Establishment of Tibetan-Sheep-Specific SNP Genetic Markers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Sample Collection
2.3. SNP BeadChip Genotyping and Quality Control
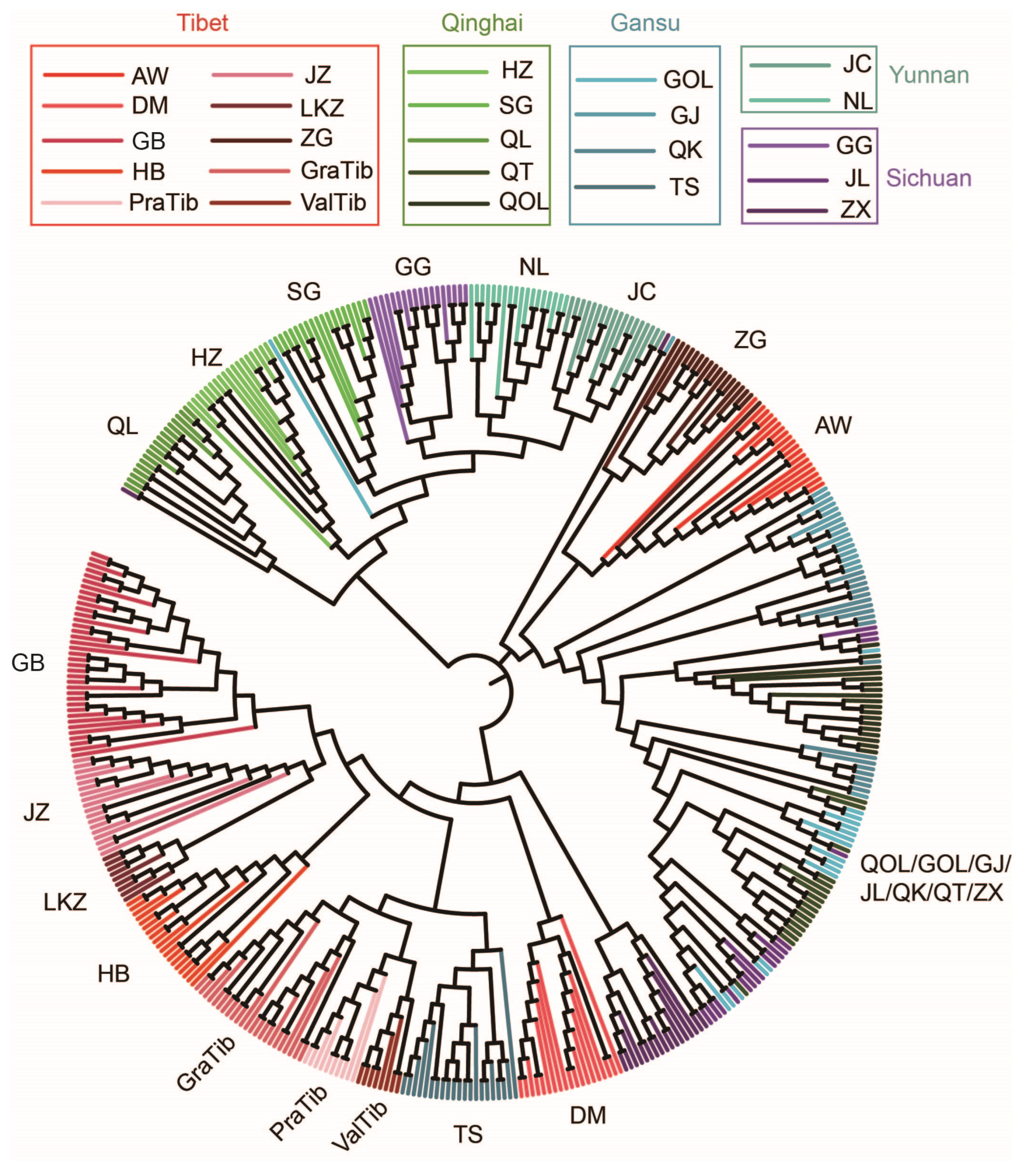
2.4. Population Structure, Phylogenetic Analysis
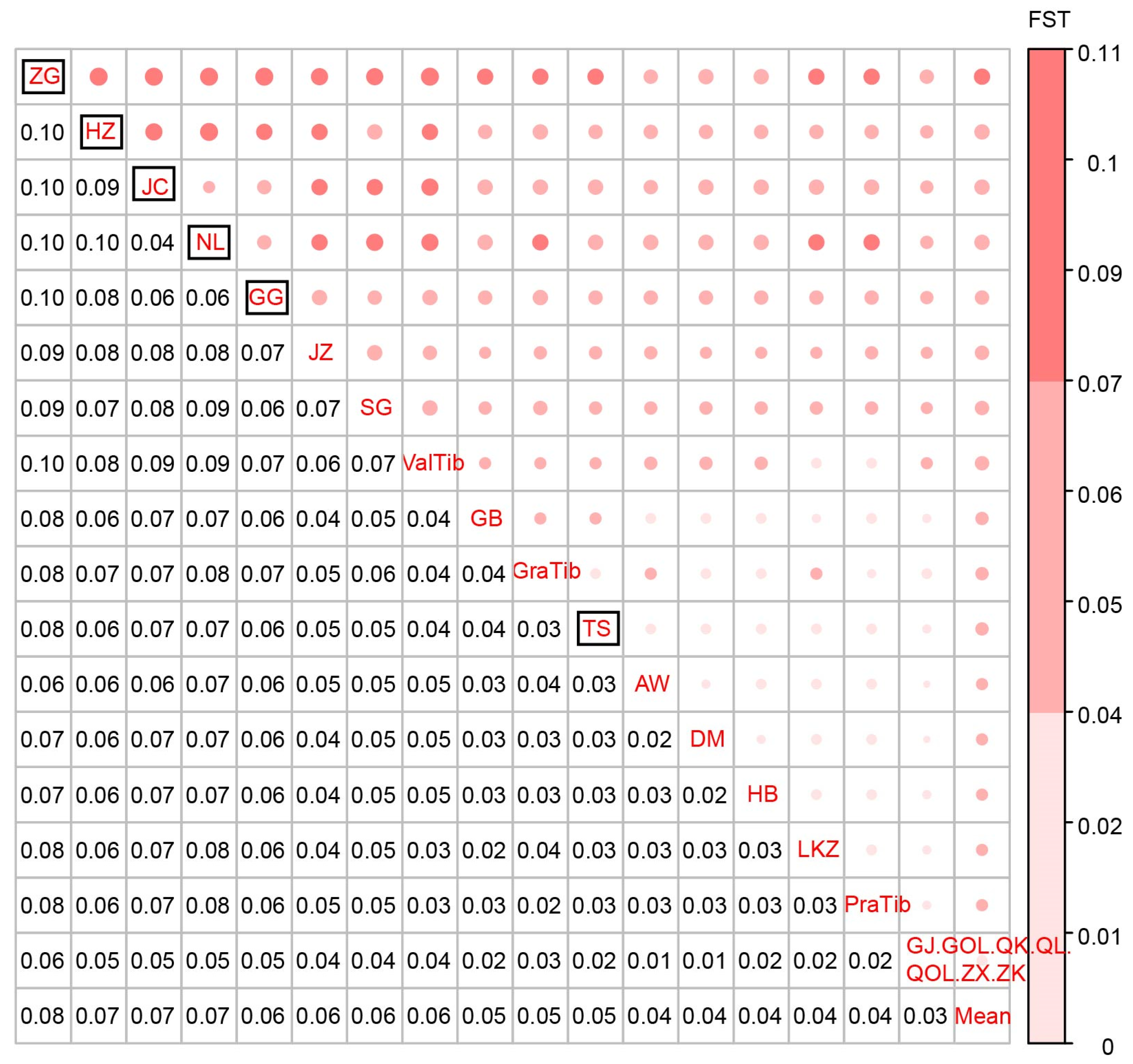
2.5. Calculation of Population Differentiation Index
3. Results
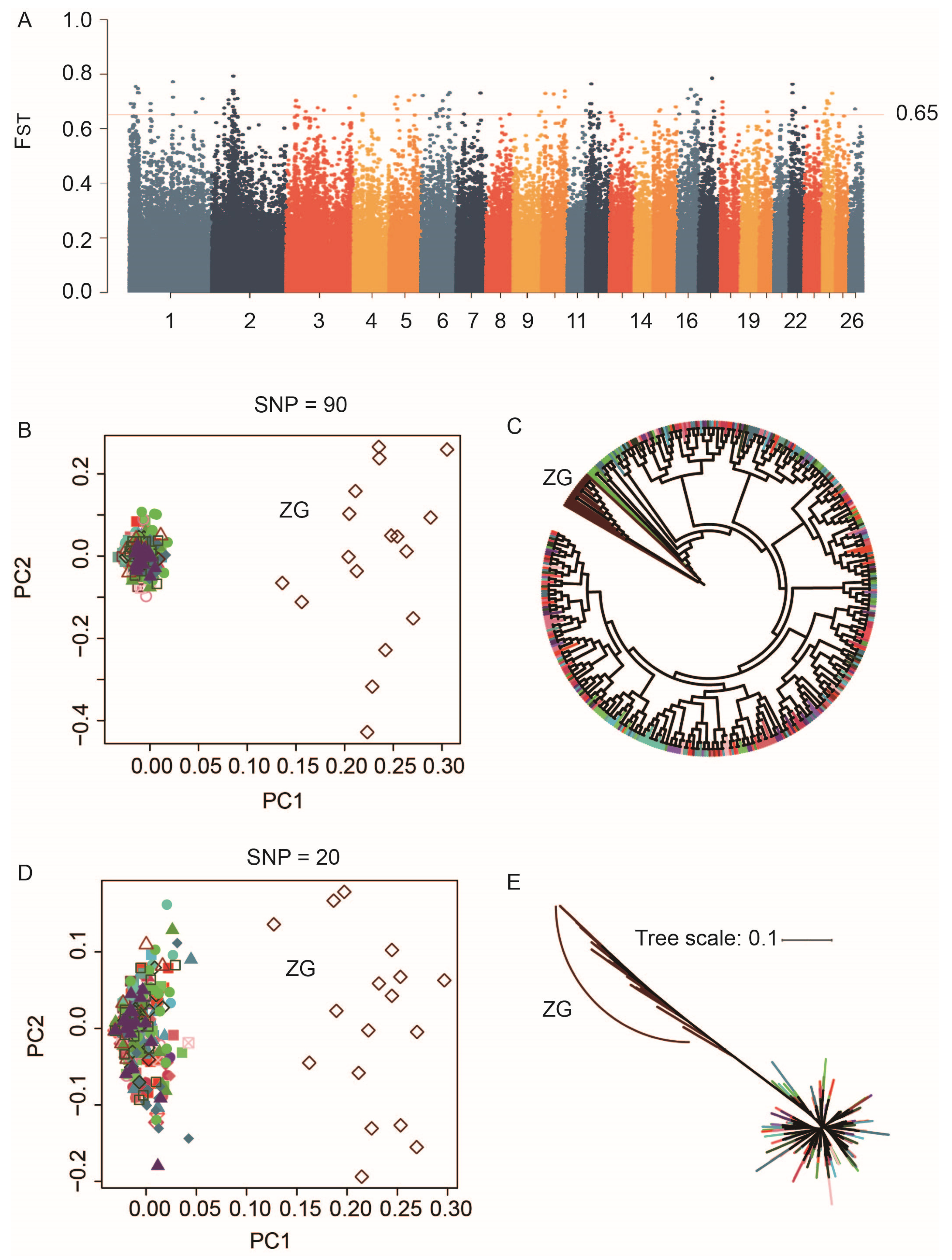
3.1. Genomic Variation and Population Structure Analysis
3.2. Population Genetic Differentiation Distance (FST)
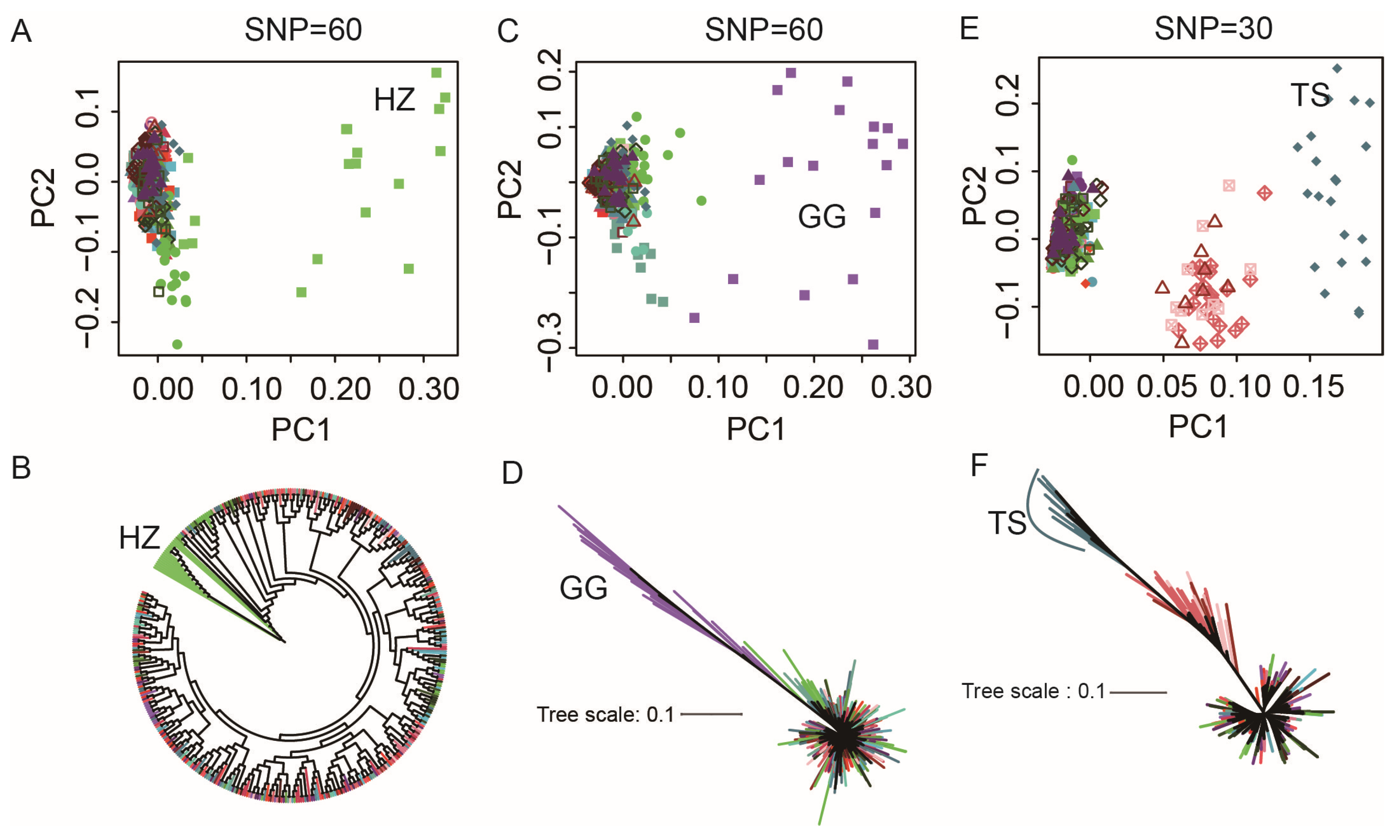
3.3. Construction of Region-Specific SNP Marker Sets
3.4. Construction of Breed-Specific SNP Marker Sets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lv, F.H.; Peng, W.F.; Yang, J.; Zhao, Y.X.; Li, W.R.; Liu, M.J.; Ma, Y.H.; Zhao, Q.J.; Yang, G.L.; Wang, F.; et al. Mitogenomic Meta-Analysis Identifies Two Phases of Migration in the History of Eastern Eurasian Sheep. Mol. Biol. Evol. 2015, 32, 2515–2533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Li, W.R.; Lv, F.H.; He, S.G.; Tian, S.L.; Peng, W.F.; Sun, Y.W.; Zhao, Y.X.; Tu, X.L.; Zhang, M.; et al. Whole-Genome Sequencing of Native Sheep Provides Insights into Rapid Adaptations to Extreme Environments. Mol. Biol. Evol. 2016, 33, 2576–2592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.X.; Yang, J.; Lv, F.H.; Hu, X.J.; Xie, X.L.; Zhang, M.; Li, W.R.; Liu, M.J.; Wang, Y.T.; Li, J.Q.; et al. Genomic Reconstruction of the History of Native Sheep Reveals the Peopling Patterns of Nomads and the Expansion of Early Pastoralism in East Asia. Mol. Biol. Evol. 2017, 34, 2380–2395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.Y.; Duan, Z.Y.; Sha, T.; Xiangyu, J.; Wu, S.F.; Zhang, Y.P. Origin, genetic diversity, and population structure of Chinese domestic sheep. Gene 2006, 376, 216–223. [Google Scholar] [CrossRef]
- Müller, K.; Lin, L.; Wang, C.; Glindemann, T.; Schiborra, A.; Schönbach, P.; Wan, H.; Dickhoefer, U.; Susenbeth, A. Effect of continuous v. daytime grazing on feed intake and growth of sheep grazing in a semi-arid grassland steppe. Animal 2012, 6, 526–534. [Google Scholar] [CrossRef]
- Liu, J.; Ding, X.; Zeng, Y.; Yue, Y.; Guo, X.; Guo, T.; Chu, M.; Wang, F.; Han, J.; Feng, R.; et al. Genetic Diversity and Phylogenetic Evolution of Tibetan Sheep Based on mtDNA D-Loop Sequences. PLoS ONE 2016, 11, e0159308. [Google Scholar] [CrossRef] [Green Version]
- Niu, L.; Chen, X.; Xiao, P.; Zhao, Q.; Zhou, J.; Hu, J.; Sun, H.; Guo, J.; Li, L.; Wang, L.; et al. Detecting signatures of selection within the Tibetan sheep mitochondrial genome. Mitochondrial DNA A 2017, 28, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Jing, X.; Wang, W.; Degen, A.; Guo, Y.; Kang, J.; Liu, P.; Ding, L.; Shang, Z.; Fievez, V.; Zhou, J.; et al. Tibetan sheep have a high capacity to absorb and to regulate metabolism of SCFA in the rumen epithelium to adapt to low energy intake. Br. J. Nutr. 2020, 123, 721–736. [Google Scholar] [CrossRef]
- Ashworth, C.J.; Ross, A.W.; Barrett, P. The use of DNA fingerprinting to assess monozygotic twinning in Meishan and Landrace × large white pigs. Reprod. Fertil. Dev. 1998, 10, 487–490. [Google Scholar] [CrossRef]
- Hebert, P.D.; Stoeckle, M.Y.; Zemlak, T.S.; Francis, C.M. Identification of Birds through DNA Barcodes. PLoS Biol. 2004, 2, e312. [Google Scholar] [CrossRef]
- Hu, Y.; Ding, Q.; Ji, H.; Wang, X.; Zhu, Z.; Zhao, Q.J. Digital Barcode Development for Single Nuclotide Polymorphism (SNP) Identification of Suzhong Swine Individuals. Anim. Husb. Feed. Sci. 2016, 8, 323–326. [Google Scholar]
- Lv, F.H.; Agha, S.; Kantanen, J.; Colli, L.; Stucki, S.; Kijas, J.W.; Joost, S.; Li, M.H.; Ajmone Marsan, P. Adaptations to climate-mediated selective pressures in sheep. Mol. Biol. Evol. 2014, 31, 3324–3343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.M.; Zhang, M.; Zhou, Z.K.; Liu, L.; Liu, X.D.; Chen, C.; Chen, X.J.; Zhu, M.; Yuan, Z.R.; Li, J.Y. Association of SCD1 Gene with Meat Quality Traits in Chinese Simmental Based on High-density SNP Chip. Acta Vet Zootech. Sin. 2012, 43, 878–886. [Google Scholar]
- Rostamzadeh Mahdabi, E.; Esmailizadeh, A.; Ayatollahi Mehrgardi, A.; Asadi Fozi, M. A genome-wide scan to identify signatures of selection in two Iranian indigenous chicken ecotypes. Genet. Sel. Evol. 2021, 53, 72. [Google Scholar] [CrossRef] [PubMed]
- Guang-Xin, E.; Zhong, T.; Ma, Y.H.; Gao, H.J.; He, J.N.; Liu, N.; Zhao, Y.J.; Zhang, J.H.; Huang, Y.F. Conservation genetics in Chinese sheep: Diversity of fourteen indigenous sheep (Ovis aries) using microsatellite markers. Ecol. Evol. 2016, 6, 810–817. [Google Scholar]
- Hu, X.J.; Yang, J.; Xie, X.L.; Lv, F.H.; Cao, Y.H.; Li, W.R.; Liu, M.J.; Wang, Y.T.; Li, J.Q.; Liu, Y.G.; et al. The Genome Landscape of Tibetan Sheep Reveals Adaptive Introgression from Argali and the History of Early Human Settlements on the Qinghai-Tibetan Plateau. Mol. Biol. Evol. 2019, 36, 283–303. [Google Scholar] [CrossRef] [Green Version]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimating F-Statistics for the Analysis of Population Structure. Evolution 1984, 38, 1358–1370. [Google Scholar]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef] [Green Version]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar] [PubMed]
- Williams, J.G.; Kubelik, A.R.; Livak, K.J.; Rafalski, J.A.; Tingey, S.V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990, 18, 6531–6535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marc, Z.; Pieter, V. Selective Restriction Fragment Amplification: A General Method for DNA Fingerprinting. EP0534858B2, 31 March 1993. [Google Scholar]
- Wang, X.; Shorgan, B.J.B.B. Progress in Detection Methods of Single Nucleotide Polymorphism. Biotechnol. Bull. 2006, 52, 64–67. [Google Scholar]
- Heaton, M.P.; Harhay, G.P.; Bennett, G.L.; Stone, R.T.; Grosse, W.M.; Casas, E.; Keele, J.W.; Smith, T.P.; Chitko-McKown, C.G.; Laegreid, W.W. Selection and use of SNP markers for animal identification and paternity analysis in U.S. beef cattle. Mamm. Genome 2002, 13, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Suekawa, Y.; Aihara, H.; Araki, M.; Hosokawa, D.; Mannen, H.; Sasazaki, S. Development of breed identification markers based on a bovine 50K SNP array. Meat Sci. 2010, 85, 285–288. [Google Scholar] [CrossRef]
- Holl, H.M.; Vanhnasy, J.; Everts, R.E.; Hoefs-Martin, K.; Cook, D.; Brooks, S.A.; Carpenter, M.L.; Bustamante, C.D.; Lafayette, C. Single nucleotide polymorphisms for DNA typing in the domestic horse. Anim. Genet. 2017, 48, 669–676. [Google Scholar] [CrossRef]
- Wu, D.D.; Yang, C.P.; Wang, M.S.; Dong, K.Z.; Yan, D.W.; Hao, Z.Q.; Fan, S.Q.; Chu, S.Z.; Shen, Q.S.; Jiang, L.P.; et al. Convergent genomic signatures of high-altitude adaptation among domestic mammals. Natl. Sci. Rev. 2020, 7, 952–963. [Google Scholar] [CrossRef] [Green Version]
- Li, L.L.; Ma, S.K.; Peng, W.; Fang, Y.G.; Duo, H.R.; Fu, H.Y.; Jia, G.X. Genetic diversity and population structure of Tibetan sheep breeds determined by whole genome resequencing. Trop. Anim. Health Prod. 2021, 53, 174. [Google Scholar] [CrossRef]
- Lv, F.H.; Cao, Y.H.; Liu, G.J.; Luo, L.Y.; Lu, R.; Liu, M.J.; Li, W.R.; Zhou, P.; Wang, X.H.; Shen, M.; et al. Whole-Genome Resequencing of Worldwide Wild and Domestic Sheep Elucidates Genetic Diversity, Introgression, and Agronomically Important Loci. Mol. Biol. Evol. 2022, 39, msab353. [Google Scholar] [CrossRef]
- Wei, C.; Wang, H.; Liu, G.; Wu, M.; Cao, J.; Liu, Z.; Liu, R.; Zhao, F.; Zhang, L.; Lu, J.; et al. Genome-wide analysis reveals population structure and selection in Chinese indigenous sheep breeds. BMC Genom. 2015, 16, 194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, X.; Chen, F.; Gao, F.; Li, L.; Liu, K.; You, L.; Hua, C.; Yang, F.; Liu, W.; Peng, C.; et al. CNSA: A data repository for archiving omics data. Database 2020, 2020, baaa055. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.Z.; You, L.J.; Yang, F.; Wang, L.N.; Guo, X.Q.; Gao, F.; Hua, C.; Tan, C.; Fang, L.; Shan, R.Q.; et al. CNGBdb: China National GeneBank DataBase. Yi Chuan = Hered. 2020, 42, 799–809. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, B.; Zhao, Y.; Pu, Y.; He, X.; Han, J.; Danzeng, B.; Ma, Y.; Liu, J.; Jiang, L. Establishment of Tibetan-Sheep-Specific SNP Genetic Markers. Agriculture 2023, 13, 322. https://doi.org/10.3390/agriculture13020322
Liang B, Zhao Y, Pu Y, He X, Han J, Danzeng B, Ma Y, Liu J, Jiang L. Establishment of Tibetan-Sheep-Specific SNP Genetic Markers. Agriculture. 2023; 13(2):322. https://doi.org/10.3390/agriculture13020322
Chicago/Turabian StyleLiang, Benmeng, Yuhetian Zhao, Yabin Pu, Xiaohong He, Jiangang Han, Baima Danzeng, Yuehui Ma, Jianfeng Liu, and Lin Jiang. 2023. "Establishment of Tibetan-Sheep-Specific SNP Genetic Markers" Agriculture 13, no. 2: 322. https://doi.org/10.3390/agriculture13020322
APA StyleLiang, B., Zhao, Y., Pu, Y., He, X., Han, J., Danzeng, B., Ma, Y., Liu, J., & Jiang, L. (2023). Establishment of Tibetan-Sheep-Specific SNP Genetic Markers. Agriculture, 13(2), 322. https://doi.org/10.3390/agriculture13020322






