Transcriptome-Wide Identification of Cytochrome P450s and GSTs from Spodoptera exigua Reveals Candidate Genes Involved in Camptothecin Detoxification
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals, Cell Line, and Culture Conditions
2.2. CPT Treatment of S. exigua Fat Body Cells
2.3. RNA Extraction, Library Construction and Sequencing
2.4. Identification of Detoxification Genes and Phylogenetic Analysis
2.5. Real-Time Quantitative PCR (RT-qPCR)
2.6. Effect of CPT on the Growth and Development of S. exigua Larvae
2.7. Effect of CPT on the Expression of Detoxification Genes in S. exigua Larvae
3. Results
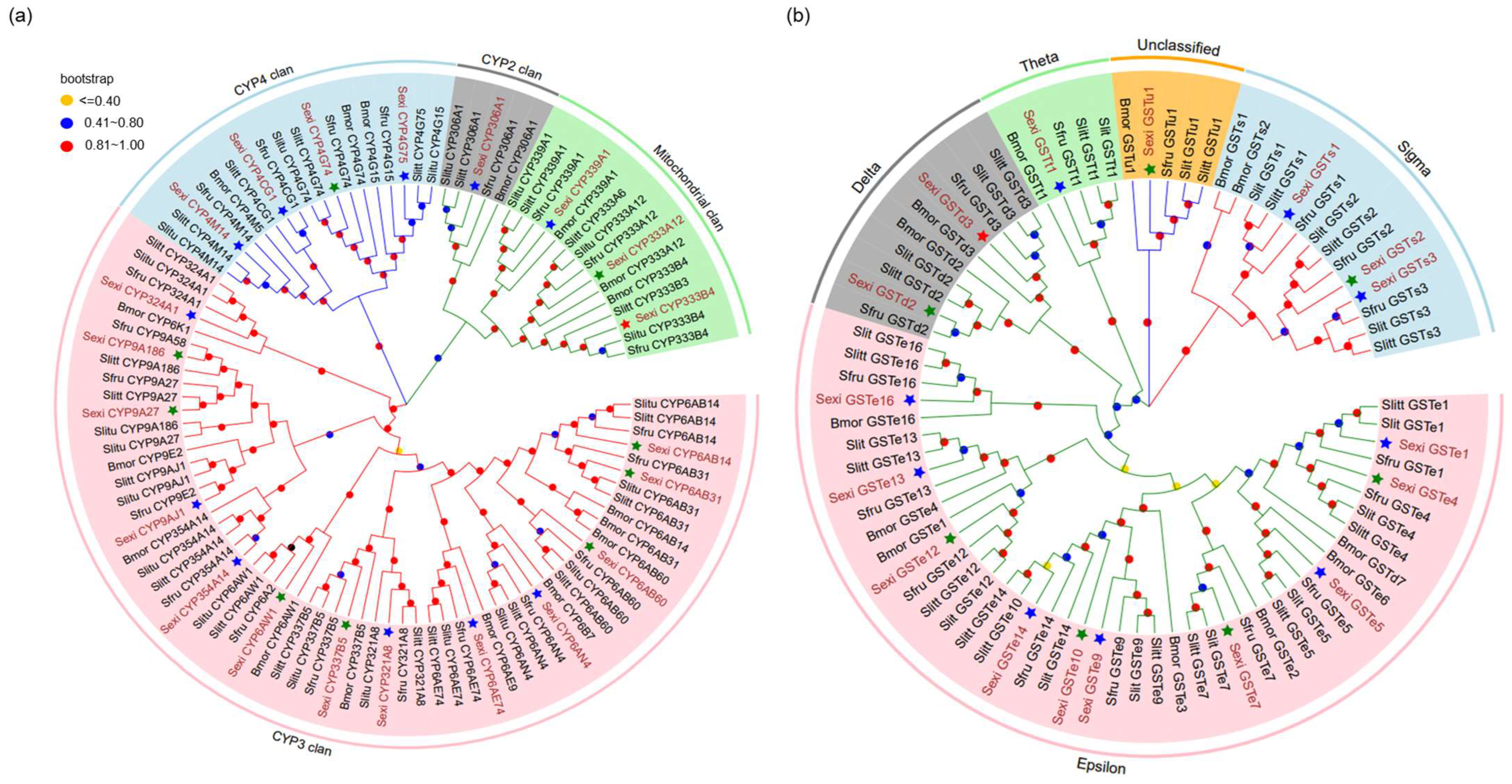
3.1. Identification of Candidate P450s and GSTs from the Fat Body Cells of S. exigua
3.2. Real-Time Quantitative PCR Validation of Candidate Genes from the Fat Body Cells of S. exigua
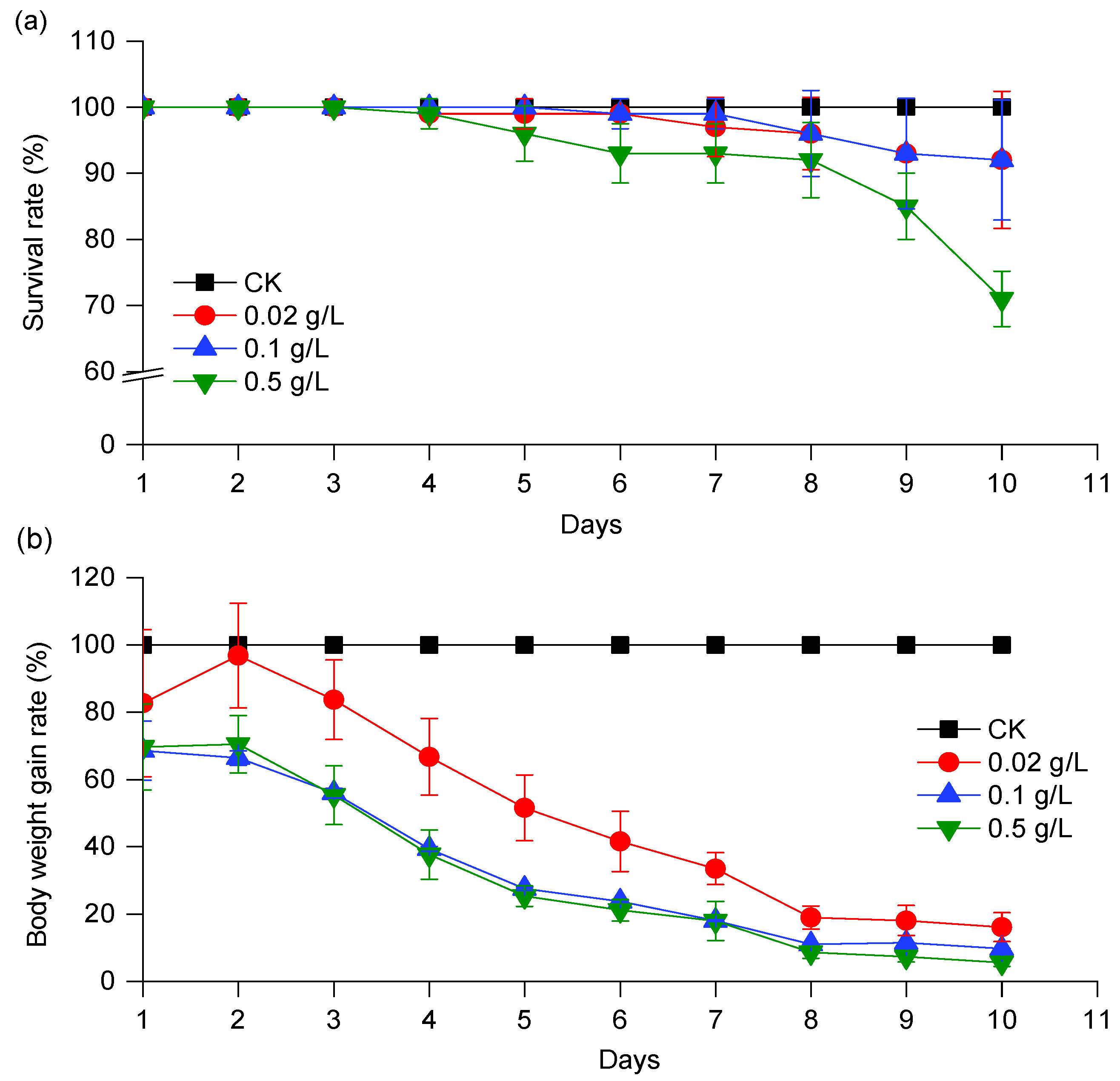
3.3. Effects of CPT on the Development of S. exigua Larvae
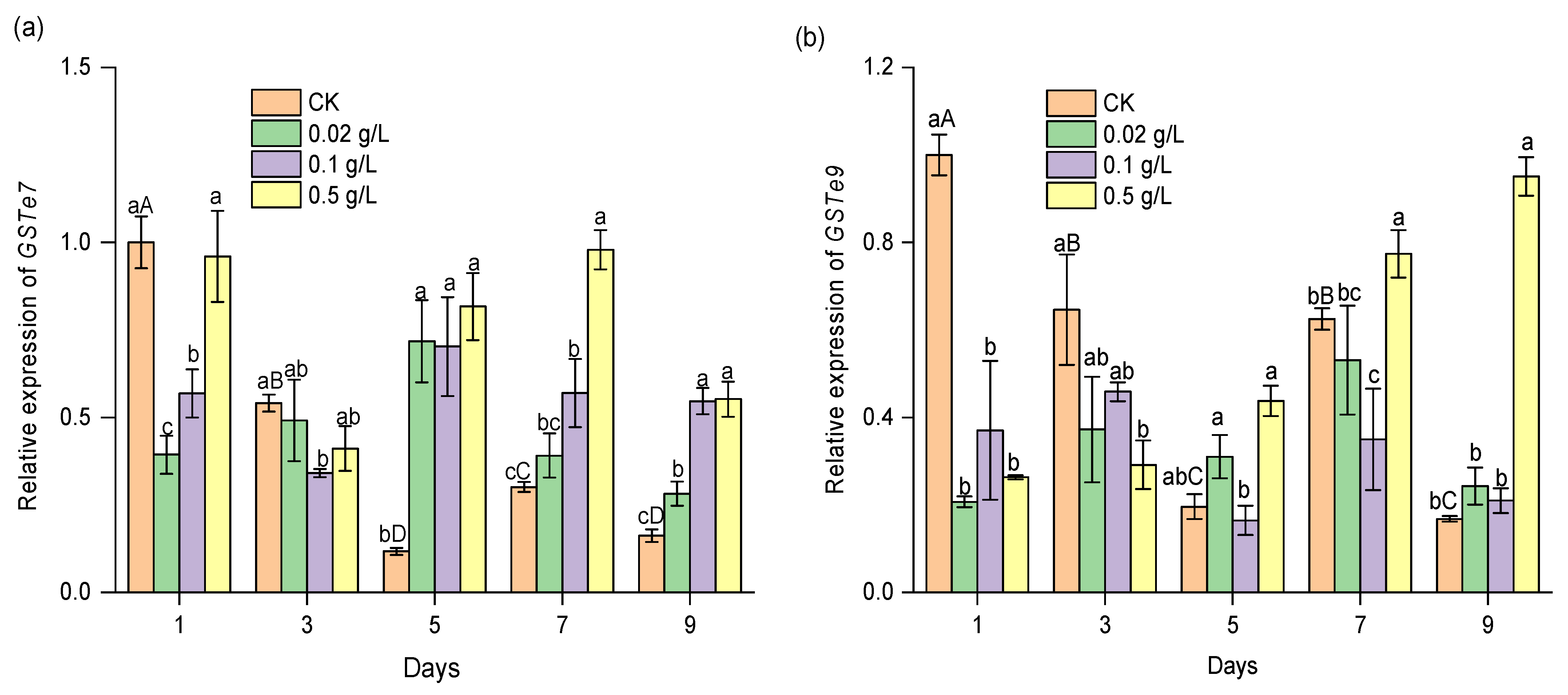
3.4. Effect of CPT on the Expression of CYP and GST Genes during the Development of S. exigua Larvae
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Toffoli, G.; Cecchin, E.; Corona, G.; Boiocchi, M. Pharmacogenetics of irinotecan. Curr. Med. Chem. Anticancer. Agents 2003, 3, 225–237. [Google Scholar] [CrossRef]
- Maier-Salamon, A.; Thalhammer, T.; Reznicek, G.; Böhmdorfer, M.; Zupkó, I.; Hartl, A.; Jaeger, W. Cytochrome P450 3A-mediated metabolism of the topoisomerase I inhibitor 9-aminocamptothecin: Impact on cancer therapy. Int. J. Oncol. 2014, 45, 877–886. [Google Scholar]
- Horn, J.; Milewska, M.; Arnold, S.M.; Leggas, M. Metabolic pathways of the camptothecin analog AR-67. Drug. Metab. Dispos. 2011, 39, 683–692. [Google Scholar]
- Bao, H.; Zhang, Q.; Yan, Z. The impact of camptothecin-encapsulated poly (lactic-co-glycolic acid) nanoparticles on the activity of cytochrome P450 in vitro. Int. J. Nanomed. 2019, 14, 383–391. [Google Scholar]
- Negoro, R.; Yamada, N.; Watanabe, K.; Kono, Y.; Fujita, T. Generation of Caco-2 cells stably expressing CYP3A4·POR·UGT1A1 and CYP3A4·POR·UGT1A1*6 using a PITCh system. Arch. Toxicol. 2022, 96, 499–510. [Google Scholar]
- Yang, F.; Wang, L.; Zhang, L.; Zhang, Y.; Mao, L.; Jiang, H. Synthesis and biological activities of two camptothecin derivatives against Spodoptera exigua. Sci. Rep. 2019, 9, 18067. [Google Scholar] [PubMed]
- Yang, F.; Zhang, L.; Zhang, Y.; Mao, L.; Zhu, L.; Liu, X.; Jiang, H. Effect of RNAi targeting SeGrx1 on the cytotoxicity and insecticide susceptibility of camptothecin in Spodoptera exigua. Agriculture 2022, 12, 930. [Google Scholar] [CrossRef]
- Dai, L.S.; Tian, H.F.; Hang, Y.; Wen, C.W.; Huang, Y.H.; Wang, B.F.; Hu, J.W.; Xu, J.P.; Deng, M.J. 1H NMR-based metabonomic evaluation of the pesticides camptothecin and matrine against larvae of Spodoptera litura. Pest Manag. Sci. 2021, 77, 208–216. [Google Scholar]
- Ngegba, P.M.; Cui, G.; Li, Y.; Zhong, G. Synergistic effects of chlorantraniliprole and camptothecin on physiological impairments, histopathological, biochemical changes, and genes responses in the larvae midgut of Spodoptera frugiperda. Pestic. Biochem. Physiol. 2023, 191, 105363. [Google Scholar]
- Ma, J.; Tong, S.; Wang, P.; Liao, W.; Liu, H.; Zhang, L. Insecticidal activity of camptothecin against Nilaparvata lugens, Brevicoryne brassicae, and Chilo suppressalis. J. Econ. Entomol. 2010, 103, 492–496. [Google Scholar]
- Zhang, L.; Zhang, Y.; He, W.; Ma, D.; Jiang, H. Effects of camptothecin and hydroxycamptothecin on insect cell lines Sf21 and IOZCAS-Spex-II. Pest Manag. Sci. 2012, 68, 652–657. [Google Scholar]
- Ren, X.; Zhang, L.; Zhang, Y.; Mao, L.; Jiang, H. Mitochondria response to camptothecin and hydroxycamptothecine-induced apoptosis in Spodoptera exigua cells. Pestic. Biochem. Physiol. 2017, 140, 97–104. [Google Scholar] [PubMed]
- Ren, X.; Zhang, L.; Zhang, Y.; Mao, L.; Jiang, H. Oxidative stress induced by camptothecin and hydroxyl-camptothecin in IOZCAS-Spex-II cells of Spodoptera exigua Hübner. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2019, 216, 52–59. [Google Scholar] [PubMed]
- Wang, L.; Li, Z.; Zhang, L.; Zhang, Y.; Mao, L.; Jiang, H. Synthesis, insecticidal activity and inhibition on topoisomerase I of 20(S)-t-Boc-amino acid derivatives of camptothecin. Pestic. Biochem. Physiol. 2017, 139, 46–52. [Google Scholar] [PubMed]
- Partridge, F.A.; Poulton, B.C.; Lake, M.A.I.; Lees, R.A.; Mann, H.J.; Lycett, G.J.; Sattelle, D.B. Actions of camptothecin derivatives on larvae and adults of the arboviral vector Aedes aegypti. Molecules 2021, 26, 6226. [Google Scholar]
- Jiang, Z.; Zhang, Z.; Cui, G.; Sun, Z.; Song, G.; Liu, Y.; Zhong, G. DNA Topoisomerase 1 structure-BASED design, synthesis, activity evaluation and molecular simulations study of new 7-amide camptothecin derivatives against Spodoptera frugiperda. Front. Chem. 2018, 6, 456. [Google Scholar]
- Zhang, Z.J.; Shang, X.F.; Yang, L.; Shi, Y.B.; Liu, Y.Q.; Li, J.C.; Yang, G.Z.; Yang, C.J. Engineering of peglayted camptothecin into nanomicelles and supramolecular hydrogels for pesticide combination control. Front. Chem. 2020, 7, 922. [Google Scholar]
- Amezian, D.; Nauen, R.; Le Goff, G. Comparative analysis of the detoxification gene inventory of four major Spodoptera pest species in response to xenobiotics. Insect Biochem. Mol. Biol. 2021, 138, 103646. [Google Scholar] [PubMed]
- Lu, K.; Song, Y.; Zeng, R. The role of cytochrome P450-mediated detoxification in insect adaptation to xenobiotics. Curr. Opin. Insect. Sci. 2021, 43, 103–107. [Google Scholar]
- Hu, C.; Liu, J.Y.; Wang, W.; Mota-Sanchez, D.; He, S.; Shi, Y.; Yang, X.Q. Glutathione S-transferase genes are involved in lambda-cyhalothrin resistance in Cydia pomonella via sequestration. J. Agric. Food. Chem. 2022, 70, 2265–2279. [Google Scholar]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [PubMed]
- Yu, H.; Yang, X.; Dai, J.; Li, Y.; Veeran, S.; Lin, J.; Shu, B. Effects of azadirachtin on detoxification-related gene expression in the fat bodies of the fall armyworm, Spodoptera frugiperda. Environ. Sci. Pollut. Res. Int. 2023, 30, 42587–42595. [Google Scholar] [PubMed]
- Dermauw, W.; Van Leeuwen, T.; Feyereisen, R. Diversity and evolution of the P450 family in arthropods. Insect Biochem. Mol. Biol. 2020, 127, 103490. [Google Scholar]
- Feyereisen, R. Origin and evolution of the CYP4G subfamily in insects, cytochrome P450 enzymes involved in cuticular hydrocarbon synthesis. Mol. Phylogenet. Evol. 2020, 143, 106695. [Google Scholar]
- Wang, Z.G.; Jiang, S.S.; Mota-Sanchez, D.; Wang, W.; Li, X.R.; Gao, Y.L.; Lu, X.P.; Yang, X.Q. Cytochrome P450-mediated λ-cyhalothrin-resistance in a field strain of Helicoverpa armigera from northeast China. J. Agric. Food Chem. 2019, 67, 3546–3553. [Google Scholar]
- Li, P.R.; Shi, Y.; Ju, D.; Liu, Y.X.; Wang, W.; He, Y.S.; Zhang, Y.Y.; Yang, X.Q. Metabolic functional redundancy of the CYP9A subfamily members leads to P450-mediated lambda-cyhalothrin resistance in Cydia pomonella. Pest Manag. Sci. 2023, 79, 1452–1466. [Google Scholar]
- Zhu, W.; Yu, R.; Wu, H.; Zhang, X.; Liu, Y.; Zhu, K.Y.; Zhang, J.; Ma, E. Identification and characterization of two CYP9A genes associated with pyrethroid detoxification in Locusta migratoria. Pestic. Biochem. Physiol. 2016, 132, 65–71. [Google Scholar]
- Teng, H.; Zuo, Y.; Yuan, J.; Fabrick, J.A.; Wu, Y.; Yang, Y. High frequency of ryanodine receptor and cytochrome P450 CYP9A186 mutations in insecticide-resistant field populations of Spodoptera exigua from China. Pestic. Biochem. Physiol. 2022, 186, 105153. [Google Scholar]
- Zhang, B.Z.; Xu, Z.; Zhen, C.A.; Lu, L.Y.; Li, Y.S.; Ge, X.; Chen, D.M.; Pei, Z.; Shi, M.W.; Chen, X.L. Silencing of cytochrome P450 in Spodoptera frugiperda (Lepidoptera: Noctuidae) by RNA interference enhances susceptibility to chlorantraniliprole. J. Insect Sci. 2020, 20, 12. [Google Scholar]
- Zuo, Y.; Shi, Y.; Zhang, F.; Guan, F.; Zhang, J.; Feyereisen, R.; Fabrick, J.A.; Yang, Y.; Wu, Y. Genome mapping coupled with CRISPR gene editing reveals a P450 gene confers avermectin resistance in the beet armyworm. PLoS Genet. 2021, 17, e1009680. [Google Scholar]
- Koirala, B.K.S.; Moural, T.; Zhu, F. Functional and structural diversity of insect glutathione S-transferases in xenobiotic adaptation. Int. J. Biol. Sci. 2022, 18, 5713–5723. [Google Scholar]
- Meng, X.; Wu, Z.; Jiang, C.; Guan, D.; Zhang, N.; Jiang, H.; Shen, Q.; Qian, K.; Wang, J. Identification and characterization of glutathione S-transferases and their potential roles in detoxification of abamectin in the rice stem borer, Chilo suppressalis. Pestic. Biochem. Physiol. 2022, 182, 105050. [Google Scholar] [PubMed]
- Zhang, J.; Ma, W.; Yin, F.; Park, Y.; Zhu, K.Y.; Zhang, X.; Qin, X.; Li, D. Evaluations of two glutathione S-transferase epsilon genes for their contributions to metabolism of three selected insecticides in Locusta migratoria. Pestic. Biochem. Physiol. 2022, 183, 105084. [Google Scholar] [PubMed]
- Kouamo, M.F.M.; Ibrahim, S.S.; Hearn, J.; Riveron, J.M.; Kusimo, M.; Tchouakui, M.; Ebai, T.; Tchapga, W.; Wondji, M.J.; Irving, H.; et al. Genome-wide transcriptional analysis and functional validation linked a cluster of epsilon glutathione S-transferases with insecticide resistance in the major malaria vector Anopheles funestus across Africa. Genes 2021, 12, 561. [Google Scholar] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Z.; Zhang, L.; Zhang, Y.; Mao, L.; Zhu, L.; Liu, X.; Jiang, H. Transcriptome-Wide Identification of Cytochrome P450s and GSTs from Spodoptera exigua Reveals Candidate Genes Involved in Camptothecin Detoxification. Agriculture 2023, 13, 1494. https://doi.org/10.3390/agriculture13081494
Zhao Z, Zhang L, Zhang Y, Mao L, Zhu L, Liu X, Jiang H. Transcriptome-Wide Identification of Cytochrome P450s and GSTs from Spodoptera exigua Reveals Candidate Genes Involved in Camptothecin Detoxification. Agriculture. 2023; 13(8):1494. https://doi.org/10.3390/agriculture13081494
Chicago/Turabian StyleZhao, Zhenzhen, Lan Zhang, Yanning Zhang, Liangang Mao, Lizhen Zhu, Xingang Liu, and Hongyun Jiang. 2023. "Transcriptome-Wide Identification of Cytochrome P450s and GSTs from Spodoptera exigua Reveals Candidate Genes Involved in Camptothecin Detoxification" Agriculture 13, no. 8: 1494. https://doi.org/10.3390/agriculture13081494
APA StyleZhao, Z., Zhang, L., Zhang, Y., Mao, L., Zhu, L., Liu, X., & Jiang, H. (2023). Transcriptome-Wide Identification of Cytochrome P450s and GSTs from Spodoptera exigua Reveals Candidate Genes Involved in Camptothecin Detoxification. Agriculture, 13(8), 1494. https://doi.org/10.3390/agriculture13081494





