Knocking out OsNAC050 Expression Causes Low-Temperature Tolerance in Rice by Regulating Photosynthesis and the Sucrose Metabolic Pathway
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Growth Conditions and Stress Treatments
2.2. Quantification of Relative Gene Expression Levels Using Quantitative Real-Time PCR (qRT-PCR)
2.3. Subcellular Localization of OsNAC050
2.4. Targeted Mutagenesis of OsNAC050
2.5. Physiological Measurements
2.6. Transcriptome and Bioinformatics Analyses
3. Results
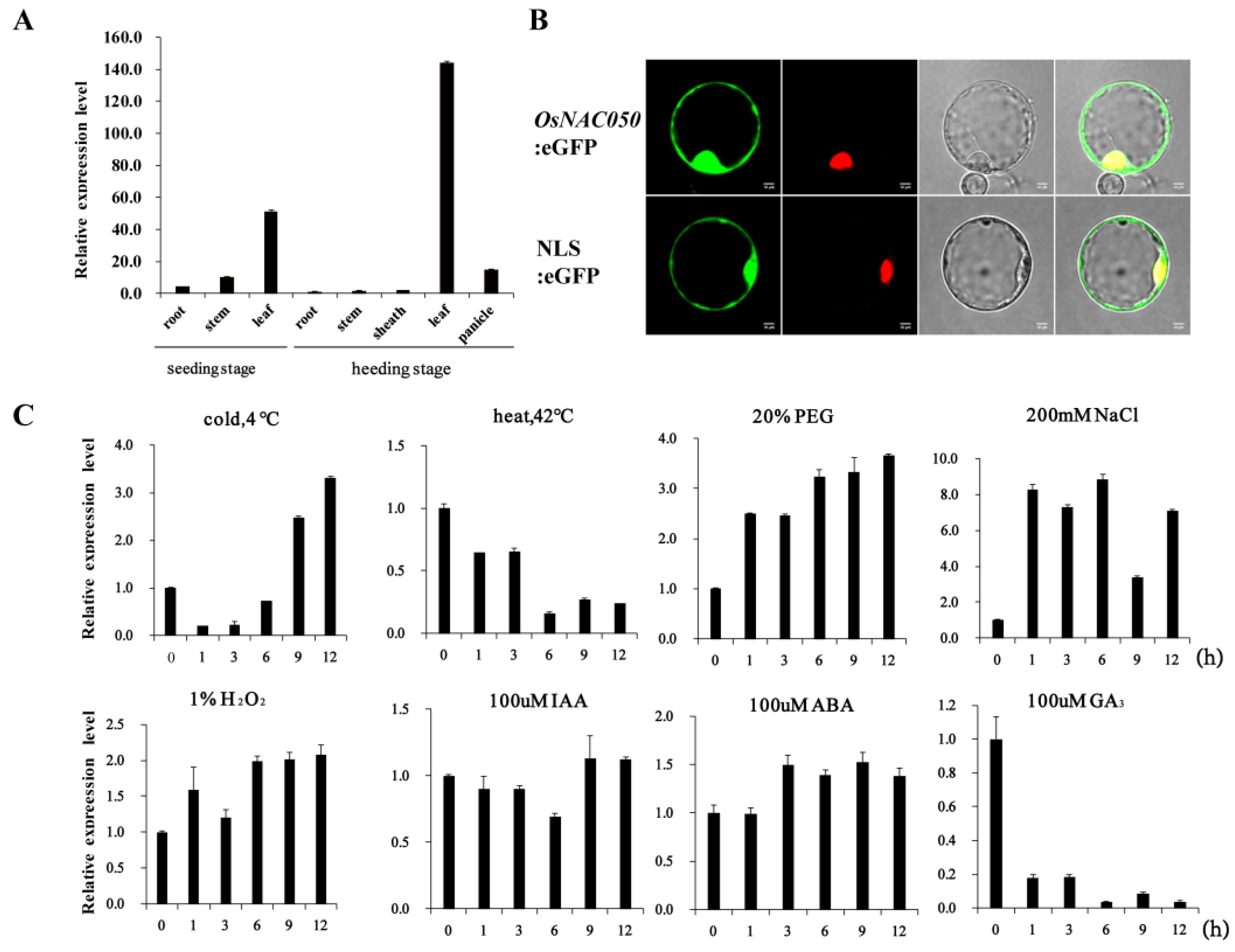
3.1. OsNAC050 Is a Cold-Inducible Transcription Factor Gene
3.2. Targeted Mutagenesis of OsNAC050
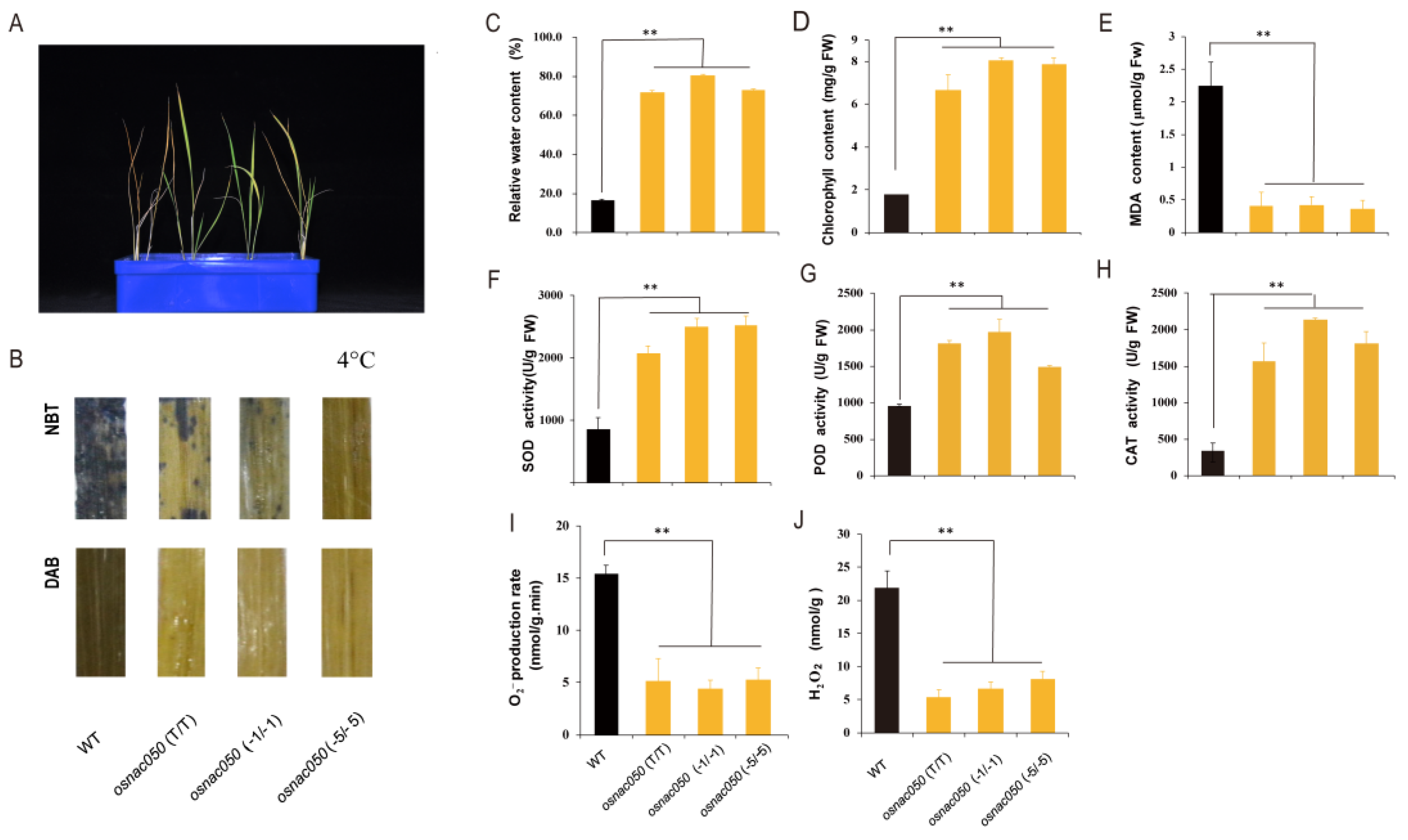
3.3. Loss of Function of OsNAC050 Increases Cold Tolerance in Rice Seedlings
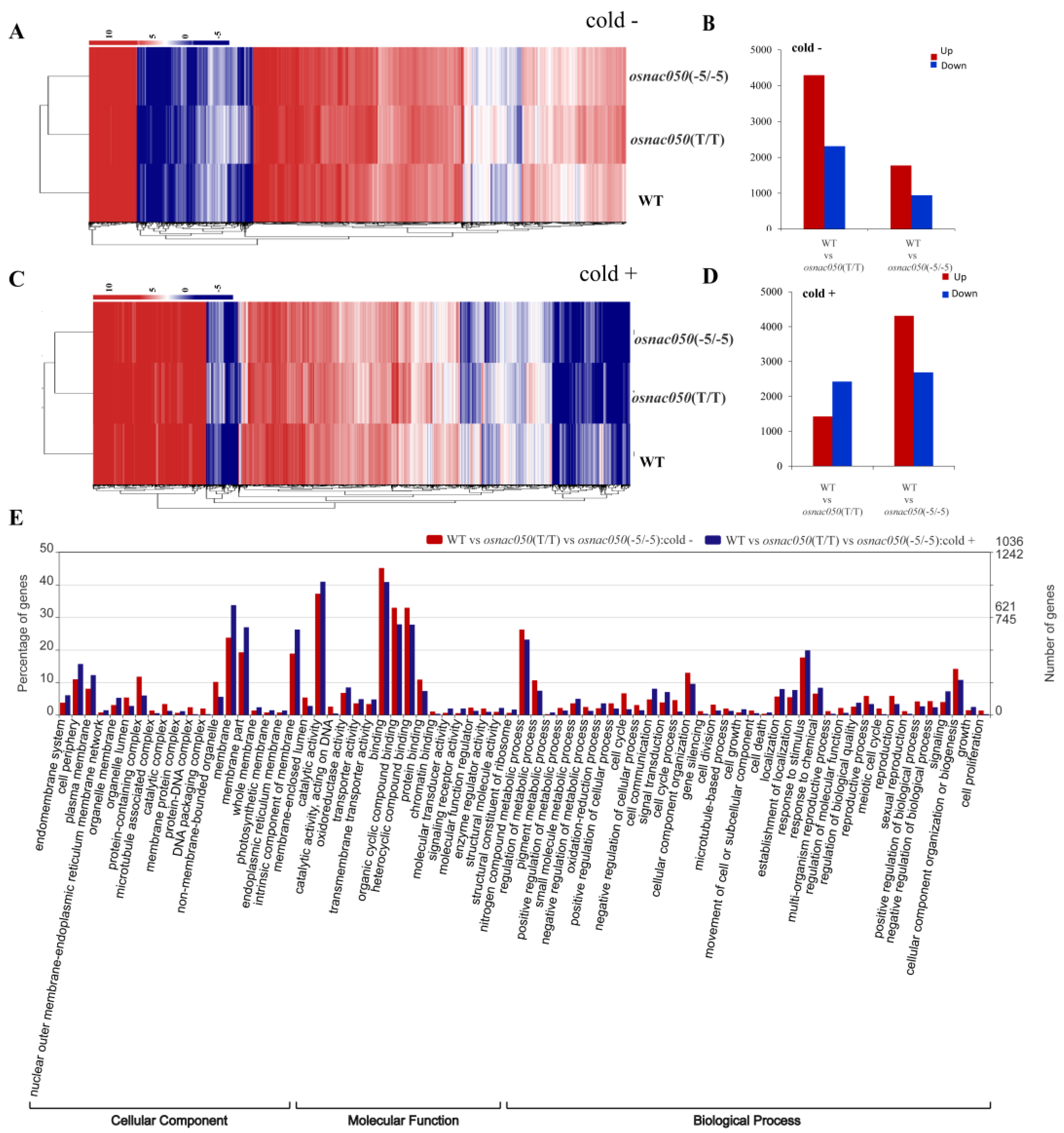
3.4. OsNAC050 Mediates Transcriptional Responses to Low-Temperature Stress
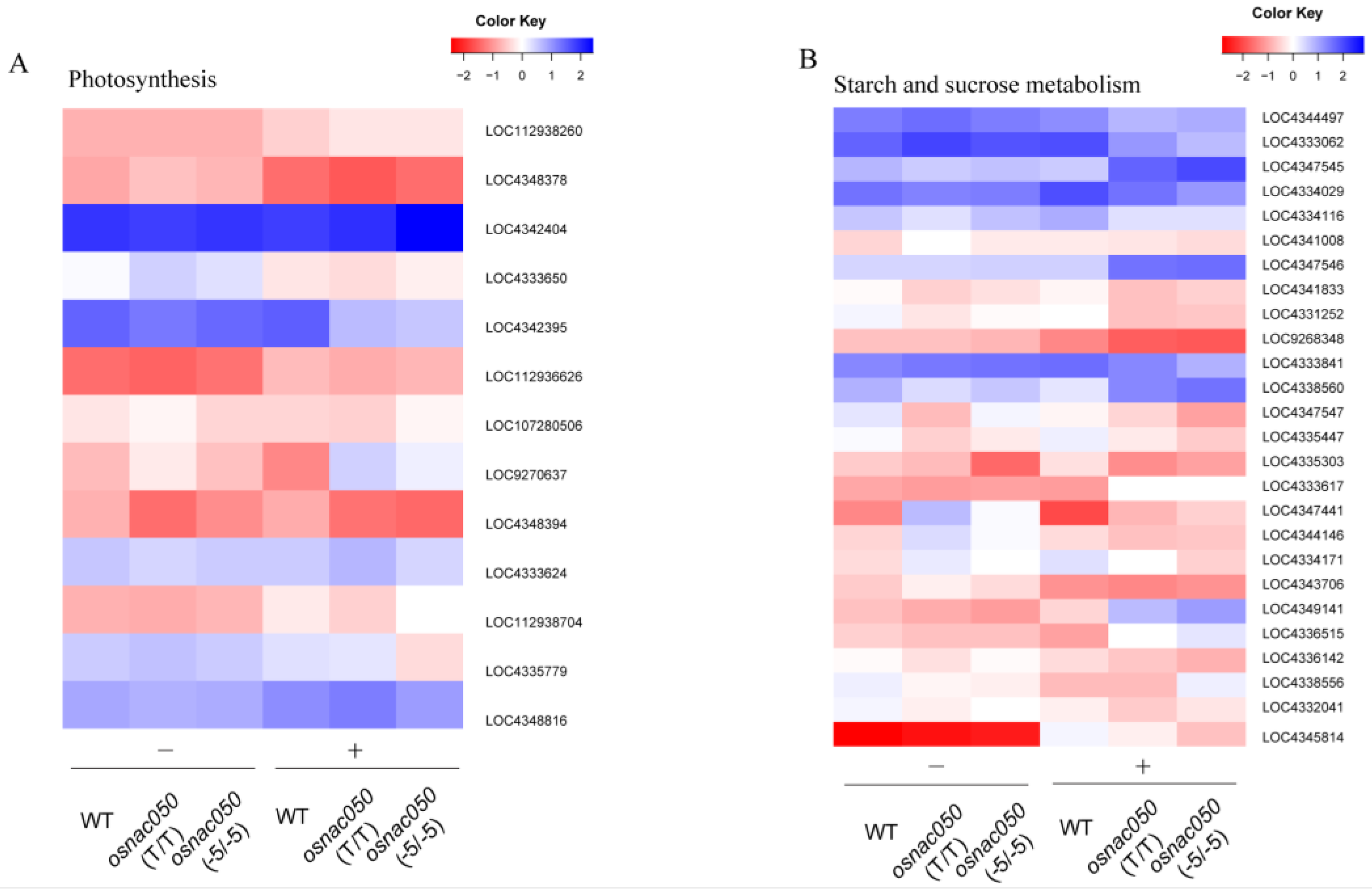
3.5. Analysis of the DEGs Identified Possible Transcription-Level Responses to Low Temperature
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, J.; Sun, Y.; Zhou, Z.; Zhang, Y.; Yang, Y.; Zan, X.; Li, X.; Wan, J.; Gao, X.; Chen, R.; et al. OsSCL30 overexpression reduces the tolerance of rice seedlings to low temperature, drought and salt. Sci. Rep. 2022, 12, 8385. [Google Scholar] [CrossRef]
- Yu, Z.; Liu, H.; Mao, S.; Zhang, J.; Zhang, J.; Yu, E.; Qu, L. Low-Temperature Thermal Desorption Effectively Mitigates Accumulation of Total Mercury and Methylmercury in Rice (Oryza sativa L.). Bull. Environ. Contam. Toxicol. 2022, 109, 757–763. [Google Scholar] [CrossRef]
- Guo, Z.; Cai, L.; Liu, C.; Chen, Z.; Guan, S.; Ma, W.; Pan, G. Low-temperature stress affects reactive oxygen species, osmotic adjustment substances, and antioxidants in rice (Oryza sativa L.) at the reproductive stage. Sci. Rep. 2022, 12, 6224. [Google Scholar] [CrossRef]
- Yang, Y.; Jiang, M.; Feng, J.; Wu, C.; Shan, W.; Kuang, J.; Chen, J.; Hu, Z.; Lu, W. Transcriptome analysis of low-temperature-affected ripening revealed MYB transcription factors-mediated regulatory network in banana fruit. Food Res. Int. 2021, 148, 110616. [Google Scholar] [CrossRef]
- Wang, W.; Du, J.; Chen, L.; Zeng, Y.; Tan, X.; Shi, Q.; Pan, X.; Wu, Z.; Zeng, Y. Transcriptomic, proteomic, and physiological comparative analyses of flooding mitigation of the damage induced by low-temperature stress in direct seeded early indica rice at the seedling stage. BMC Genom. 2021, 22, 176. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, X.; Cao, R.; Jiao, G.; Hu, S.; Shao, G.; Sheng, Z.; Xie, L.; Tang, S.; Wei, X.; et al. CDE4 encodes a pentatricopeptide repeat protein involved in chloroplast RNA splicing and affects chloroplast development under low-temperature conditions in rice. J. Integr. Plant Biol. 2021, 63, 1724–1739. [Google Scholar] [CrossRef]
- Sun, Y.; Song, K.; Guo, M.; Wu, H.; Ji, X.; Hou, L.; Liu, X.; Lu, S. A NAC Transcription Factor from ‘Sea Rice 86’ Enhances Salt Tolerance by Promoting Hydrogen Sulfide Production in Rice Seedlings. Int. J. Mol. Sci. 2022, 23, 6435. [Google Scholar] [CrossRef]
- Zhang, X.; Long, Y.; Chen, X.; Zhang, B.; Xin, Y.; Li, L.; Cao, S.; Liu, F.; Wang, Z.; Huang, H.; et al. A NAC transcription factor OsNAC3 positively regulates ABA response and salt tolerance in rice. BMC Plant Biol. 2021, 21, 546. [Google Scholar] [CrossRef]
- Liu, G.; Li, X.; Jin, S.; Liu, X.; Zhu, L.; Nie, Y.; Zhang, X. Overexpression of rice NAC gene SNAC1 improves drought and salt tolerance by enhancing root development and reducing transpiration rate in transgenic cotton. PLoS ONE 2014, 9, e86895. [Google Scholar] [CrossRef] [Green Version]
- Shen, J.; Lv, B.; Luo, L.; He, J.; Mao, C.; Xi, D.; Ming, F. The NAC-type transcription factor OsNAC2 regulates ABA-dependent genes and abiotic stress tolerance in rice. Sci. Rep. 2017, 7, 40641. [Google Scholar] [CrossRef]
- Huang, L.; Hong, Y.; Zhang, H.; Li, D.; Song, F. Rice NAC transcription factor ONAC095 plays opposite roles in drought and cold stress tolerance. BMC Plant Biol. 2016, 16, 203. [Google Scholar] [CrossRef] [Green Version]
- El Mannai, Y.; Akabane, K.; Hiratsu, K.; Satoh-Nagasawa, N.; Wabiko, H. The NAC Transcription Factor Gene OsY37 (ONAC011) Promotes Leaf Senescence and Accelerates Heading Time in Rice. Int. J. Mol. Sci. 2017, 18, 2165. [Google Scholar] [CrossRef] [Green Version]
- Takasaki, H.; Maruyama, K.; Kidokoro, S.; Ito, Y.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K.; Nakashima, K. The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol. Genet. Genom. 2010, 284, 173–183. [Google Scholar] [CrossRef]
- Sun, X.; Xiong, H.; Jiang, C.; Zhang, D.; Yang, Z.; Huang, Y.; Zhu, W.; Ma, S.; Duan, J.; Wang, X.; et al. Natural variation of DROT1 confers drought adaptation in upland rice. Nat. Commun. 2022, 13, 4265. [Google Scholar] [CrossRef]
- Ganie, S.A.; Reddy, A.S.N. Stress-Induced Changes in Alternative Splicing Landscape in Rice: Functional Significance of Splice Isoforms in Stress Tolerance. Biology 2021, 10, 309. [Google Scholar] [CrossRef]
- Li, N.; Liu, H.; Sun, J.; Zheng, H.; Wang, J.; Yang, L.; Zhao, H.; Zou, D. Transcriptome analysis of two contrasting rice cultivars during alkaline stress. Sci. Rep. 2018, 8, 9586. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Li, J.; Liu, Y.; Noman, A.; Chen, L.; Liu, J. Transcriptome profiling in rice reveals a positive role for OsNCED3 in defense against the brown planthopper, Nilaparvata lugens. BMC Genom. 2022, 23, 634. [Google Scholar] [CrossRef]
- Yang, D.; Li, S.; Xiao, Y.; Lu, L.; Zheng, Z.; Tang, D.; Cui, H. Transcriptome analysis of rice response to blast fungus identified core genes involved in immunity. Plant Cell Environ. 2021, 44, 3103–3121. [Google Scholar] [CrossRef]
- Qian, G.; Wang, M.; Wang, X.; Liu, K.; Li, Y.; Bu, Y.; Li, L. Integrated Transcriptome and Metabolome Analysis of Rice Leaves Response to High Saline-Alkali Stress. Int. J. Mol. Sci. 2023, 24, 4062. [Google Scholar] [CrossRef]
- Illa-Berenguer, E.; LaFayette, P.R.; Parrott, W.A. Editing efficiencies with Cas9 orthologs, Cas12a endonucleases, and temperature in rice. Front. Genome Ed. 2023, 5, 1074641. [Google Scholar] [CrossRef]
- Tang, X.; Zheng, X.; Qi, Y.; Zhang, D.; Cheng, Y.; Tang, A.; Voytas, D.F.; Zhang, Y. A Single Transcript CRISPR-Cas9 System for Efficient Genome Editing in Plants. Mol. Plant 2016, 9, 1088–1091. [Google Scholar] [CrossRef] [Green Version]
- Zhong, Z.; Zhang, Y.; You, Q.; Tang, X.; Ren, Q.; Liu, S.; Yang, L.; Wang, Y.; Liu, X.; Liu, B.; et al. Plant Genome Editing Using FnCpf1 and LbCpf1 Nucleases at Redefined and Altered PAM Sites. Mol. Plant 2018, 11, 999–1002. [Google Scholar] [CrossRef] [Green Version]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [Green Version]
- Zhao, F.; Zhao, T.; Deng, L.; Lv, D.; Zhang, X.; Pan, X.; Xu, J.; Long, G. Visualizing the Essential Role of Complete Virion Assembly Machinery in Efficient Hepatitis C Virus Cell-to-Cell Transmission by a Viral Infection-Activated Split-Intein-Mediated Reporter System. J. Virol. 2017, 91, 10–1128. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Zhong, Z.; Wang, X.; Han, X.; Yu, D.; Wang, C.; Song, W.; Zheng, X.; Chen, C.; Zhang, Y. Knockout of the OsNAC006 Transcription Factor Causes Drought and Heat Sensitivity in Rice. Int. J. Mol. Sci. 2020, 21, 2288. [Google Scholar] [CrossRef] [Green Version]
- Yarra, R.; Wei, W. The NAC-type transcription factor GmNAC20 improves cold, salinity tolerance, and lateral root formation in transgenic rice plants. Funct. Integr. Genom. 2021, 21, 473–487. [Google Scholar] [CrossRef]
- Ren, Y.; Huang, Z.; Jiang, H.; Wang, Z.; Wu, F.; Xiong, Y.; Yao, J. A heat stress responsive NAC transcription factor heterodimer plays key roles in rice grain filling. J. Exp. Bot. 2021, 72, 2947–2964. [Google Scholar] [CrossRef]
- Mao, C.; Lu, S.; Lv, B.; Zhang, B.; Shen, J.; He, J.; Luo, L.; Xi, D.; Chen, X.; Ming, F. A Rice NAC Transcription Factor Promotes Leaf Senescence via ABA Biosynthesis. Plant Physiol. 2017, 174, 1747–1763. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.H.; Lyu, Y.S.; Yang, W.; Yang, Z.T.; Lu, S.J.; Liu, J.X. A membrane-associated NAC transcription factor OsNTL3 is involved in thermotolerance in rice. Plant Biotechnol. J. 2020, 18, 1317–1329. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.; Shi, Z.; Zhang, S.; Wang, Y.; Xia, X.; Jiang, Y.; Gull, S.; Chen, L.; Guo, H.; Wu, T.; et al. Methylation and expression of rice NLR genes after low temperature stress. Gene 2022, 845, 146830. [Google Scholar] [CrossRef]
- Kang, M.; Liu, G.; Zeng, Y.; Zhou, J.; Shi, J.; Tang, L.; Liu, L.; Cao, W.; Zhu, Y.; Liu, B. Extreme Low-Temperature Stress Affects Nutritional Quality of Amino Acids in Rice. Front. Plant Sci. 2022, 13, 905348. [Google Scholar] [CrossRef]
- Joo, J.Y.; Kim, M.S.; Cho, Y.G.; Fernie, A.R.; Sung, J. Transcriptional Comparison of Genes Associated with Photosynthesis, Photorespiration, and Photo-Assimilate Allocation and Metabolic Profiling of Rice Species. Int. J. Mol. Sci. 2022, 23, 8901. [Google Scholar] [CrossRef]
- Xiong, H.; Hua, L.; Reyna-Llorens, I.; Shi, Y.; Chen, K.M.; Smirnoff, N.; Kromdijk, J.; Hibberd, J.M. Photosynthesis-independent production of reactive oxygen species in the rice bundle sheath during high light is mediated by NADPH oxidase. Proc. Natl. Acad. Sci. USA 2021, 118, e2022702118. [Google Scholar] [CrossRef]
- Gong, B.; Li, X.; van den Langenberg, K.M.; Wen, D.; Sun, S.; Wei, M.; Li, Y.; Yang, F.; Shi, Q.; Wang, X. Overexpression of S-adenosyl-L-methionine synthetase increased tomato tolerance to alkali stress through polyamine metabolism. Plant Biotechnol. J. 2014, 12, 694–708. [Google Scholar] [CrossRef]
- Liu, X.; Quan, W.; Bartels, D. Stress memory responses and seed priming correlate with drought tolerance in plants: An overview. Planta 2022, 255, 45. [Google Scholar] [CrossRef]
- Bao, G.; Zhuo, C.; Qian, C.; Xiao, T.; Guo, Z.; Lu, S. Co-expression of NCED and ALO improves vitamin C level and tolerance to drought and chilling in transgenic tobacco and stylo plants. Plant Biotechnol. J. 2016, 14, 206–214. [Google Scholar] [CrossRef]
- Ramanjulu, S.; Bartels, D. Drought- and desiccation-induced modulation of gene expression in plants. Plant Cell Environ. 2002, 25, 141–151. [Google Scholar] [CrossRef]
- Mohamadi Esboei, M.; Ebrahimi, A.; Amerian, M.R.; Alipour, H. Melatonin confers fenugreek tolerance to salinity stress by stimulating the biosynthesis processes of enzymatic, non-enzymatic antioxidants, and diosgenin content. Front. Plant Sci. 2022, 13, 890613. [Google Scholar] [CrossRef]
- Jiang, Z.; Chen, Q.; Chen, L.; Yang, H.; Zhu, M.; Ding, Y.; Li, W.; Liu, Z.; Jiang, Y.; Li, G. Efficiency of Sucrose to Starch Metabolism Is Related to the Initiation of Inferior Grain Filling in Large Panicle Rice. Front. Plant Sci. 2021, 12, 732867. [Google Scholar] [CrossRef]
- Toczewska, J.; Zalewska, A.; Konopka, T.; Maciejczyk, M. Enzymatic Antioxidants Activity in Gingival Crevicular Fluid and Saliva in Advanced Periodontitis. In Oral Diseases; Wiley Online Library: Hoboken, NJ, USA, 2022. [Google Scholar]
- Di, Q.; Li, Y.; Li, S.; Shi, A.; Zhou, M.; Ren, H.; Yan, Y.; He, C.; Wang, J.; Sun, M.; et al. Photosynthesis Mediated by RBOH-Dependent Signaling Is Essential for Cold Stress Memory. Antioxidants 2022, 11, 969. [Google Scholar] [CrossRef]
- Wang, Z.; Sun, J.; Zu, X.; Gong, J.; Deng, H.; Hang, R.; Zhang, X.; Liu, C.; Deng, X.; Luo, L.; et al. Pseudouridylation of chloroplast ribosomal RNA contributes to low temperature acclimation in rice. New Phytol. 2022, 236, 1708–1720. [Google Scholar] [CrossRef]
- Li, F.; Deng, H.; Wang, Y.; Li, X.; Chen, X.; Liu, L.; Zhang, H. Potato growth, photosynthesis, yield, and quality response to regulated deficit drip irrigation under film mulching in a cold and arid environment. Sci. Rep. 2021, 11, 15888. [Google Scholar] [CrossRef]
- Szechynska-Hebda, M.; Wasek, I.; Golebiowska-Pikania, G.; Dubas, E.; Zur, I.; Wedzony, M. Photosynthesis-dependent physiological and genetic crosstalk between cold acclimation and cold-induced resistance to fungal pathogens in triticale (Triticosecale Wittm.). J. Plant Physiol. 2015, 177, 30–43. [Google Scholar] [CrossRef]
- Lu, G.; Wang, L.; Zhou, L.; Su, X.; Guo, H.; Cheng, H. Overexpression of AmCBF1 enhances drought and cold stress tolerance, and improves photosynthesis in transgenic cotton. PeerJ 2022, 10, e13422. [Google Scholar] [CrossRef]
- Gai, Z.; Liu, L.; Zhang, J.; Liu, J.; Cai, L. Effects of exogenous alpha-oxoglutarate on proline accumulation, ammonium assimilation and photosynthesis of soybean seedling (Glycine max (L.) Merr.) exposed to cold stress. Sci. Rep. 2020, 10, 17017. [Google Scholar] [CrossRef]
- Yamori, W.; Sakata, N.; Suzuki, Y.; Shikanai, T.; Makino, A. Cyclic electron flow around photosystem I via chloroplast NAD(P)H dehydrogenase (NDH) complex performs a significant physiological role during photosynthesis and plant growth at low temperature in rice. Plant J. 2011, 68, 966–976. [Google Scholar] [CrossRef]
- Lopez-Gonzalez, C.; Juarez-Colunga, S.; Trachsel, S.; Marsch-Martinez, N.; Gillmor, C.S.; Tiessen, A. Analysis of Global Gene Expression in Maize (Zea mays) Vegetative and Reproductive Tissues That Differ in Accumulation of Starch and Sucrose. Plants 2022, 11, 238. [Google Scholar] [CrossRef]
- Apriyanto, A.; Compart, J.; Zimmermann, V.; Alseekh, S.; Fernie, A.R.; Fettke, J. Indication that starch and sucrose are biomarkers for oil yield in oil palm (Elaeis guineensis Jacq.). Food Chem. 2022, 393, 133361. [Google Scholar] [CrossRef]
- Yu, J.; Xu, S.; Liu, X.; Li, T.; Zhang, D.; Teng, N.; Wu, Z. Starch Degradation and Sucrose Accumulation of Lily Bulbs after Cold Storage. Int. J. Mol. Sci. 2022, 23, 4366. [Google Scholar] [CrossRef]
- Xie, F.; Zhang, H.; Xiong, Z.; Wu, Y.; Ai, L. Effects and mechanism of sucrose on retrogradation, freeze-thaw stability, and texture of corn starch-tamarind seed polysaccharide complexes. J. Food Sci. 2022, 87, 623–635. [Google Scholar] [CrossRef]
- Shi, W.; Ma, Q.; Yin, W.; Liu, T.; Song, Y.; Chen, Y.; Song, L.; Sun, H.; Hu, S.; Liu, T.; et al. The transcription factor StTINY3 enhances cold-induced sweetening resistance by coordinating starch resynthesis and sucrose hydrolysis in potato. J. Exp. Bot. 2022, 73, 4968–4980. [Google Scholar] [CrossRef]
- Nilholm, C.; Manoharan, L.; Roth, B.; D’Amato, M.; Ohlsson, B. A starch- and sucrose-reduced dietary intervention in irritable bowel syndrome patients produced a shift in gut microbiota composition along with changes in phylum, genus, and amplicon sequence variant abundances, without affecting the micro-RNA levels. United Eur. Gastroenterol. J. 2022, 10, 363–375. [Google Scholar] [CrossRef]
- Roth, B.; Myllyvainio, J.; D’Amato, M.; Larsson, E.; Ohlsson, B. A Starch- and Sucrose-Reduced Diet in Irritable Bowel Syndrome Leads to Lower Circulating Levels of PAI-1 and Visfatin: A Randomized Controlled Study. Nutrients 2022, 14, 1688. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, B.; Wang, Y.; Xie, L.; Yu, W.; Lan, Q.; Wang, Y.; Chen, C.; Zhang, Y. Knocking out OsNAC050 Expression Causes Low-Temperature Tolerance in Rice by Regulating Photosynthesis and the Sucrose Metabolic Pathway. Agriculture 2023, 13, 1378. https://doi.org/10.3390/agriculture13071378
Wang B, Wang Y, Xie L, Yu W, Lan Q, Wang Y, Chen C, Zhang Y. Knocking out OsNAC050 Expression Causes Low-Temperature Tolerance in Rice by Regulating Photosynthesis and the Sucrose Metabolic Pathway. Agriculture. 2023; 13(7):1378. https://doi.org/10.3390/agriculture13071378
Chicago/Turabian StyleWang, Bo, Yiheng Wang, Likun Xie, Wancong Yu, Qingkuo Lan, Yong Wang, Chengbin Chen, and Yong Zhang. 2023. "Knocking out OsNAC050 Expression Causes Low-Temperature Tolerance in Rice by Regulating Photosynthesis and the Sucrose Metabolic Pathway" Agriculture 13, no. 7: 1378. https://doi.org/10.3390/agriculture13071378
APA StyleWang, B., Wang, Y., Xie, L., Yu, W., Lan, Q., Wang, Y., Chen, C., & Zhang, Y. (2023). Knocking out OsNAC050 Expression Causes Low-Temperature Tolerance in Rice by Regulating Photosynthesis and the Sucrose Metabolic Pathway. Agriculture, 13(7), 1378. https://doi.org/10.3390/agriculture13071378





