Patterns of CsasCSPs
3.3. Bacterial Expression and Purification of CsasCSPs
3.4. Binding Properties of Recombinant CsasCSPs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, Y.P.; Zhao, J.L.; Su, T.J.; He, Q.S.; Xie, J.L.; Zhu, C.D. The complete mitochondrial genome of Carposina sasakii (Lepidoptera: Carposinidae). Mitochondrial DNA 2016, 27, 1432–1434. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.J.; Song, W.; Yue, L.; Guo, S.K.; Chen, J.C.; Gong, Y.J.; Hoffmann, A.A.; Wei, S.J. Chromosome-level genome of the peach fruit moth Carposina sasakii (Lepidoptera: Carposinidae) provides a resource for evolutionary studies on moths. Mol. Ecol. Resour. 2021, 21, 834–848. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.Q.; Qiu, G.S.; Li, Y.Y.; Zhang, H.J.; Yan, W.T.; Yue, Q.; Sun, L. N Molecular characterization and functional analysis of pheromone binding proteins and general odorant binding proteins from Carposina sasakii matsumura (Lepidoptera: Carposinidae). Pest. Manag. Sci. 2019, 75, 234–245. [Google Scholar] [CrossRef] [PubMed]
- Han, K.S.; Jin, K.J.; Choi, K.H.; Sun, W.L.; Boo, K.S. Sex pheromone composition and male trapping of the peach fruit moth, Carposina sasakii (matsumura) (Lepidoptera: Carposinidae) in korea. J. Asia-Pac. Entomol. 2000, 3, 83–88. [Google Scholar] [CrossRef]
- Gadenne, C.; Barrozo, R.B.; Anton, S. Plasticity in insect olfaction: To smell or not to smell? Annu. Rev. Entomol. 2016, 61, 317–333. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Liu, Y.; Wang, B.; Wang, G.R. Research status, opportunities and challenges of pest olfactory behavior regulation technology. Chin. Sci. Found. 2020, 4, 441–446. (In Chinese) [Google Scholar]
- Brezolin, A.N.; Martinazzo, J.; Muenchen, D.K.; De Cezaro, A.M.; Rigo, A.A.; Steffens, C.; Steffens, J.; Blassioli-Moraes, M.C.; Borges, M. Tools for detecting insect semiochemicals: A review. Anal. Bioanal. Chem. 2018, 410, 4091–4108. [Google Scholar] [CrossRef]
- Kalske, A.; Shiojiri, K.; Uesugi, A.; Sakata, Y.; Morrell, K.; Kessler, A. 2019. Insect herbivory selects for volatile-mediated plant-plant communication. Curr. Biol. 2005, 29, 3128–3133. [Google Scholar] [CrossRef]
- Xu, H.; Turlings, T.C.J. Plant volatiles as mate-finding cues for insects. Trends Plant Sci. 2018, 23, 100–111. [Google Scholar] [CrossRef]
- Bruce, T.J.; Pickett, J.A. Perception of plant volatile blends by herbivorous insects--finding the right mix. Phytochemistry 2011, 72, 1605–1611. [Google Scholar] [CrossRef]
- Koutroumpa, F.A.; Jacquin-Joly, E. Sex in the night: Fatty acid-derived sex pheromones and corresponding membrane pheromone receptors in insects. Biochimie 2014, 107, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Regnier, F.E.; Law, J.H. Insect pheromones. J. Lipid Res. 1968, 9, 541–551. [Google Scholar] [CrossRef]
- Gregg, P.C.; Del Socorro, A.P.; Landolt, P.J. Advances in attract-and-kill for agricultural pests: Beyond pheromones. Annu. Rev. Entomol. 2018, 63, 453–470. [Google Scholar] [CrossRef] [PubMed]
- Ruther, J.; Reinecke, A.; Tolasch, T.; Hilker, M. Make love not war: A common arthropod defence compound as sex pheromone in the forest cockchafer Melolontha hippocastani. Oecologia 2001, 128, 44–47. [Google Scholar] [CrossRef] [PubMed]
- McKenna, M.P.; Hekmat-Scafe, D.S.; Gaines, P.; Carlson, J.R. Putative drosophila pheromone-binding proteins expressed in a subregion of the olfactory system. J. Biol. Chem. 1994, 269, 16340–16347. [Google Scholar] [CrossRef]
- Pikielny, C.W.; Hasan, G.; Rouyer, F.; Rosbash, M. Members of a family of drosophila putative odorant-binding proteins are expressed in different subsets of olfactory hairs. Neuron 1994, 12, 35–49. [Google Scholar] [CrossRef]
- Mameli, M.; Tuccini, A.; Mazza, M.; Petacchi, R.; Pelosi, P. Soluble proteins in chemosensory organs of phasmids. Insect Biochem. Mol. Biol. 1996, 26, 875–882. [Google Scholar] [CrossRef]
- Tuccini, A.; Maida, R.; Rovero, P.; Mazza, M.; Pelosi, P. Putative odorant-binding protein in antennae and legs of Carausius morosus (insecta, phasmatodea). Insect Biochem. Mol. Biol. 1996, 26, 19–24. [Google Scholar] [CrossRef]
- Maleszka, R.; Stange, G. Molecular cloning, by a novel approach, of a cdna encoding a putative olfactory protein in the labial palps of the moth Cactoblastis cactorum. Gene 1997, 202, 39–43. [Google Scholar] [CrossRef]
- Biessmann, H.; Nguyen, Q.K.; Le, D.; Walter, M.F. Microarray-based survey of a subset of putative olfactory genes in the mosquito Anopheles gambiae. Insect Mol. Biol. 2005, 14, 575–589. [Google Scholar] [CrossRef]
- Biessmann, H.; Walter, M.F.; Dimitratos, S.; Woods, D. Isolation of cdna clones encoding putative odourant binding proteins from the antennae of the malaria-transmitting mosquito, Anopheles gambiae. Insect Mol. Biol. 2002, 11, 123–132. [Google Scholar] [CrossRef]
- Angeli, S.; Ceron, F.; Scaloni, A.; Monti, M.; Monteforti, G.; Minnocci, A.; Petacchi, R.; Pelosi, P. Purification, structural characterization, cloning and immunocytochemical localization of chemoreception proteins from Schistocerca gregaria. Eur. J. Biochem. 1999, 262, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, P.; Zhou, J.J.; Ban, L.P.; Calvello, M. Soluble proteins in insect chemical communication. Cell. Mol. Life Sci. 2006, 63, 1658–1676. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, P.; Iovinella, I.; Zhu, J.; Wang, G.; Dani, F.R. Beyond chemoreception: Diverse tasks of soluble olfactory proteins in insects. Biol. Rev. Camb. Philos. Soc. 2018, 93, 184–200. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.Y.; Zhang, N.X.; Shu, Z.Q.; Huang, K.X. Observation of mating and egg-laying habits of adult worms (carposina niponensis wal.). Insect Knowl. 1964, 6, 271–273. (In Chinese) [Google Scholar]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Younas, A.; Waris, M.I.; Shaaban, M.; Ul Qamar, M.T.; Wang, M.Q. Appraisal of MsepCSP14 for chemosensory functions in Mythimna separata. Insect Sci. 2022, 29, 162–176. [Google Scholar] [CrossRef]
- Zhang, Z.K.; Lei, Z.R. Identification, expression profiling and fluorescence-based binding assays of a chemosensory protein gene from the western flower thrips, Frankliniella occidentalis. PLoS ONE 2015, 10, e0117726. [Google Scholar] [CrossRef]
- Zhang, C.; Tang, B.; Zhou, T.; Yu, X.; Hu, M.; Dai, W. Involvement of chemosensory protein bodocsp1 in perception of host plant volatiles in Bradysia odoriphaga. J. Agric. Food Chem. 2021, 69, 10797–10806. [Google Scholar] [CrossRef]
- Li, C.; Sun, K.; Li, D.; Liu, D. Functional characterization of chemosensory protein AmalCSP5 from apple buprestid beetle, Agrilus mali (Coleoptera: Buprestidae). J. Econ. Entomol. 2021, 114, 348–359. [Google Scholar] [CrossRef]
- Yi, X.; Qi, J.; Zhou, X.; Hu, M.Y.; Zhong, G.H. Differential expression of chemosensory-protein genes in midguts in response to diet of Spodoptera litura. Sci. Rep. 2017, 7, 296. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Wang, S.N.; Li, K.M.; Liu, J.T.; Zheng, Y.; Shan, S.; Yang, Y.Q.; Li, R.J.; Zhang, Y.J.; Guo, Y.Y. Identification of odorant binding proteins and chemosensory proteins in Microplitis mediator as well as functional characterization of chemosensory protein 3. PLoS ONE 2017, 12, e0180775. [Google Scholar] [CrossRef] [PubMed]
- Hua, J.F.; Zhang, S.; Cui, J.J.; Wang, D.J.; Wang, C.Y.; Luo, J.Y.; Lv, L.M.; Ma, Y. Functional characterizations of one odorant binding protein and three chemosensory proteins from Apolygus lucorum (meyer-dur) (Hemiptera: Miridae) legs. J. Insect Physiol. 2013, 59, 690–696. [Google Scholar] [CrossRef] [PubMed]
- Waris, M.I.; Younas, A.; Ameen, A.; Rasool, F.; Wang, M.Q. Expression profiles and biochemical analysis of chemosensory protein 3 from Nilaparvata lugens (Hemiptera: Delphacidae). J. Chem. Ecol. 2020, 46, 363–377. [Google Scholar] [CrossRef]
- Jacquin-Joly, E.; Vogt, R.G.; François, M.C.; Nagnan-Le Meillour, P. Functional and expression pattern analysis of chemosensory proteins expressed in antennae and pheromonal gland of Mamestra brassicae. Chem. Senses 2001, 26, 833–844. [Google Scholar] [CrossRef]
- Nagnan-Le Meillour, P.; Cain, A.H.; Jacquin-Joly, E.; François, M.C.; Ramachandran, S.; Maida, R.; Steinbrecht, R.A. Chemosensory proteins from the proboscis of Mamestra brassicae. Chem. Senses 2000, 25, 541–553. [Google Scholar] [CrossRef]
- Zhou, X.H.; Ban, L.P.; Iovinella, I.; Zhao, L.J.; Gao, Q.; Felicioli, A.; Sagona, S.; Pieraccini, G.; Pelosi, P.; Zhang, L.; et al. Diversity, abundance, and sex-specific expression of chemosensory proteins in the reproductive organs of the locust Locusta migratoria manilensis. Biol. Chem. 2013, 394, 43–54. [Google Scholar] [CrossRef]
- Baer, B.; Zareie, R.; Paynter, E.; Poland, V.; Millar, A.H. Seminal fluid proteins differ in abundance between genetic lineages of honeybees. J. Proteom. 2012, 75, 5646–5653. [Google Scholar] [CrossRef]
- Kitabayashi, A.N.; Arai, T.; Kubo, T.; Natori, S. Molecular cloning of cdna for p10, a novel protein that increases in the regenerating legs of periplaneta americana (American cockroach). Insect Biochem. Mol. Biol. 1998, 28, 785–790. [Google Scholar] [CrossRef]
- Nomura, A.; Kawasaki, K.; Kubo, T.; Natori, S. Purification and localization of p10, a novel protein that increases in nymphal regenerating legs of periplaneta americana (American cockroach). Int. J. Dev. Biol. 1992, 36, 391–398. [Google Scholar]
- Forêt, S.; Wanner, K.W.; Maleszka, R. Chemosensory proteins in the honey bee: Insights from the annotated genome, comparative analyses and expressional profiling. Insect Biochem. Mol. Biol. 2007, 37, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Maleszka, J.; Forêt, S.; Saint, R.; Maleszka, R. Rnai-induced phenotypes suggest a novel role for a chemosensory protein csp5 in the development of embryonic integument in the honeybee (Apis mellifera). Dev. Genes. Evol. 2007, 217, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.; Lu, Y.; Zeng, L.; Liang, G.; He, X. Si-csp9 regulates the integument and moulting process of larvae in the red imported fire ant, Solenopsis invicta. Sci. Rep. 2015, 5, 9245. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.L.; Guo, H.; Huang, L.Q.; Pelosi, P.; Wang, C.Z. Unique function of a chemosensory protein in the proboscis of two helicoverpa species. J. Exp. Biol. 2014, 217, 1821–1826. [Google Scholar] [CrossRef]
- Jiang, X.; Xu, H.; Zheng, N.; Yin, X.; Zhang, L. A chemosensory protein detects antifeedant in locust (Locusta migratoria). Insects 2020, 12, 1. [Google Scholar] [CrossRef]
- Zeng, Y.; Merchant, A.; Wu, Q.; Wang, S.; Kong, L.; Zhou, X.; Xie, W.; Zhang, Y. A chemosensory protein btabcsp11 mediates reproduction in Bemisia tabaci. Front. Physiol. 2020, 11, 709. [Google Scholar] [CrossRef]
- Younas, A.; Waris, M.I.; Chang, X.Q.; Shaaban, M.; Abdelnabby, H.; Ul Qamar, M.T.; Wang, M.Q. A chemosensory protein MsepCSP5 involved in chemoreception of oriental armyworm Mythimna separata. Int. J. Biol. Sci. 2018, 14, 1935–1949. [Google Scholar] [CrossRef]
- Ali, S.; Ahmed, M.Z.; Li, N.; Ali, S.A.I.; Wang, M.Q. Functional characteristics of chemosensory proteins in the sawyer beetle Monochamus alternatus hope. Bull. Entomol. Res. 2019, 109, 34–42. [Google Scholar] [CrossRef]
- Chénier, J.V.; Philogène, B.J. Field responses of certain forest coleoptera to conifer monoterpenes and ethanol. J. Chem. Ecol. 1989, 15, 1729–1745. [Google Scholar] [CrossRef]
- Hassaballa, I.B.; Torto, B.; Sole, C.L.; Tchouassi, D.P. Exploring the influence of different habitats and their volatile chemistry in modulating sand fly population structure in a leishmaniasis endemic foci, kenya. PLOS Neglected Trop. Dis. 2021, 15, e0009062. [Google Scholar] [CrossRef]
- Jumean, Z.; Gries, R.; Unruh, T.; Rowland, E.; Gries, G. Identification of the larval aggregation pheromone of codling moth, Cydia pomonella. J. Chem. Ecol. 2005, 31, 911–924. [Google Scholar] [CrossRef]
- Siljander, E.; Gries, R.; Khaskin, G.; Gries, G. Identification of the airborne aggregation pheromone of the common bed bug, cimex lectularius. J. Chem. Ecol. 2008, 34, 708–718. [Google Scholar] [CrossRef] [PubMed]
- Farré-Armengol, G.; Fernández-Martínez, M.; Filella, I.; Junker, R.R.; Peñuelas, J. Deciphering the biotic and climatic factors that influence floral scents: A systematic review of floral volatile emissions. Front. Plant Sci. 2020, 11, 1154. [Google Scholar] [CrossRef] [PubMed]
- Lohonyai, Z.; Vuts, J.; Kárpáti, Z.; Koczor, S.; Domingue, M.J.; Fail, J.; Birkett, M.A.; Tóth, M.; Imrei, Z. Benzaldehyde: An alfalfa-related compound for the spring attraction of the pest weevil Sitona humeralis (Coleoptera: Curculionidae). Pest. Manag. Sci. 2019, 75, 3153–3159. [Google Scholar] [CrossRef] [PubMed]
- De Fouchier, A.; Sun, X.; Caballero-Vidal, G.; Travaillard, S.; Jacquin-Joly, E.; Montagné, N. Behavioral effect of plant volatiles binding to Spodoptera littoralis larval odorant receptors. Front. Behav. Neurosci. 2018, 12, 264. [Google Scholar] [CrossRef] [PubMed]
- Pope, T.W.; Campbell, C.A.; Hardie, J.; Pickett, J.A.; Wadhams, L.J. Interactions between host-plant volatiles and the sex pheromones of the bird cherry-oat aphid, Rhopalosiphum padi and the damson-hop aphid. Phorodon humuli. J. Chem. Ecol. 2007, 33, 157–165. [Google Scholar] [CrossRef]
- Piñero, J.C.; Prokopy, R.J. Field evaluation of plant odor and pheromonal combinations for attracting plum curculios. J. Chem. Ecol. 2003, 29, 2735–2748. [Google Scholar] [CrossRef]
- Ullah, I.; Khan, A.L.; Ali, L.; Khan, A.R.; Waqas, M.; Hussain, J.; Lee, I.J.; Shin, J.H. Benzaldehyde as an insecticidal, antimicrobial, and antioxidant compound produced by photorhabdus temperata M1021. J. Microbiol. 2015, 53, 127–133. [Google Scholar] [CrossRef]
- Cooperband, M.F.; McElfresh, J.S.; Millar, J.G.; Cardé, R.T. Attraction of female Culex quinquefasciatus say (Diptera: Culicidae) to odors from chicken feces. J. Insect Physiol. 2008, 54, 1184–1192. [Google Scholar] [CrossRef]
- Bezerra-Silva, P.C.; Dutra, K.A.; Santos, G.K.; Silva, R.C.; Iulek, J.; Milet-Pinheiro, P.; Navarro, D.M. Evaluation of the activity of the essential oil from an ornamental flower against Aedes aegypti: Electrophysiology, molecular dynamics and behavioral assays. PLoS ONE 2016, 11, e0150008. [Google Scholar] [CrossRef]
- Collatz, J.; Tolasch, T.; Steidle, J.L. Mate finding in the parasitic wasp Cephalonomia tarsalis (ashmead): More than one way to a female’s heart. J. Chem. Ecol. 2009, 35, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Jayanthi, K.; Saravan, P.D.; Kumar, P.; Vyas, M. Odour cues from fruit arils of Artocarpus heterophyllus attract both sexes of oriental fruit flies. J. Chem. Ecol. 2021, 47, 552–563. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.Y.; Liu, Z.J.; Wang, X.Y.; Zhang, Z.L.; Lu, W.; Zheng, X.L. Electroantennographic and olfactory responses of Quadrastichus mendeli to eucalyptus volatiles induced by the gall-forming insect Leptocybe invasa. Pest. Manag. Sci. 2022, 78, 2405–2416. [Google Scholar] [CrossRef] [PubMed]
- Magalhães-Junior, J.T.; Oliva-Filho, A.A.; Novais, H.O.; Mesquita, P.R.R.; Rodrigues, M.; Pinto, M.C.; Barrouin-Melo, S.M. Attraction of the sandfly lutzomyia longipalpis to possible biomarker compounds from dogs infected with Leishmania infantum. Med. Vet. Entomol. 2019, 33, 322–325. [Google Scholar] [CrossRef]
- Laznik, Ž.; Trdan, S. Attraction behaviors of entomopathogenic nematodes (steinernematidae and heterorhabditidae) to synthetic volatiles emitted by insect damaged potato tubers. J. Chem. Ecol. 2016, 42, 314–322. [Google Scholar] [CrossRef]
- Li, J.; Zhang, L.; Wang, X. An odorant-binding protein involved in perception of host plant odorants in locust Locusta migratoria. Arch. Insect Biochem. Physiol. 2016, 91, 221–229. [Google Scholar] [CrossRef]
- Ullah, R.M.K.; Quershi, S.R.; Adeel, M.M.; Abdelnabby, H.; Waris, M.I.; Duan, S.G.; Wang, M.Q. An odorant binding protein (saveobp9) involved in chemoreception of the wheat aphid Sitobion avenae. Int. J. Mol. Sci. 2020, 21, 8331. [Google Scholar] [CrossRef]
- Molnár, B.P.; Tóth, Z.; Fejes-Tóth, A.; Dekker, T.; Kárpáti, Z. Electrophysiologically-active maize volatiles attract gravid female european corn borer, Ostrinia nubilalis. J. Chem. Ecol. 2015, 41, 997–1005. [Google Scholar] [CrossRef]
- Morawo, T.; Burrows, M.; Fadamiro, H. Electroantennogram response of the parasitoid, Microplitis croceipes to host-related odors: The discrepancy between relative abundance and level of antennal responses to volatile compound. F1000Res 2016, 5, 2725. [Google Scholar] [CrossRef]
- Du, Y.; Zhang, J.; Yan, Z.; Ma, Y.; Yang, M.; Zhang, M.; Zhang, Z.; Qin, L.; Cao, Q. Host preference and performance of the yellow peach moth (Conogethes punctiferalis) on chestnut cultivars. PLoS ONE 2016, 11, e0157609. [Google Scholar] [CrossRef]
- Liu, X.F.; Chen, H.H.; Li, J.K.; Zhang, R.; Turlings, T.C.; Chen, L. Volatiles released by chinese liquorice roots mediate host location behaviour by neonate Porphyrophora sophorae (Hemiptera: Margarodidae). Pest. Manag. Sci. 2016, 72, 1959–1964. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.; Liu, Y.; Si, H.; Xiao, Z.; Fan, G.; Chen, S.; Wang, P.; Wang, Z. Hydronopylformamides: Modification of the naturally occurring compound (-)-β-pinene to produce insect repellent candidates against Blattella germanica. Molecules 2017, 22, 1004. [Google Scholar] [CrossRef] [PubMed]
- Dekel, A.; Yakir, E.; Bohbot, J.D. The sulcatone receptor of the strict nectar-feeding mosquito toxorhynchites amboinensis. Insect Biochem. Mol. Biol. 2019, 111, 103174. [Google Scholar] [CrossRef] [PubMed]
- May-Concha, I.; Rojas, J.C.; Cruz-López, L.; Millar, J.G.; Ramsey, J.M. Volatile compounds emitted by Triatoma dimidiata, a vector of chagas disease: Chemical analysis and behavioural evaluation. Med. Vet. Entomol. 2013, 27, 165–174. [Google Scholar] [CrossRef]
- Gu, D.; Yang, D.R. Utilisation of chemical signals by inquiline wasps in entering their host figs. J. Insect Physiol. 2013, 59, 1065–1068. [Google Scholar] [CrossRef]
- Olaide, O.Y.; Tchouassi, D.P.; Yusuf, A.A.; Pirk, C.W.W.; Masiga, D.K.; Saini, R.K.; Torto, B. Zebra skin odor repels the savannah tsetse fly, Glossina pallidipes (Diptera: Glossinidae). PLOS Neglected Trop. Dis. 2019, 13, e0007460. [Google Scholar] [CrossRef]
- Logan, J.G.; Stanczyk, N.M.; Hassanali, A.; Kemei, J.; Santana, A.E.; Ribeiro, K.A.; Pickett, J.A.; Mordue Luntz, A.J. Arm-in-cage testing of natural human-derived mosquito repellents. Malar. J. 2010, 9, 239. [Google Scholar] [CrossRef]
- Logan, J.G.; Seal, N.J.; Cook, J.I.; Stanczyk, N.M.; Birkett, M.A.; Clark, S.J.; Gezan, S.A.; Wadhams, L.J.; Pickett, J.A.; Mordue, A.J. Identification of human-derived volatile chemicals that interfere with attraction of the scottish biting midge and their potential use as repellents. J. Med. Entomol. 2009, 46, 208–219. [Google Scholar] [CrossRef]
- Birkett, M.A.; Agelopoulos, N.; Jensen, K.M.; Jespersen, J.B.; Pickett, J.A.; Prijs, H.J.; Thomas, G.; Trapman, J.J.; Wadhams, L.J.; Woodcock, C.M. The role of volatile semiochemicals in mediating host location and selection by nuisance and disease-transmitting cattle flies. Med. Vet. Entomol. 2004, 18, 313–322. [Google Scholar] [CrossRef]
- Malcicka, M.; Bezemer, T.M.; Visser, B.; Bloemberg, M.; Snart, C.J.; Hardy, I.C.; Harvey, J.A. Multi-trait mimicry of ants by a parasitoid wasp. Sci. Rep. 2015, 5, 8043. [Google Scholar] [CrossRef]
- Wu, F.; Feng, Y.; Han, B.; Hu, H.; Feng, M.; Meng, L.; Ma, C.; Yu, L.; Li, J. Mechanistic insight into binding interaction between chemosensory protein 4 and volatile larval pheromones in honeybees (Apis mellifera). Int. J. Biol. Macromol. 2019, 141, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Jing, T.; Qian, X.; Du, W.; Gao, T.; Li, D.; Guo, D.; He, F.; Yu, G.; Li, S.; Schwab, W.; et al. Herbivore-induced volatiles influence moth preference by increasing the β-ocimene emission of neighbouring tea plants. Plant Cell Environ. 2021, 44, 3667–3680. [Google Scholar] [CrossRef] [PubMed]
- Cozzolino, S.; Fineschi, S.; Litto, M.; Scopece, G.; Trunschke, J.; Schiestl, F.P. Herbivory increases fruit set in silene latifolia: A consequence of induced pollinator-attracting floral volatiles. J. Chem. Ecol. 2015, 41, 622–630. [Google Scholar] [CrossRef]
- Tang, R.; Zhang, J.P.; Zhang, Z.N. Electrophysiological and behavioral responses of male fall webworm moths (Hyphantria cunea) to herbivory-induced mulberry (Morus alba) leaf volatiles. PLoS ONE 2012, 7, e49256. [Google Scholar] [CrossRef] [PubMed]
- Kang, Z.W.; Liu, F.H.; Zhang, Z.F.; Tian, H.G.; Liu, T.X. Volatile β-ocimene can regulate developmental performance of peach aphid Myzus persicae through activation of defense responses in chinese cabbage brassica pekinensis. Front. Plant Sci. 2018, 9, 708. [Google Scholar] [CrossRef]
- Cascone, P.; Iodice, L.; Maffei, M.E.; Bossi, S.; Arimura, G.; Guerrieri, E. Tobacco overexpressing β-ocimene induces direct and indirect responses against aphids in receiver tomato plants. J. Plant Physiol. 2015, 173, 28–32. [Google Scholar] [CrossRef]
- Jacob, J.W.; Tchouassi, D.P.; Lagat, Z.O.; Mathenge, E.M.; Mweresa, C.K.; Torto, B. Independent and interactive effect of plant- and mammalian- based odors on the response of the malaria vector, Anopheles gambiae. Acta Trop. 2018, 185, 98–106. [Google Scholar] [CrossRef]
- Koczor, S.; Vuts, J.; Caulfield, J.C.; Withall, D.M.; Sarria, A.; Pickett, J.A.; Birkett, M.A.; Csonka, É.B.; Tóth, M. Sex pheromone of the alfalfa plant bug, Adelphocoris lineolatus: Pheromone composition and antagonistic effect of 1-hexanol (Hemiptera: Miridae). J. Chem. Ecol. 2021, 47, 525–533. [Google Scholar] [CrossRef]
- Sun, L.; Gu, S.H.; Xiao, H.J.; Zhou, J.J.; Guo, Y.Y.; Liu, Z.W.; Zhang, Y.J. The preferential binding of a sensory organ specific odorant binding protein of the alfalfa plant bug Adelphocoris lineolatus alinobp10 to biologically active host plant volatiles. J. Chem. Ecol. 2013, 39, 1221–1231. [Google Scholar] [CrossRef]
- Faccoli, M.; Blazenec, M.; Schlyter, F. Feeding response to host and nonhost compounds by males and females of the spruce bark beetle Ips typographus in a tunneling microassay. J. Chem. Ecol. 2005, 31, 745–759. [Google Scholar] [CrossRef]





| Primer Name | Primer Sequence (5′-3′) | Length (bp) | Purpose |
|---|---|---|---|
| CsasCSP7-F | AGGTTATTGAGCATCTGATTAAG | 95 | Fluorescence quantification |
| CsasCSP7-R | TTCATACTTCTTTCTCCACTTG | ||
| CsasCSP9-F | GTTATGGAGTACATCATAGATC | 102 | |
| CsasCSP9-R | TTTCTTCTCTTCATACTTACTC | ||
| CsasCSP11-F | CAAGTAGTCCGATACATTAGG | 125 | |
| CsasCSP11-R | TAATCATCAGAAGCGAAGAAT | ||
| CsasCSP7-F | CGGGATCCATGGAAGAAAAGTATTCGGACAAATA | 342 | Prokaryotic expression |
| CsasCSP7-R | TGGAATTCCTATTTTTCAGGTATTTCAATACCCCT | ||
| CsasCSP9-F | CGGGATCCATGCGCCCCGAAGAGCACT | 432 | |
| CsasCSP9-R | TGGAATTCTTATGGCCTTGACGGTGCG | ||
| CsasCSP11-F | CGGGATCCATGGATGAGGAGCAGTATACAGATAGAT | 333 | |
| CsasCSP11-R | TGGAATTCTTAATCATCAGAAGCGAAGAATG |
| Code | Prospective Ligand | CAS | Molecular Weight /g·mol−1 | Purity/% |
|---|---|---|---|---|
| L1 | 2-Methylbutyl acetate | 624-41-9 | 130.18 | 99 |
| L2 | Butyl butyrate | 109-21-7 | 144.21 | >99 |
| L3 | Butyl heptanoate | 5454-28-4 | 186.29 | >99 |
| L4 | Butyl octanoate | 589-75-3 | 200.32 | >99 |
| L5 | Ethyl butyrate | 105-54-4 | 116.16 | 99 |
| L6 | Ethyl heptanoate | 106-30-9 | 158.24 | >98 |
| L7 | Ethyl hexanoate | 123-66-0 | 144.21 | 99 |
| L8 | Hexyl hexanoate | 6378-65-0 | 200.32 | 98 |
| L9 | Isoamyl acetate | 123-92-2 | 130.19 | 99 |
| L10 | Methyl jasmonate | 39,924-52-2 | 224.3 | 95 |
| L11 | Propyl octanoate | 624-13-5 | 186.29 | 98 |
| L12 | Tert-butyl acetate | 540-88-5 | 116.15 | 99 |
| L13 | (Z)-3-Hexenyl acetate | 3681-71-8 | 142.2 | 98 |
| L14 | Benzaldehyde | 100-52-7 | 106.12 | >99 |
| L15 | Decanal | 112-31-2 | 156.27 | 97 |
| L16 | Dodecanal | 112-54-9 | 184.32 | >95 |
| L17 | (E)-Hex-2-enal | 6728-26-3 | 98.14 | 98 |
| L18 | Hexanal | 66-25-1 | 100.16 | 97 |
| L19 | Honanal | 124-19-6 | 142.24 | 96 |
| L20 | Octanal | 124-13-0 | 128.215 | 97 |
| L21 | 1-Hexanol | 111-27-3 | 102.18 | >98 |
| L22 | 2-Ethylhexanol | 104-76-7 | 130.22 | 99 |
| L23 | 3-Methyl-1-butanol | 123-51-3 | 88.15 | 99 |
| L24 | (E)-2-Hexen-1-ol | 928-95-0 | 100.16 | 97 |
| L25 | (Z)-Hex-3-en-1-ol | 928-96-1 | 100.16 | ≥98 |
| L26 | Decane | 124-18-5 | 142.29 | >99 |
| L27 | Hexadecane | 544-76-3 | 226.45 | >98 |
| L28 | Octadecane | 593-45-3 | 254.49 | 98 |
| L29 | Pentadecane | 629-62-9 | 212.41 | 98 |
| L30 | Tetradecane | 629-59-4 | 198.39 | ≥99 |
| L31 | alpha-Farnesene | 502-61-4 | 204.25 | >99 |
| L32 | Beta-Ocimene | 13,877-91-3 | 136.23 | >90 |
| L33 | (-)-beta-Pinene | 18,172-67-3 | 136.24 | >94 |
| L34 | Myrcene | 123-35-3 | 136.236 | >80 |
| L35 | 6-Methyl-5-hepten-2-one | 110-93-0 | 126.2 | >98 |
| L36 | Benzonitrile | 100-47-0 | 103.12 | >99 |
| L37 | Z-7-Eicosene-11-one | 63,408-44-6 | 294.5 | >99 |
| L38 | Z-7-Nonadecen-11-one | 63,408-45-7 | 280.5 | >99 |
| Ligand | Ki | IC50 | Ligand | Ki | IC50 | Ligand | Ki | IC50 |
|---|---|---|---|---|---|---|---|---|
| 2-Methylbutyl acetate | - | >50 | Benzaldehyde | 7.25 ± 0.23 | 8.00 ± 0.59 | Hexadecane | - | >50 |
| Butyl butyrate | - | >50 | Decanal | - | >50 | Octadecane | - | >50 |
| Butyl heptanoate | - | >50 | Dodecanal | 13.61 ± 0.54 | 14.97 ± 0.60 | Pentadecane | - | >50 |
| Butyl octanoate | - | >50 | (E)-Hex-2-enal | - | >50 | Tetradecane | - | >50 |
| Ethyl butyrate | - | >50 | Hexanal | - | >50 | alpha-Farnesene | - | >50 |
| Ethyl heptanoate | - | >50 | Honanal | - | >50 | Beta-Ocimene | - | >50 |
| Ethyl hexanoate | - | >50 | Octanal | - | >50 | (-)-beta-Pinene | - | >50 |
| Hexyl hexanoate | - | >50 | 1-Hexanol | - | >50 | Myrcene | - | >50 |
| Isoamyl acetate | - | >50 | 2-Ethylhexanol | - | >50 | 6-Methyl-5-hepten-2-one | - | >50 |
| Methyl jasmonate | - | >50 | 3-Methyl-1-butanol | - | >50 | Benzonitrile | - | >50 |
| Propyl octanoate | - | >50 | (E)-2-Hexen-1-ol | - | >50 | Z-7-Eicosene-11-one | - | >50 |
| Tert-butyl acetate | - | >50 | (Z)-Hex-3-en-1-ol | - | >50 | Z-7-Nonadecen-11-one | - | >50 |
| (Z)-3-Hexenyl acetate | - | >50 | Decane | - | >50 |
| Ligand | Ki | IC50 | Ligand | Ki | IC50 | Ligand | Ki | IC50 |
|---|---|---|---|---|---|---|---|---|
| 2-Methylbutyl acetate | - | >50 | Benzaldehyde | - | >50 | Hexadecane | - | >50 |
| Butyl butyrate | - | >50 | Decanal | 1.65 ± 0.31 | 1.92 ± 0.34 | Octadecane | - | >50 |
| Butyl heptanoate | - | >50 | Dodecanal | - | >50 | Pentadecane | - | >50 |
| Butyl octanoate | 1.47 ± 0.13 | 1.71 ± 0.16 | (E)-Hex-2-enal | - | >50 | Tetradecane | - | >50 |
| Ethyl butyrate | - | >50 | Hexanal | - | >50 | alpha-Farnesene | - | >50 |
| Ethyl heptanoate | - | >50 | Honanal | - | >50 | Beta-Ocimene | - | >50 |
| Ethyl hexanoate | - | >50 | Octanal | - | >50 | (-)-beta-Pinene | 14.26 ± 0.62 | 16.56 ± 0.24 |
| Hexyl hexanoate | - | >50 | 1-Hexanol | - | >50 | Myrcene | - | >50 |
| Isoamyl acetate | - | >50 | 2-Ethylhexanol | - | >50 | 6-Methyl-5-hepten-2-one | - | >50 |
| Methyl jasmonate | - | >50 | 3-Methyl-1-butanol | - | >50 | Benzonitrile | - | >50 |
| Propyl octanoate | - | >50 | (E)-2-Hexen-1-ol | - | >50 | Z-7-Eicosene-11-one | - | >50 |
| Tert-butyl acetate | - | >50 | (Z)-Hex-3-en-1-ol | - | >50 | Z-7-Nonadecen-11-one | - | >50 |
| (Z)-3-Hexenyl acetate | - | >50 | Decane | - | >50 |
| Ligand | Ki | IC50 | Ligand | Ki | IC50 | Ligand | Ki | IC50 |
|---|---|---|---|---|---|---|---|---|
| 2-Methylbutyl acetate | - | >50 | Benzaldehyde | - | >50 | Hexadecane | - | >50 |
| Butyl butyrate | - | >50 | Decanal | - | >50 | Octadecane | - | >50 |
| Butyl heptanoate | - | >50 | Dodecanal | - | >50 | Pentadecane | - | >50 |
| Butyl octanoate | - | >50 | (E)-Hex-2-enal | - | >50 | Tetradecane | - | >50 |
| Ethyl butyrate | - | >50 | Hexanal | - | >50 | alpha-Farnesene | - | >50 |
| Ethyl heptanoate | - | >50 | Honanal | - | >50 | Beta-Ocimene | 1.83 ± 0.66 | 2.09 ± 0.76 |
| Ethyl hexanoate | - | >50 | Octanal | - | >50 | (-)-beta-Pinene | - | >50 |
| Hexyl hexanoate | - | >50 | 1-Hexanol | 8.13 ± 0.78 | 9.31 ± 0.39 | Myrcene | - | >50 |
| Isoamyl acetate | - | >50 | 2-Ethylhexanol | - | >50 | 6-Methyl-5-hepten-2-one | 0.71 ± 0.07 | 0.81 ± 0.08 |
| Methyl jasmonate | - | >50 | 3-Methyl-1-butanol | - | >50 | Benzonitrile | - | >50 |
| Propyl octanoate | - | >50 | (E)-2-Hexen-1-ol | - | >50 | Z-7-Eicosene-11-one | - | >50 |
| Tert-butyl acetate | - | >50 | (Z)-Hex-3-en-1-ol | - | >50 | Z-7-Nonadecen-11-one | - | >50 |
| (Z)-3-Hexenyl acetate | - | >50 | Decane | - | >50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Qiu, G.; Zhang, H.; Yue, Q.; Yan, W.; Sun, L. Molecular Characterization of Three Chemosensory Proteins from Carposina sasakii. Agriculture 2023, 13, 2066. https://doi.org/10.3390/agriculture13112066
Liu L, Qiu G, Zhang H, Yue Q, Yan W, Sun L. Molecular Characterization of Three Chemosensory Proteins from Carposina sasakii. Agriculture. 2023; 13(11):2066. https://doi.org/10.3390/agriculture13112066
Chicago/Turabian StyleLiu, Liu, Guisheng Qiu, Huaijiang Zhang, Qiang Yue, Wentao Yan, and Lina Sun. 2023. "Molecular Characterization of Three Chemosensory Proteins from Carposina sasakii" Agriculture 13, no. 11: 2066. https://doi.org/10.3390/agriculture13112066
APA StyleLiu, L., Qiu, G., Zhang, H., Yue, Q., Yan, W., & Sun, L. (2023). Molecular Characterization of Three Chemosensory Proteins from Carposina sasakii. Agriculture, 13(11), 2066. https://doi.org/10.3390/agriculture13112066





