Unraveling the Function of Stress Kinase in the Progeny of Soybean Plants Grown from Low-Temperature Pretreated Seeds
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Growing Conditions
2.2. Abiotic Stress Treatment of Seedlings
2.3. Gene Expression Analyses
2.4. Analyses of Antioxidant Activity TAC and MDA
2.5. Statistical Analyses
3. Results
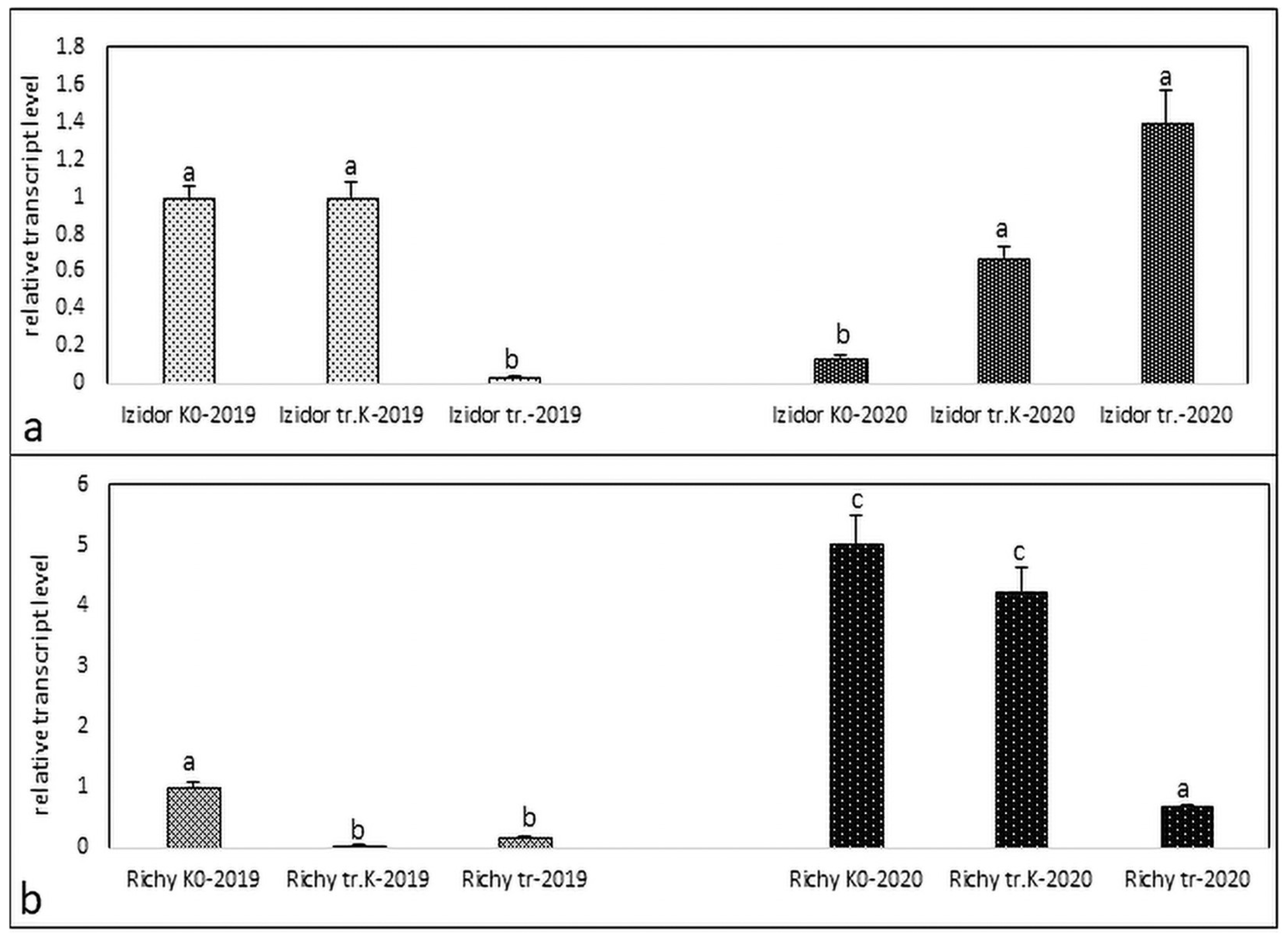
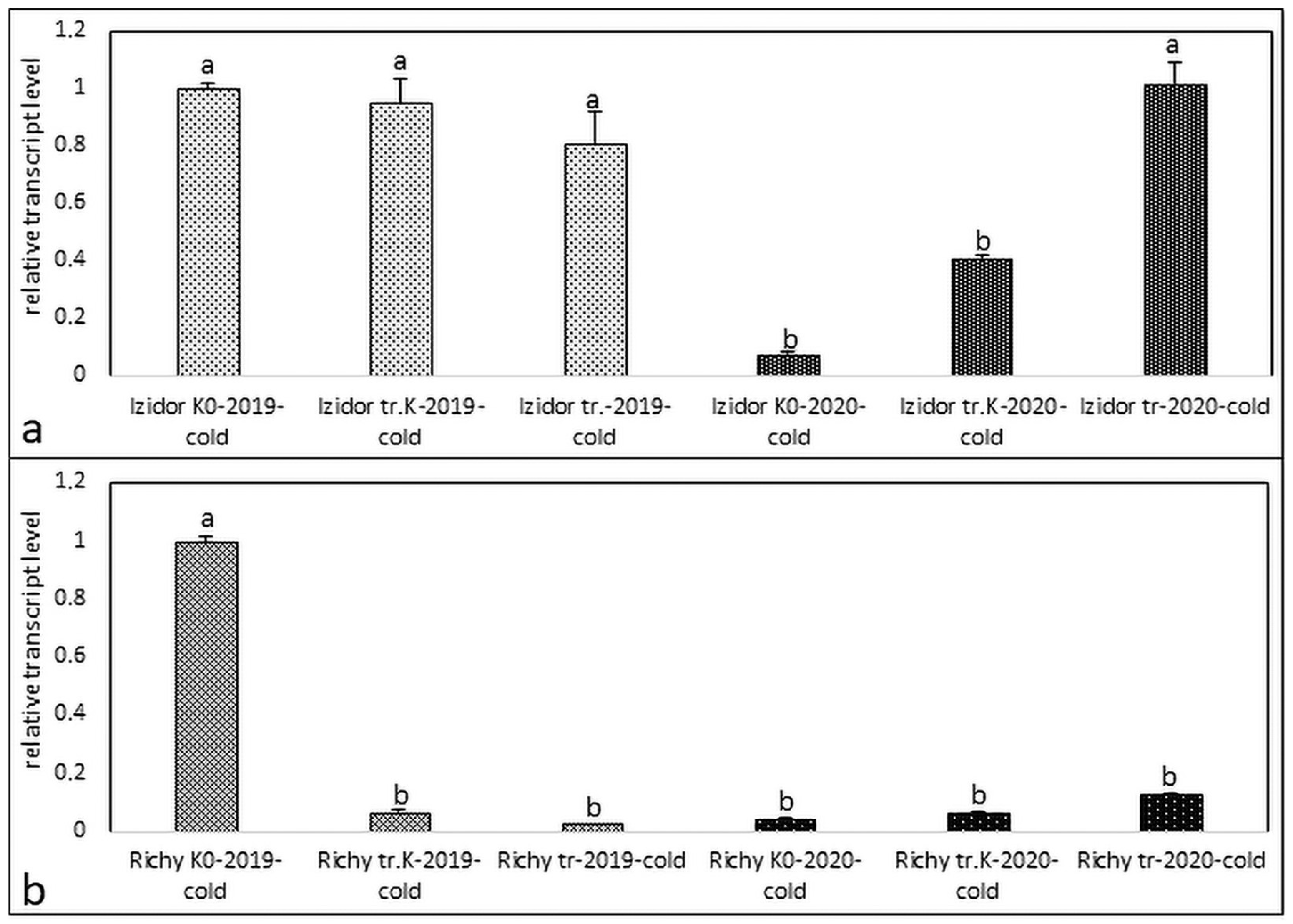
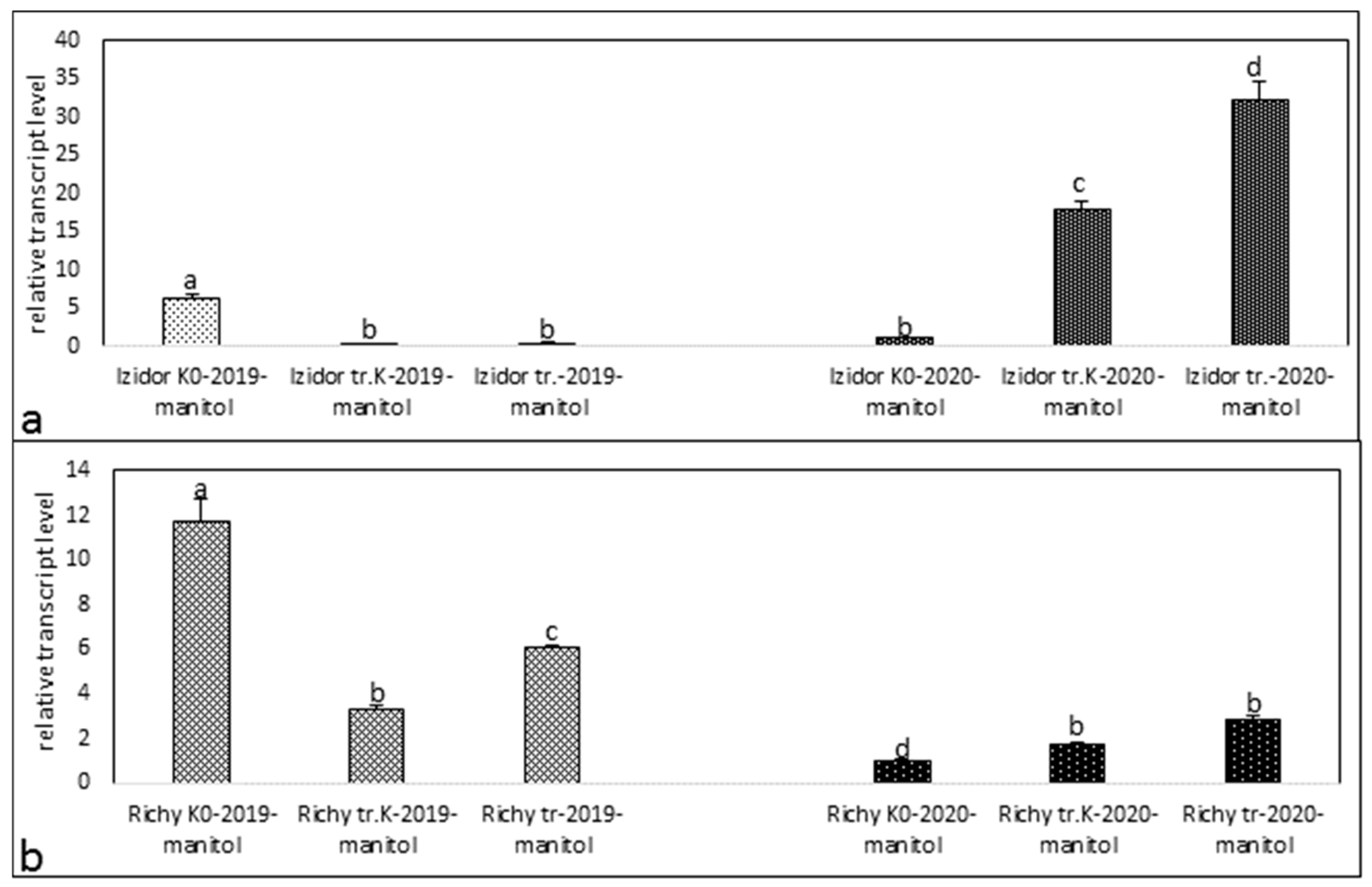
3.1. Expression Profiles of Protein Kinase in the Stressed and Control Samples of Progeny of 2019 and 2020 of Treated Control, Treated, and Non-Treated Groups
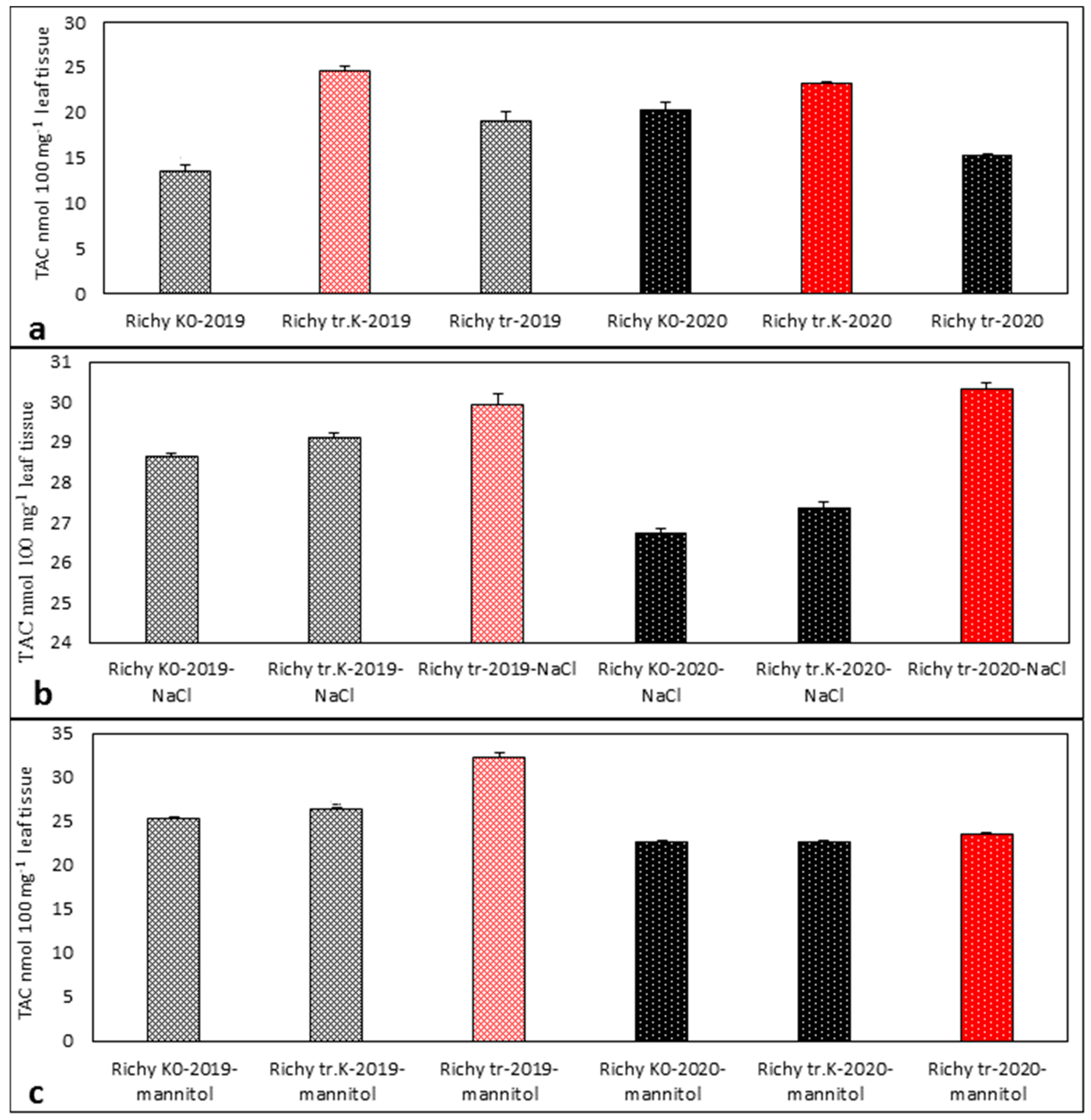
3.2. Evaluation of TAC in the Stressed and Control Samples of Progeny of 2019 and 2020 of “Treated Control”, “Treated”, and “Non-Treated” Groups
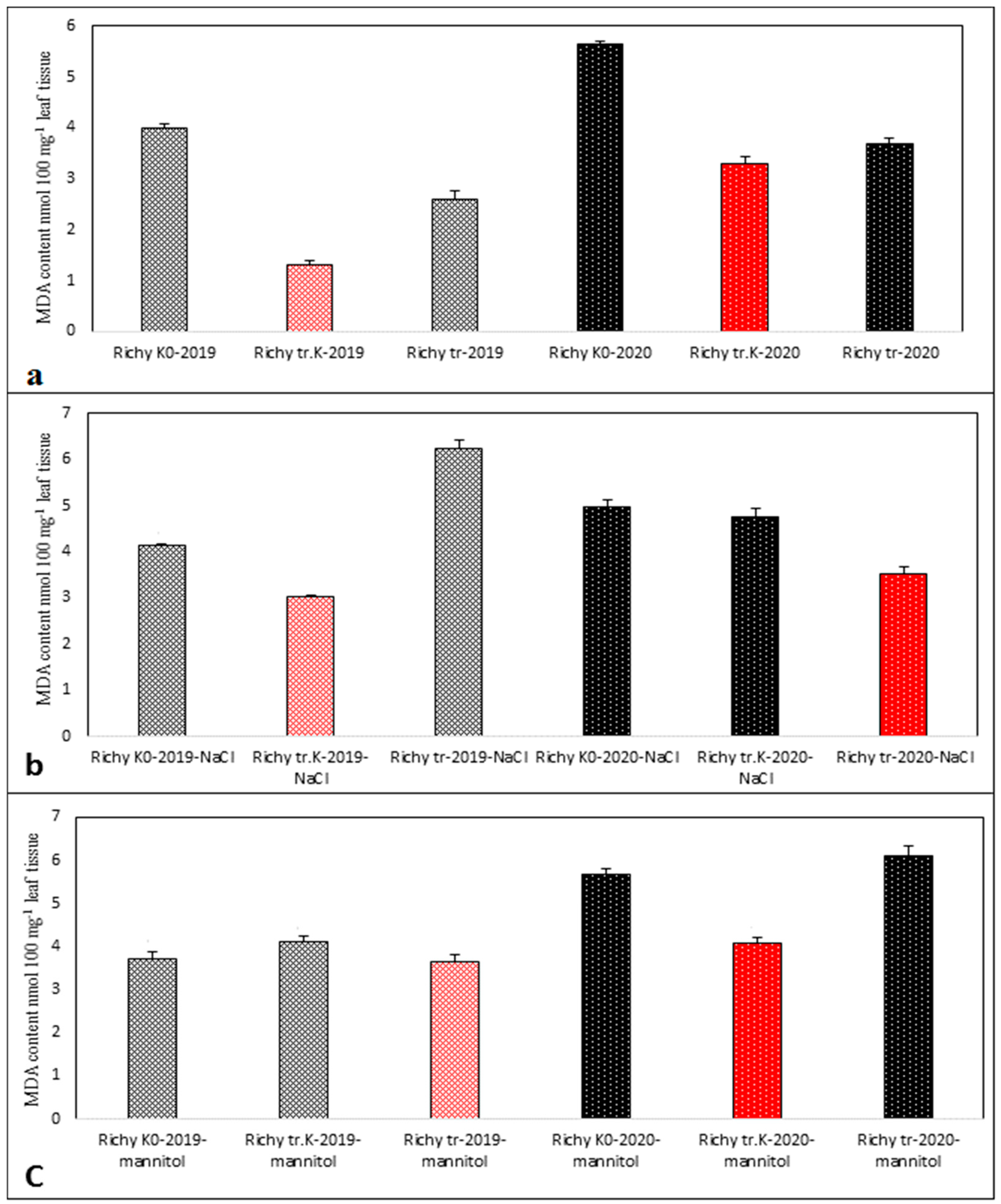
3.3. Evaluation of the Quantity of MDA in the Stressed and Control Samples of Progeny of 2019 and 2020 of “Treated Control”, “Treated”, and “Non-Treated” Groups
4. Discussion
4.1. Disclosure the Function of Investigated Stress Protein Kinase
4.2. Dependence of the Expression on Type of Sample, Genotype, and Environment
4.3. Enhanced Antioxidant Activity Depends on Type of the Sample
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ge, L.; Yu, J.; Wang, H.; Luth, D.; Bai, G.; Wang, K.; Chen, R. Increasing seed size and quality by manipulating BIG SEEDS1 in legume species. Proc. Natl. Acad. Sci. USA 2016, 113, 12414–12419. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, M.; Feng, F.; Tian, Z. Toward a “green revolution” for soybean. Mol. Plant 2020, 3, 688–697. [Google Scholar] [CrossRef] [PubMed]
- Staniak, M.; Szpunar-Krok, E.; Kocira, A. Responses of Soybean to Selected Abiotic Stresses—Photoperiod, Temperature and Water. Agriculture 2023, 13, 146. [Google Scholar] [CrossRef]
- Chaves, M.M.; Oliveira, M.M. Mechanisms underlying plant resilience to water deficits: Prospects for water-saving agriculture. J. Exp. Bot. 2004, 407, 2365–2379. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Asaf, S.; Khan, A.L.; Ullah, I.; Ali, S.; Kang, S.M.; Lee, I.J. Alleviation of salt stress response in soybean plants with the endophytic bacterial isolate Curtobacterium sp. SAK1. Ann. Microbiol. 2019, 69, 797–808. [Google Scholar] [CrossRef]
- Grainger, C.M.; Rajcan, I. Characterization of the genetic changes in a multi-generational pedigree of an elite Canadian soybean cultivar. Theor. Appl. Genet. 2013, 127, 211–229. [Google Scholar] [CrossRef] [PubMed]
- Mandic, V.; Ðordevic, S.; Ðordevic, N.; Bijelic, Z.; Krnjaja, V.; Petricevic, M.; Brankov, M. Genotype and Sowing Time effects on Soybean Yield and Quality. Agriculture 2020, 10, 502. [Google Scholar] [CrossRef]
- Zając, T.; Oleksy, A.; Ślizowska, A.; Śliwa, J.; Klimek-Kopyra, A.; Kulig, B. Aboveground dry biomass partitioning and nitrogen accumulation in early maturing soybean ‘Merlin’. Acta Agrobot. 2017, 70, 1728. [Google Scholar] [CrossRef]
- Naydenova, G.; Radkova, M.; Iantcheva, A. Moldovan soybean varieties testing in the condition of North Bulgaria. Bulg. J. Agric. Sci. 2022, 28, 299–304. [Google Scholar]
- Deshmukh, R.; Sonah, H.; Patil, G.; Chen, W.; Prince, S.; Mutava, R.; Vuong, T.; Valliyodan, B.; Nguyen, H.T. Integrating omic approaches for abiotic stress tolerance in soybean. Front Plant Sci. 2014, 5, 244. [Google Scholar] [CrossRef]
- Bashir, K.; Matsui, A.; Rasheed, S.; Seki, M. Recent Advances in the Characterization of Plant Transcriptomes in Response to Drought, Salinity, Heat, and Cold Stress. F1000Research 2019, 8, 658. [Google Scholar] [CrossRef] [PubMed]
- Brás, T.A.; Seixas, J.; Carvalhais, N.; Jägermeyr, J. Severity of Drought and Heatwave Crop Losses Tripled over the Last Five Decades in Europe. Environ. Res. Lett. 2021, 16, 065012. [Google Scholar] [CrossRef]
- Alharby, H.F.; Hasanuzzaman, M.; Al-Zahrani, H.S.; Hakeem, K.R. Exogenous selenium mitigates salt stress in soybean by improving growth, physiology, glutathione homeostasis and antioxidant defense. Phyton-Int. J. Exp. Bot. 2021, 90, 373–388. [Google Scholar] [CrossRef]
- Inupakutika, M.; Devireddy, A.R.; Willmon, D.; Puppala, N.; Cho, Y. Genome-wide comparative analysis of genes encoding core components of ABA signaling pathway in legume family. Int. J. Comput. Bioinform. Silico Model. 2016, 5, 828–843. [Google Scholar]
- Shukla, V.; Mattoo, A.K. Sucrose non-fermenting 1-related protein kinase 2 (SnRK2): A family of protein kinases involved in hyperosmotic stress signaling. Physiol. Mol. Biol. Plants 2008, 14, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Raihan, M.R.H.; Masud, A.A.C.; Rahman, K.; Nowroz, F.; Rahman, M.; Nahar, K.; Fujita, M. Regulation of reactive oxygen species and antioxidant defense in plants under salinity. Int. J. Mol. Sci. 2021, 22, 9326. [Google Scholar] [CrossRef]
- Das, K.; Roychoudhury, A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2014, 2, 53. [Google Scholar] [CrossRef]
- Wang, S.; Cheng, H.; Wei, Y. Supplemental Silicon and Boron Alleviates Aluminum-Induced Oxidative Damage in Soybean Roots. Plants 2024, 13, 821. [Google Scholar] [CrossRef]
- Choi, Y.M.; Yoon, H.; Lee, S.; Ko, H.C.; Shin, M.J.; Lee, M.C.; Hur, O.S.; Ro, N.Y.; Desta, K.T. Isofavones, anthocyanins, phenolic content, and antioxidant activities of black soybeans (Glycine max (L.) Merrill) as afected by seed weight. Sci. Rep. 2020, 10, 19960. [Google Scholar] [CrossRef]
- Pinheiro, D.T.; dos Santos Dias, D.C.F.; da Silva, L.J.; Martins, M.S.; Finger, F.L. Oxidative stress, protein metabolism, and physiological potential of soybean seeds under weathering deterioration in the pre-harvest phase. Acta Sci. Agron. 2023, 45, e56910. [Google Scholar] [CrossRef]
- Porcel, R.; Ruiz-Lozano, J.M. Arbuscular mycorrhizal influence on leaf water potential,solute accumulation, and oxidative stress in soybean plants subjected to drought stress. J. Exp. Bot. 2004, 55, 1743–1750. [Google Scholar] [CrossRef] [PubMed]
- Iantcheva, A.; Dincheva, I.; Nedeva, R.; Naydenova, G.; Badjakov, I.; Radkova, M.; Revalska, M.; Apostolov, A. An innovative approach for assessment of Bulgarian soybean cultivars. Biotechnol. Biotechnol. Equip. 2021, 35, 1099–1117. [Google Scholar] [CrossRef]
- Naydenova, G.; Dincheva, I.; Badjakov, I.; Radkova, M.; Revalska, M.; Iantcheva, A. Long-lasting low temperature pretreatment of soybean seeds enhance plant field performance and content of free metabolites. Bulg. J. Agric. Sci. 2022, 28, 1063–1074. [Google Scholar]
- Liu, S.; Lv, Z.; Liu, Y.; Li, L.; Zhang, L. Network analysis of ABA-dependent and ABA-independent drought responsive genes in Arabidopsis thaliana. Genet. Mol. Biol. 2018, 41, 624–637. [Google Scholar] [CrossRef]
- Chourasia, K.N.; Lal, M.K.; Tiwari, R.K.; Dev, D.; Kardile, H.B.; Patil, V.U.; Kumar, A.; Vanishree, G.; Kumar, D.; Bhardwaj, V.; et al. Salinity Stress in Potato: Understanding Physiological, Biochemical and Molecular Responses. Life 2021, 11, 545. [Google Scholar] [CrossRef]
- Shu, K.; Qi, Y.; Chen, F.; Meng, Y.; Luo, X.; Shuai, H.; Zhou, W.; Ding, J.; Du, J.; Liu, J.; et al. Salt stress represses soybean seed germination by negatively regulating GA biosynthesis while positively mediating ABA biosynthesis. Front. Plant Sci. 2017, 8, 1372. [Google Scholar] [CrossRef]
- Płazek, A.; Dubert, F.; Kopec, P.; Dziurka, M.; Kalandyk, A.; Pastuszak, J.; Waligórski, P.; Wolko, B. Long-term effects of cold on growth, development and yield of narrow-leaf lupine may be alleviated by seed hydropriming or butenolide. Int. J. Mol. Sci. 2018, 19, 2416. [Google Scholar] [CrossRef]
- Vieira, B.G.T.L.; Vieira, R.D.; Krzyzanowski, F.C.; Franca-Neto, J.d.B. Alternative procedure for the cold test for soybean seeds. Sci. Agric. 2010, 67, 540–545. [Google Scholar] [CrossRef]
- Vinkovic, T.; Paradikovic, N.; Plavisic, H.; Guberac, V.; Lavai, L. Maize and soybean seed vigor under influence of seed age, seed treatment and temperature in cold stress test. Cereal Res. Commun. 2007, 35, 1213–1216. [Google Scholar] [CrossRef]
- Joshi-Paneri, J.; Sharma, S.; Guruprasad, K.N.; Kataria, S. Enhancing the Yield Potential of Soybean after Magneto-Priming: Detailed Study on Its Relation to Underlying Physiological Processes. Seeds 2023, 2, 60–84. [Google Scholar] [CrossRef]
- Radhakrishnan, R. Seed pretreatment with magnetic field alters the storage proteins and lipid profiles in harvested soybean seeds. Physiol. Mol. Biol. Plants 2018, 24, 343–347. [Google Scholar] [CrossRef] [PubMed]
- Ďurčányová, S.; Slováková, L.; Klas, M.; Tomeková, J.; Ďurina, P.; Stupavská, M.; Kováčik, D.; Zahoranová, A. Efcacy Comparison of Three Atmospheric Pressure Plasma Sources for Soybean Seed Treatment: Plasma Characteristics, Seed Properties, Germination. Plasma Chem. Plasma Process. 2023, 43, 1863–1885. [Google Scholar] [CrossRef]
- Švubová, R.; Slováková, L.; Holubová, L.; Rovnanová, D.; Gálová, E.; Tomeková, J. Evaluation of the Impact of Cold Atmospheric Pressure Plasma on Soybean Seed Germination. Plants 2021, 10, 177. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.; Mazhar, K.; Ijaz, M.; Ali, Q.; Ahmad, S. Seedling pretreatment: Methods and protocols. In Priming and Pretreatment of Seeds and Seedlings; Hasanuzzaman, M., Fotopoulos, V., Eds.; Springer: Singapore, 2019; pp. 117–134. [Google Scholar]
- Agawane, R.B.; Parhe, S.D. Effect of seed priming on crop growth and seed yield of soybean [Glycine max (L.) MERILL]. Bioscan 2015, 10, 265–270. [Google Scholar]
- Lewandowska, S.; Łoziński, M.; Marczewski, K.; Kozak, M.; Schmidtke, K. Influence of priming on germination, development, and yield of soybean varieties. Open Agric. 2020, 5, 930–935. [Google Scholar] [CrossRef]
- Sadeghi, H.; Khazaei, F.; Yari, L.; Sheidaei, S. Effect of seed osmopriming on seed germination behavior and vigor of soybean (Glycine max L.). J. Agric. Biol. Sci. 2011, 6, 39–43. [Google Scholar]
- Elkelish, A.A.; Alnusaire, T.S.; Soliman, M.H.; Gowayed, S.; Senousy, H.H.; Fahad, S. Calcium availability regulates antioxidant system, physio-biochemical activities and alleviates salinity stress mediated oxidative damage in soybean seedlings. J. Appl. Bot. Food Qual. 2019, 92, 258–266. [Google Scholar] [CrossRef]
- Alharby, H.F.; Nahar, K.; Al-Zahrani, H.S.; Hakeem, K.R.; Hasanuzzaman, M. Enhancing salt tolerance in soybean by exogenous boron: Intrinsic study of the ascorbate-glutathione and glyoxalase pathways. Plants 2021, 10, 2085. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Radkova, M.; Revalska, M.; Iantcheva, A. Unraveling the Function of Stress Kinase in the Progeny of Soybean Plants Grown from Low-Temperature Pretreated Seeds. Agriculture 2024, 14, 1731. https://doi.org/10.3390/agriculture14101731
Radkova M, Revalska M, Iantcheva A. Unraveling the Function of Stress Kinase in the Progeny of Soybean Plants Grown from Low-Temperature Pretreated Seeds. Agriculture. 2024; 14(10):1731. https://doi.org/10.3390/agriculture14101731
Chicago/Turabian StyleRadkova, Mariana, Miglena Revalska, and Anelia Iantcheva. 2024. "Unraveling the Function of Stress Kinase in the Progeny of Soybean Plants Grown from Low-Temperature Pretreated Seeds" Agriculture 14, no. 10: 1731. https://doi.org/10.3390/agriculture14101731




