Adaptation Mechanisms of Olive Tree under Drought Stress: The Potential of Modern Omics Approaches
Abstract
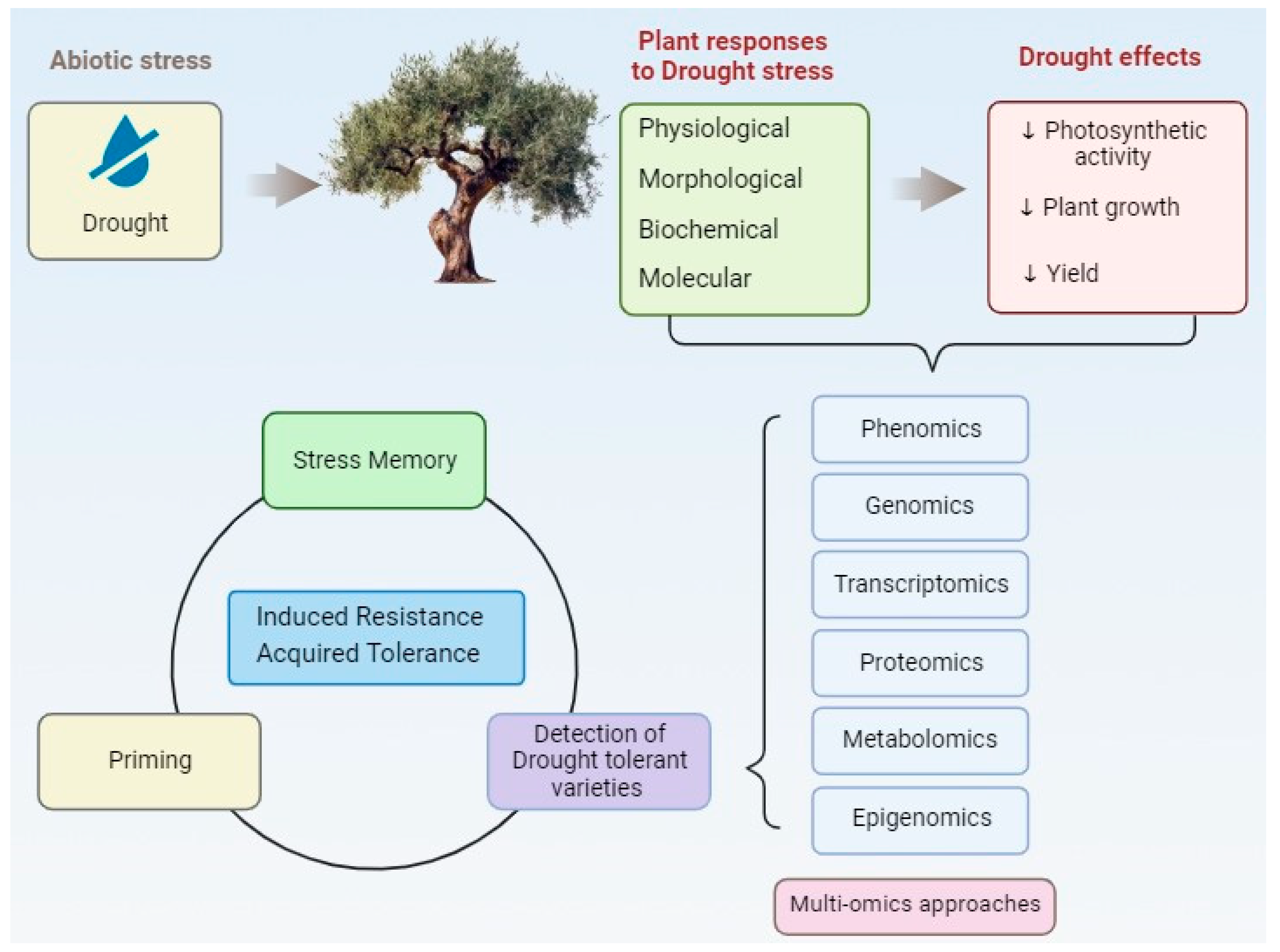
:1. Introduction
1.1. Olives and Drought in a Changing Climate
1.2. Drought Stress Tolerance and Adaptation Mechanisms
2. Morphophysiological and Biochemical Responses of Olive Tree under Drought Stress
3. Omics Approaches in Regulation of Drought Tolerance
3.1. Genomics
3.2. Transcriptomics
3.3. Proteomics
3.4. Metabolomics
3.5. Epigenomics
3.6. Multi-Omic Approaches
4. Future Perspectives and Challenges
Author Contributions
Funding
Conflicts of Interest
References
- Diez:, C.M.; Trujillo, I.; Martinez-Urdiroz, N.; Barranco, D.; Rallo, L.; Marfil, P.; Gaut, B.S. Olive Domestication and Diversification in the Mediterranean Basin. New Phytol. 2015, 206, 436–447. [Google Scholar] [CrossRef] [PubMed]
- Fraga, H.; Moriondo, M.; Leolini, L.; Santos, J.A. Mediterranean Olive Orchards under Climate Change: A Review of Future Impacts and Adaptation Strategies. Agronomy 2021, 11, 56. [Google Scholar] [CrossRef]
- Medda, S.; Fadda, A.; Mulas, M. Influence of Climate Change on Metabolism and Biological Characteristics in Perennial Woody Fruit Crops in the Mediterranean Environment. Horticulturae 2022, 8, 273. [Google Scholar] [CrossRef]
- Moriondo, M.; Trombi, G.; Ferrise, R.; Brandani, G.; Dibari, C.; Ammann, C.M.; Lippi, M.M.; Bindi, M. Olive Trees as Bio-Indicators of Climate Evolution in the Mediterranean Basin. Glob. Ecol. Biogeogr. 2013, 22, 818–833. [Google Scholar] [CrossRef]
- Meng, L.S. Compound Synthesis or Growth and Development of Roots/Stomata Regulate Plant Drought Tolerance or Water Use Efficiency/Water Uptake Efficiency. J. Agric. Food Chem. 2018, 66, 3595–3604. [Google Scholar] [CrossRef] [PubMed]
- Bacelar, E.A.; Santos, D.L.; Moutinho-Pereira, J.M.; Gonçalves, B.C.; Ferreira, H.F.; Correia, C.M. Immediate Responses and Adaptative Strategies of Three Olive Cultivars under Contrasting Water Availability Regimes: Changes on Structure and Chemical Composition of Foliage and Oxidative Damage. Plant Sci. 2006, 170, 596–605. [Google Scholar] [CrossRef]
- Angelopoulos, K.; Dichio, B.; Xiloyannis, C. Inhibition of Photosynthesis in Olive Trees (Olea europaea L.) during Water Stress and Rewatering. J. Exp. Bot. 1996, 47, 1093–1100. [Google Scholar] [CrossRef]
- Moriana, A.; Villalobos, F.J.; Fereres, E. Stomatal and Photosynthetic Responses of Olive (Olea europaea L.) Leaves to Water Deficits. Plant Cell Environ. 2002, 25, 395–405. [Google Scholar] [CrossRef]
- Ben Ahmed, C.; Ben Rouina, B.; Sensoy, S.; Boukhriss, M.; Abdullah, F.B. Saline Water Irrigation Effects on Antioxidant Defense System and Proline Accumulation in Leaves and Roots of Field-Grown Olive. J. Agric. Food Chem. 2009, 57, 11484–11490. [Google Scholar] [CrossRef]
- Hu, H.; Xiong, L. Genetic Engineering and Breeding of Drought-Resistant Crops. Annu. Rev. Plant Biol. 2014, 65, 715–741. [Google Scholar] [CrossRef]
- Haworth, M.; Marino, G.; Brunetti, C.; Killi, D.; De Carlo, A.; Centritto, M. The Impact of Heat Stress and Water Deficit on the Photosynthetic and Stomatal Physiology of Olive (Olea europaea L.)—A Case Study of the 2017 Heat Wave. Plants 2018, 7, 76. [Google Scholar] [CrossRef]
- Iglesias, M.A.; Rousseaux, M.C.; Agüero Alcaras, L.M.; Hamze, L.; Searles, P.S. Influence of Deficit Irrigation and Warming on Plant Water Status during the Late Winter and Spring in Young Olive Trees. Agric. Water Manag. 2023, 275, 108030. [Google Scholar] [CrossRef]
- Inague, G.M.; Zwiener, V.P.; Marques, M.C.M. Climate Change Threatens the Woody Plant Taxonomic and Functional Diversities of the Restinga Vegetation in Brazil. Perspect. Ecol. Conserv. 2021, 19, 53–60. [Google Scholar] [CrossRef]
- Shanker, A.K.; Maheswari, M.; Yadav, S.K.; Desai, S.; Bhanu, D.; Attal, N.B.; Venkateswarlu, B. Drought Stress Responses in Crops. Funct. Integr. Genom. 2014, 14, 11–22. [Google Scholar] [CrossRef]
- Seneviratne, S.I.; Zhang, X.; Adnan, M.; Badi, W.; Dereczynski, C.; Di Luca, A.; Ghosh, S.; Iskandar, I.; Kossin, J.; Lewis, S.; et al. Weather and Climate Extreme in a Changing Climate. In Climate Change 2021: The Physical Science Basis. Contribution of Working Group to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2021; ISBN 9781009157896. [Google Scholar]
- Provisional State of the Global Climate 2023; United Nations: New York, NY, USA, 2023; ISBN 9789213586891.
- United Nations: Department of Economic and Social Affairs. The Sustainable Development Goals Report 2022–July 2022; UN DESA: New York, NY, USA, 2022. [Google Scholar]
- Karamatlou, I.; Navabpour, S.; Nezhad, K.Z.; Mariotti, R.; Mousavi, S.; Hosseini-Mazinani, M. Cold Stress Resilience of Iranian Olive Genetic Resources: Evidence from Autochthonous Genotypes Diversity. Front. Plant Sci. 2023, 14, 1140270. [Google Scholar] [CrossRef] [PubMed]
- Brito, C.; Dinis, L.-T.T.; Moutinho-Pereira, J.; Correia, C.M. Drought Stress Effects and Olive Tree Acclimation under a Changing Climate. Plants 2019, 8, 232. [Google Scholar] [CrossRef] [PubMed]
- Jaremba, U. European Commission. In Research Handbook on the Enforcement of EU Law; Edward Elgar Publishing: Cheltenham, UK; Northampton, MA, USA, 2023; Volume 2023, pp. 107–122. ISBN 9781802208030. [Google Scholar]
- Alfieri, S.M.; Riccardi, M.; Menenti, M.; Basile, A.; Bonfante, A.; De Lorenzi, F. Adaptability of Global Olive Cultivars to Water Availability under Future Mediterranean Climate. Mitig. Adapt. Strateg. Glob. Chang. 2019, 24, 435–466. [Google Scholar] [CrossRef]
- Arenas-Castro, S.; Gonçalves, J.F.; Moreno, M.; Villar, R. Projected Climate Changes Are Expected to Decrease the Suitability and Production of Olive Varieties in Southern Spain. Sci. Total Environ. 2020, 709, 136161. [Google Scholar] [CrossRef]
- Connor, D.J.; Fereres, E. The Physiology of Adaptation and Yield Expression in Olive; Horticultural Reviews; Janick, J., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2010; Volume 31, ISBN 9780470650882. [Google Scholar]
- Fang, Y.; Xiong, L. General Mechanisms of Drought Response and Their Application in Drought Resistance Improvement in Plants. Cell. Mol. Life Sci. 2015, 72, 673–689. [Google Scholar] [CrossRef]
- Lagiotis, G.; Madesis, P.; Stavridou, E. Echoes of a Stressful Past: Abiotic Stress Memory in Crop Plants towards Enhanced Adaptation. Agriculture 2023, 13, 2090. [Google Scholar] [CrossRef]
- Fleta-Soriano, E.; Munné-Bosch, S. Stress Memory and the Inevitable Effects of Drought: A Physiological Perspective. Front. Plant Sci. 2016, 7, 171549. [Google Scholar] [CrossRef] [PubMed]
- Harris, C.J.; Amtmann, A.; Ton, J. Epigenetic Processes in Plant Stress Priming: Open Questions and New Approaches. Curr. Opin. Plant Biol. 2023, 75, 102432. [Google Scholar] [CrossRef]
- Bandurska, H. Drought Stress Responses: Coping Strategy and Resistance. Plants 2022, 11, 922. [Google Scholar] [CrossRef] [PubMed]
- Conrath, U.; Beckers, G.J.M.; Langenbach, C.J.G.; Jaskiewicz, M.R. Priming for Enhanced Defense. Annu. Rev. Phytopathol. 2015, 53, 97–119. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Hernández, M.C.; Parola-Contreras, I.; Montoya-Gómez, L.M.; Torres-Pacheco, I.; Schwarz, D.; Guevara-González, R.G. Eustressors: Chemical and Physical Stress Factors Used to Enhance Vegetables Production. Sci. Hortic. 2019, 250, 223–229. [Google Scholar] [CrossRef]
- Ben Abdallah, M.; Methenni, K.; Nouairi, I.; Zarrouk, M.; Youssef, N. Ben Drought Priming Improves Subsequent More Severe Drought in a Drought-Sensitive Cultivar of Olive Cv. Chétoui. Sci. Hortic. 2017, 221, 43–52. [Google Scholar] [CrossRef]
- Ben Abdallah, M.; Methenni, K.; Taamalli, W.; Hessini, K.; Ben Youssef, N. Cross-Priming Approach Induced Beneficial Metabolic Adjustments and Repair Processes during Subsequent Drought in Olive. Water 2022, 14, 4050. [Google Scholar] [CrossRef]
- Yang, M.; He, J.; Sun, Z.; Li, Q.; Cai, J.; Zhou, Q.; Wollenweber, B.; Jiang, D.; Wang, X. Drought Priming Mechanisms in Wheat Elucidated by In-Situ Determination of Dynamic Stomatal Behavior. Front. Plant Sci. 2023, 14, 1138494. [Google Scholar] [CrossRef]
- Wang, X.; Ge, J.; He, M.; Li, Q.; Cai, J.; Zhou, Q.; Zhong, Y.; Wollenweber, B.; Jiang, D. Enhancing Crop Resilience: Understanding the Role of Drought Priming in Wheat Stress Response. Field Crops Res. 2023, 302, 109083. [Google Scholar] [CrossRef]
- Sintaha, M.; Man, C.-K.; Yung, W.-S.; Duan, S.; Li, M.-W.; Lam, H.-M. Drought Stress Priming Improved the Drought Tolerance of Soybean. Plants 2022, 11, 2954. [Google Scholar] [CrossRef]
- Marthandan, V.; Geetha, R.; Kumutha, K.; Renganathan, V.G.; Karthikeyan, A.; Ramalingam, J. Seed Priming: A Feasible Strategy to Enhance Drought Tolerance in Crop Plants. Int. J. Mol. Sci. 2020, 21, 8258. [Google Scholar] [CrossRef] [PubMed]
- Aswathi, K.P.R.; Kalaji, H.M.; Puthur, J.T. Seed Priming of Plants Aiding in Drought Stress Tolerance and Faster Recovery: A Review. Plant Growth Regul. 2022, 97, 235–253. [Google Scholar] [CrossRef]
- Saha, D.; Choyal, P.; Mishra, U.N.; Dey, P.; Bose, B.; MD, P.; Gupta, N.K.; Mehta, B.K.; Kumar, P.; Pandey, S.; et al. Drought Stress Responses and Inducing Tolerance by Seed Priming Approach in Plants. Plant Stress 2022, 4, 100066. [Google Scholar] [CrossRef]
- Georgii, E.; Kugler, K.; Pfeifer, M.; Vanzo, E.; Block, K.; Domagalska, M.A.; Jud, W.; Elgawad, H.A.; Asard, H.; Reinhardt, R.; et al. The Systems Architecture of Molecular Memory in Poplar after Abiotic Stress. Plant Cell 2019, 31, 346–367. [Google Scholar] [CrossRef] [PubMed]
- Rosso, L.; Cantamessa, S.; Bergante, S.; Biselli, C.; Fricano, A.; Chiarabaglio, P.M.; Gennaro, M.; Nervo, G.; Secchi, F.; Carra, A. Responses to Drought Stress in Poplar: What Do We Know and What Can We Learn? Life 2023, 13, 533. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, A.; Punzo, P.; Landi, S.; Costa, A.; Van Oosten, M.J.; Grillo, S. Improving Plant Water Use Efficiency through Molecular Genetics. Horticulturae 2017, 3, 31. [Google Scholar] [CrossRef]
- Sonkar, I.; Kotnoor, H.P.; Sen, S. Estimation of Root Water Uptake and Soil Hydraulic Parameters from Root Zone Soil Moisture and Deep Percolation. Agric. Water Manag. 2019, 222, 38–47. [Google Scholar] [CrossRef]
- Yoon, H.I.; Zhang, W.; Son, J.E. Optimal Duration of Drought Stress near Harvest for Promoting Bioactive Compounds and Antioxidant Capacity in Kale with or without UV-B Radiation in Plant Factories. Plants 2020, 9, 295. [Google Scholar] [CrossRef] [PubMed]
- Gholami, R.; Zahedi, S.M. Identifying Superior Drought-Tolerant Olive Genotypes and Their Biochemical and Some Physiological Responses to Various Irrigation Levels. J. Plant Nutr. 2019, 42, 2057–2069. [Google Scholar] [CrossRef]
- Razouk, R.; Hssaini, L.; Alghoum, M.; Adiba, A.; Hamdani, A. Phenotyping Olive Cultivars for Drought Tolerance Using Leaf Macro-Characteristics. Horticulturae 2022, 8, 939. [Google Scholar] [CrossRef]
- Trentacoste, E.R.; Contreras-Zanessi, O.; Beyá-Marshall, V.; Puertas, C.M. Genotypic Variation of Physiological and Morphological Traits of Seven Olive Cultivars under Sustained and Cyclic Drought in Mendoza, Argentina. Agric. Water Manag. 2018, 196, 48–56. [Google Scholar] [CrossRef]
- Tugendhaft, Y.; Eppel, A.; Kerem, Z.; Barazani, O.; Ben-Gal, A.; Kadereit, J.W.; Dag, A. Drought Tolerance of Three Olive Cultivars Alternatively Selected for Rain Fed or Intensive Cultivation. Sci. Hortic. 2016, 199, 158–162. [Google Scholar] [CrossRef]
- Parri, S.; Romi, M.; Hoshika, Y.; Giovannelli, A.; Dias, M.C.; Piritore, F.C.; Cai, G.; Cantini, C. Morpho-Physiological Responses of Three Italian Olive Tree (Olea europaea L.) Cultivars to Drought Stress. Horticulturae 2023, 9, 830. [Google Scholar] [CrossRef]
- Sofo, A.; Manfreda, S.; Fiorentino, M.; Dichio, B.; Xiloyannis, C. Hydrology and Earth System Sciences The Olive Tree: A Paradigm for Drought Tolerance in Mediterranean Climates. Hydrol. Earth Syst. Sci. 2008, 12, 293–301. [Google Scholar] [CrossRef]
- Saradadevi, R.; Bramley, H.; Palta, J.A.; Siddique, K.H.M. Stomatal Behaviour under Terminal Drought Affects Post-Anthesis Water Use in Wheat. Funct. Plant Biol. 2017, 44, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Chaves, M.M.; Costa, J.M.; Zarrouk, O.; Pinheiro, C.; Lopes, C.M.; Pereira, J.S. Controlling Stomatal Aperture in Semi-Arid Regions—The Dilemma of Saving Water or Being Cool? Plant Sci. 2016, 251, 54–64. [Google Scholar] [CrossRef]
- Ahumada-Orellana, L.; Ortega-Farías, S.; Poblete-Echeverría, C.; Searles, P.S. Estimation of Stomatal Conductance and Stem Water Potential Threshold Values for Water Stress in Olive Trees (Cv. Arbequina). Irrig. Sci. 2019, 37, 461–467. [Google Scholar] [CrossRef]
- Ennajeh, M.; Vadel, A.M.; Cochard, H.; Khemira, H. Comparative Impacts of Water Stress on the Leaf Anatomy of a Drought-Resistant and a Drought-Sensitive Olive Cultivar. J. Hortic. Sci. Biotechnol. 2010, 85, 289–294. [Google Scholar] [CrossRef]
- Brito, C.; Dinis, L.T.; Ferreira, H.; Moutinho-Pereira, J.; Correia, C. The Role of Nighttime Water Balance on Olea europaea Plants Subjected to Contrasting Water Regimes. J. Plant Physiol. 2018, 226, 56–63. [Google Scholar] [CrossRef]
- Ma, Y.; Dias, M.C.; Freitas, H. Drought and Salinity Stress Responses and Microbe-Induced Tolerance in Plants. Front. Plant Sci. 2020, 11, 591911. [Google Scholar] [CrossRef]
- Baccari, S.; Elloumi, O.; Chaari-Rkhis, A.; Fenollosa, E.; Morales, M.; Drira, N.; Ben Abdallah, F.; Fki, L.; Munné-Bosch, S. Linking Leaf Water Potential, Photosynthesis and Chlorophyll Loss With Mechanisms of Photo- and Antioxidant Protection in Juvenile Olive Trees Subjected to Severe Drought. Front. Plant Sci. 2020, 11, 614144. [Google Scholar] [CrossRef] [PubMed]
- Ennajeh, M.; Vadel, A.M.; Khemira, H. Osmoregulation and Osmoprotection in the Leaf Cells of Two Olive Cultivars Subjected to Severe Water Deficit. Acta Physiol. Plant. 2009, 31, 711–721. [Google Scholar] [CrossRef]
- Martins, J.P.R.; Schimildt, E.R.; Alexandre, R.S.; Falqueto, A.R.; Otoni, W.C. Chlorophyll a Fluorescence and Growth of Neoregelia Concentrica (Bromeliaceae) during Acclimatization in Response to Light Levels. Vitr. Cell. Dev. Biol.-Plant 2015, 51, 471–481. [Google Scholar] [CrossRef]
- Melaouhi, A.; Baraza, E.; Escalona, J.M.; El-AouOuad, H.; Mahjoub, I.; Bchir, A.; Braham, M.; Bota, J. Physiological and Biochemical Responses to Water Deficit and Recovery of Two Olive Cultivars (Olea europaea L., Arbequina and Empeltre Cvs.) under Mediterranean Conditions. Theor. Exp. Plant Physiol. 2021, 33, 369–383. [Google Scholar] [CrossRef]
- Karimi, S.; Rahemi, M.; Rostami, A.A.; Sedaghat, S. Drought Effects on Growth, Water Content and Osmoprotectants in Four Olive Cultivars with Different Drought Tolerance. Int. J. Fruit Sci. 2018, 18, 254–267. [Google Scholar] [CrossRef]
- Santos, M.; Barros, V.; Lima, L.; Frosi, G.; Santos, M.G. Whole Plant Water Status and Non-structural Carbohydrates under Progressive Drought in a Caatinga Deciduous Woody Species. Trees-Struct. Funct. 2021, 35, 1257–1266. [Google Scholar] [CrossRef]
- Zhang, X.; Lei, L.; Lai, J.; Zhao, H.; Song, W. Effects of Drought Stress and Water Recovery on Physiological Responses and Gene Expression in Maize Seedlings. BMC Plant Biol. 2018, 18, 68. [Google Scholar] [CrossRef] [PubMed]
- Mubarik, M.S.; Khan, S.H.; Sajjad, M.; Raza, A.; Hafeez, M.B.; Yasmeen, T.; Rizwan, M.; Ali, S.; Arif, M.S. A Manipulative Interplay between Positive and Negative Regulators of Phytohormones: A Way Forward for Improving Drought Tolerance in Plants. Physiol. Plant. 2021, 172, 1269–1290. [Google Scholar] [CrossRef]
- Ullah, A.; Manghwar, H.; Shaban, M.; Khan, A.H.; Akbar, A.; Ali, U.; Ali, E.; Fahad, S. Phytohormones Enhanced Drought Tolerance in Plants: A Coping Strategy. Environ. Sci. Pollut. Res. 2018, 25, 33103–33118. [Google Scholar] [CrossRef]
- Raza, A.; Tabassum, J.; Kudapa, H.; Varshney, R.K. Can Omics Deliver Temperature Resilient Ready-to-Grow Crops? Crit. Rev. Biotechnol. 2021, 41, 1209–1232. [Google Scholar] [CrossRef]
- Varshney, R.K.; Barmukh, R.; Roorkiwal, M.; Qi, Y.; Kholova, J.; Tuberosa, R.; Reynolds, M.P.; Tardieu, F.; Siddique, K.H.M. Breeding Custom-designed Crops for Improved Drought Adaptation. Adv. Genet. 2021, 2, e202100017. [Google Scholar] [CrossRef] [PubMed]
- Abdurakhmonov, I.Y. Genotyping, Abdurakhmonov, I., Ed.; IntechOpen: London, UK; Rijeka, Croatia, 2018; ISBN 9781789232806.
- Cruz, F.; Julca, I.; Gómez-Garrido, J.; Loska, D.; Marcet-Houben, M.; Cano, E.; Galán, B.; Frias, L.; Ribeca, P.; Derdak, S.; et al. Genome Sequence of the Olive Tree, Olea europaea. Gigascience 2016, 5, s13742-016. [Google Scholar] [CrossRef] [PubMed]
- Unver, T.; Wu, Z.; Sterck, L.; Turktas, M.; Lohaus, R.; Li, Z.; Yang, M.; He, L.; Deng, T.; Escalante, F.J.; et al. Genome of Wild Olive and the Evolution of Oil Biosynthesis. Proc. Natl. Acad. Sci. USA 2017, 114, E9413–E9422. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Ruiz, J.; Ramírez-Tejero, J.A.; Fernández-Pozo, N.; Leyva-Pérez, M.D.L.O.; Yan, H.; de la Rosa, R.; Belaj, A.; Montes, E.; Rodríguez-Ariza, M.O.; Navarro, F.; et al. Transposon Activation Is a Major Driver in the Genome Evolution of Cultivated Olive Trees (Olea europaea L.). Plant Genome 2020, 13, e20010. [Google Scholar] [CrossRef]
- Rao, G.; Zhang, J.; Liu, X.; Lin, C.; Xin, H.; Xue, L.; Wang, C. De Novo Assembly of a New Olea europaea Genome Accession Using Nanopore Sequencing. Hortic. Res. 2021, 8, 64. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Zeng, Y.; Li, J.; Deng, Y.; Su, G.; Zhang, J. One Hundred Single-Copy Nuclear Sequence Markers for Olive Variety Identification: A Case of Fingerprinting Database Construction in China. Mol. Breed. 2023, 43, 86. [Google Scholar] [CrossRef] [PubMed]
- Koubouris, G.C.; Avramidou, E.V.; Metzidakis, I.T.; Petrakis, P.V.; Sergentani, C.K.; Doulis, A.G. Phylogenetic and Evolutionary Applications of Analyzing Endocarp Morphological Characters by Classification Binary Tree and Leaves by SSR Markers for the Characterization of Olive Germplasm. Tree Genet. Genomes 2019, 15, 26. [Google Scholar] [CrossRef]
- Belaj, A.; de la Rosa, R.; Lorite, I.J.; Mariotti, R.; Cultrera, N.G.M.; Beuzón, C.R.; González-Plaza, J.J.; Muñoz-Mérida, A.; Trelles, O.; Baldoni, L. Usefulness of a New Large Set of High Throughput Est-Snp Markers as a Tool for Olive Germplasm Collection Management. Front. Plant Sci. 2018, 9, 377691. [Google Scholar] [CrossRef] [PubMed]
- Omri, A.; Abdelhamid, S.; Benincasa, C.; Araouki, A.; Ayadi, M.; Gharsallaoui, M.; Gouiaa, M. Genetic Diversity and Association of Molecular Markers with Biochemical Traits in Tunisian Olive Cultivars. Genet. Resour. Crop Evol. 2021, 68, 1181–1197. [Google Scholar] [CrossRef]
- Biton, I.; Doron-Faigenboim, A.; Jamwal, M.; Mani, Y.; Eshed, R.; Rosen, A.; Sherman, A.; Ophir, R.; Lavee, S.; Avidan, B.; et al. Development of a Large Set of SNP Markers for Assessing Phylogenetic Relationships between the Olive Cultivars Composing the Israeli Olive Germplasm Collection. Mol. Breed. 2015, 35, 107. [Google Scholar] [CrossRef]
- Bazakos, C.; Alexiou, K.G.; Ramos-Onsins, S.; Koubouris, G.; Tourvas, N.; Xanthopoulou, A.; Mellidou, I.; Moysiadis, T.; Vourlaki, I.T.; Metzidakis, I.; et al. Whole Genome Scanning of a Mediterranean Basin Hotspot Collection Provides New Insights into Olive Tree Biodiversity and Biology. Plant J. 2023, 116, 303–319. [Google Scholar] [CrossRef] [PubMed]
- De Ollas, C.; Morillón, R.; Fotopoulos, V.; Puértolas, J.; Ollitrault, P.; Gómez-Cadenas, A.; Arbona, V. Facing Climate Change: Biotechnology of Iconic Mediterranean Woody Crops. Front. Plant Sci. 2019, 10, 431851. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Tejero, J.A.; Jiménez-Ruiz, J.; Leyva-Pérez, M.D.L.O.; Barroso, J.B.; Luque, F. Gene Expression Pattern in Olive Tree Organs (Olea europaea L.). Genes 2020, 11, 544. [Google Scholar] [CrossRef] [PubMed]
- Julca, I.; Marcet-Houben, M.; Cruz, F.; Gómez-Garrido, J.; Gaut, B.S.; Díez, C.M.; Gut, I.G.; Alioto, T.S.; Vargas, P.; Gabaldón, T. Genomic Evidence for Recurrent Genetic Admixture during the Domestication of Mediterranean Olive Trees (Olea europaea L.). BMC Biol. 2020, 18, 148. [Google Scholar] [CrossRef] [PubMed]
- Vatansever, R.; Hernandez, P.; Escalante, F.J.; Dorado, G.; Unver, T. Genome-Wide Exploration of Oil Biosynthesis Genes in Cultivated Olive Tree Varieties (Olea europaea): Insights into Regulation of Oil Biosynthesis. Funct. Integr. Genom. 2022, 22, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Lowe, R.; Shirley, N.; Bleackley, M.; Dolan, S.; Shafee, T. Transcriptomics Technologies. PLoS Comput. Biol. 2017, 13, e1005457. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, S.; Bashir, K.; Matsui, A.; Tanaka, M.; Seki, M. Transcriptomic Analysis of Soil-Grown Arabidopsis Thaliana Roots and Shoots in Response to a Drought Stress. Front. Plant Sci. 2016, 7, 183475. [Google Scholar] [CrossRef]
- Öztürk Gökçe, Z.N.; Gökçe, A.F.; Junaid, M.D.; Chaudhry, U.K. Comparative Transcriptomics of Drought Stress Response of Taproot Meristem Region of Contrasting Purple Carrot Breeding Lines Supported by Physio-Biochemical Parameters. Funct. Integr. Genom. 2022, 22, 697–710. [Google Scholar] [CrossRef]
- Filichkin, S.A.; Hamilton, M.; Dharmawardhana, P.D.; Singh, S.K.; Sullivan, C.; Ben-Hur, A.; Reddy, A.S.N.; Jaiswal, P. Abiotic Stresses Modulate Landscape of Poplar Transcriptome via Alternative Splicing, Differential Intron Retention, and Isoform Ratio Switching. Front. Plant Sci. 2018, 9, 5. [Google Scholar] [CrossRef]
- Gros-Balthazard, M.; Besnard, G.; Sarah, G.; Holtz, Y.; Leclercq, J.; Santoni, S.; Wegmann, D.; Glémin, S.; Khadari, B. Evolutionary Transcriptomics Reveals the Origins of Olives and the Genomic Changes Associated with Their Domestication. Plant J. 2019, 100, 143–157. [Google Scholar] [CrossRef]
- İpek, A.; İpek, M.; Ercişli, S.; Tangu, N.A. Transcriptome-Based SNP Discovery by GBS and the Construction of a Genetic Map for Olive. Funct. Integr. Genom. 2017, 17, 493–501. [Google Scholar] [CrossRef]
- Jiménez-Ruiz, J.; de la O Leyva-Pérez, M.; Vidoy-Mercado, I.; Barceló, A.; Luque, F. Transcriptomic Time-Series Analysis of Early Development in Olive from Germinated Embryos to Juvenile Tree. BMC Genom. 2018, 19, 824. [Google Scholar] [CrossRef]
- Ferrari, M.; Muto, A.; Bruno, L.; Muzzalupo, I.; Chiappetta, A. Modulation of Anthocyanin Biosynthesis-Related Genes during the Ripening of Olea europaea L. Cvs Carolea and Tondina Drupes in Relation to Environmental Factors. Int. J. Mol. Sci. 2023, 24, 8770. [Google Scholar] [CrossRef]
- Skodra, C.; Titeli, V.S.; Michailidis, M.; Bazakos, C.; Ganopoulos, I.; Molassiotis, A.; Tanou, G. Olive Fruit Development and Ripening: Break on through to the “-omics” Side. Int. J. Mol. Sci. 2021, 22, 5806. [Google Scholar] [CrossRef]
- González-Plaza, J.J.; Ortiz-Martín, I.; Muñoz-Mérida, A.; García-López, C.; Sánchez-Sevilla, J.F.; Luque, F.; Trelles, O.; Bejarano, E.R.; De La Rosa, R.; Valpuesta, V.; et al. Transcriptomic Analysis Using Olive Varieties and Breeding Progenies Identifies Candidate Genes Involved in Plant Architecture. Front. Plant Sci. 2016, 7, 177573. [Google Scholar] [CrossRef]
- Bullones, A.; Castro, A.J.; Lima-Cabello, E.; Fernandez-Pozo, N.; Bautista, R.; Alché, J.D.D.; Claros, M.G. Transcriptomic Insight into the Pollen Tube Growth of Olea europaea L. Subsp. Europaea Reveals Reprogramming and Pollen-Specific Genes Including New Transcription Factors. Plants 2023, 12, 2894. [Google Scholar] [CrossRef]
- Dastkar, E.; Soleimani, A.; Jafary, H.; de Dios Alche, J.; Bahari, A.; Zeinalabedini, M.; Salami, S.A. Differential Expression of Genes in Olive Leaves and Buds of ON- versus OFF-Crop Trees. Sci. Rep. 2020, 10, 15762. [Google Scholar] [CrossRef]
- Marchese, A.; Balan, B.; Trippa, D.A.; Bonanno, F.; Caruso, T.; Imperiale, V.; Marra, F.P.; Giovino, A. NGS Transcriptomic Analysis Uncovers the Possible Resistance Mechanisms of Olive to Spilocea oleagina Leaf Spot Infection. Front. Plant Sci. 2023, 14, 1219580. [Google Scholar] [CrossRef]
- Leyva-Pérez, M.D.L.O.; Jiménez-Ruiz, J.; Gómez-Lama Cabanás, C.; Valverde-Corredor, A.; Barroso, J.B.; Luque, F.; Mercado-Blanco, J. Tolerance of Olive (Olea europaea) Cv Frantoio to Verticillium dahliae Relies on Both Basal and Pathogen-Induced Differential Transcriptomic Responses. New Phytol. 2018, 217, 671–686. [Google Scholar] [CrossRef]
- Bazakos, C.; Manioudaki, M.E.; Therios, I.; Voyiatzis, D.; Kafetzopoulos, D.; Awada, T.; Kalaitzis, P. Comparative Transcriptome Analysis of Two Olive Cultivars in Response to NaCl-Stress. PLoS ONE 2012, 7, e42931. [Google Scholar] [CrossRef]
- Bazakos, C.; Manioudaki, M.E.; Sarropoulou, E.; Spano, T.; Kalaitzis, P. 454 Pyrosequencing of Olive (Olea europaea L.) Transcriptome in Response to Salinity. PLoS ONE 2015, 10, e0143000. [Google Scholar] [CrossRef]
- Mousavi, S.; Mariotti, R.; Valeri, M.C.; Regni, L.; Lilli, E.; Albertini, E.; Proietti, P.; Businelli, D.; Baldoni, L. Characterization of Differentially Expressed Genes under Salt Stress in Olive. Int. J. Mol. Sci. 2022, 23, 154. [Google Scholar] [CrossRef]
- Rossi, L.; Borghi, M.; Francini, A.; Lin, X.; Xie, D.Y.; Sebastiani, L. Salt Stress Induces Differential Regulation of the Phenylpropanoid Pathway in Olea europaea Cultivars Frantoio (Salt-Tolerant) and Leccino (Salt-Sensitive). J. Plant Physiol. 2016, 204, 8–15. [Google Scholar] [CrossRef]
- Galicia-Campos, E.; García-Villaraco, A.; Montero-Palmero, M.B.; Gutiérrez-Mañero, F.J.; Ramos-Solano, B. Bacillus G7 Improves Adaptation to Salt Stress in Olea europaea L. Plantlets, Enhancing Water Use Efficiency and Preventing Oxidative Stress. Sci. Rep. 2023, 13, 22507. [Google Scholar] [CrossRef]
- Guerra, D.; Lamontanara, A.; Bagnaresi, P.; Orrù, L.; Rizza, F.; Zelasco, S.; Beghè, D.; Ganino, T.; Pagani, D.; Cattivelli, L.; et al. Transcriptome Changes Associated with Cold Acclimation in Leaves of Olive Tree (Olea europaea L.). Tree Genet. Genomes 2015, 11, 113. [Google Scholar] [CrossRef]
- De La O Leyva-Pérez, M.; Valverde-Corredor, A.; Valderrama, R.; Jiménez-Ruiz, J.; Muñoz-Merida, A.; Trelles, O.; Barroso, J.B.; Mercado-Blanco, J.; Luque, F. Early and Delayed Long-Term Transcriptional Changes and Short-Term Transient Responses during Cold Acclimation in Olive Leaves. DNA Res. 2015, 22, 1–11. [Google Scholar] [CrossRef]
- D’Angeli, S.; Falasca, G.; Matteucci, M.; Altamura, M.M. Cold Perception and Gene Expression Differ in Olea europaea Seed Coat and Embryo during Drupe Cold Acclimation. New Phytol. 2013, 197, 123–138. [Google Scholar] [CrossRef]
- Benny, J.; Marchese, A.; Giovino, A.; Marra, F.P.; Perrone, A.; Caruso, T.; Martinelli, F. Gaining Insight into Exclusive and Common Transcriptomic Features Linked to Drought and Salinity Responses across Fruit Tree Crops. Plants 2020, 9, 1059. [Google Scholar] [CrossRef]
- Tsamir-Rimon, M.; Ben-Dor, S.; Feldmesser, E.; Oppenhimer-Shaanan, Y.; David-Schwartz, R.; Samach, A.; Klein, T. Rapid Starch Degradation in the Wood of Olive Trees under Heat and Drought Is Permitted by Three Stress-Specific Beta Amylases. New Phytol. 2021, 229, 1398–1414. [Google Scholar] [CrossRef]
- Bullones, A.; Castro, A.J.; Lima-Cabello, E.; Alché, J.D.D.; Luque, F.; Claros, M.G.; Fernandez-Pozo, N. OliveAtlas: A Gene Expression Atlas Tool for Olea europaea. Plants 2023, 12, 1274. [Google Scholar] [CrossRef]
- Skodra, C.; Michailidis, M.; Moysiadis, T.; Stamatakis, G.; Ganopoulou, M.; Adamakis, I.D.S.; Angelis, L.; Ganopoulos, I.; Tanou, G.; Samiotaki, M.; et al. Disclosing the Molecular Basis of Salinity Priming in Olive Trees Using Proteogenomic Model Discovery. Plant Physiol. 2023, 191, 1913–1933. [Google Scholar] [CrossRef]
- Ben Abdallah, M.; Trupiano, D.; Polzella, A.; De Zio, E.; Sassi, M.; Scaloni, A.; Zarrouk, M.; Ben Youssef, N.; Scippa, G.S. Unraveling Physiological, Biochemical and Molecular Mechanisms Involved in Olive (Olea europaea L. Cv. Chétoui) Tolerance to Drought and Salt Stresses. J. Plant Physiol. 2018, 220, 83–95. [Google Scholar] [CrossRef]
- Dias, M.C.; Figueiredo, C.; Pinto, D.C.G.A.; Freitas, H.; Santos, C.; Silva, A.M.S. Heat Shock and UV-B Episodes Modulate Olive Leaves Lipophilic and Phenolic Metabolite Profiles. Ind. Crops Prod. 2019, 133, 269–275. [Google Scholar] [CrossRef]
- Koubouris, G.C.; Kavroulakis, N.; Metzidakis, I.T.; Vasilakakis, M.D.; Sofo, A. Ultraviolet-B Radiation or Heat Cause Changes in Photosynthesis, Antioxidant Enzyme Activities and Pollen Performance in Olive Tree. Photosynthetica 2015, 53, 279–287. [Google Scholar] [CrossRef]
- Araújo, M.; Prada, J.; Mariz-ponte, N.; Santos, C.; Pereira, J.A.; Pinto, D.C.G.A.; Silva, A.M.S.; Dias, M.C. Antioxidant Adjustments of Olive Trees (Olea europaea) under Field Stress Conditions. Plants 2021, 10, 684. [Google Scholar] [CrossRef]
- Mousavi, S.; Regni, L.; Bocchini, M.; Mariotti, R.; Cultrera, N.G.M.; Mancuso, S.; Googlani, J.; Chakerolhosseini, M.R.; Guerrero, C.; Albertini, E.; et al. Physiological, Epigenetic and Genetic Regulation in Some Olive Cultivars under Salt Stress. Sci. Rep. 2019, 9, 1093. [Google Scholar] [CrossRef]
- Wang, W.; Tai, F.; Hu, X. Chapter 3—Current Initiatives in Proteomics of the Olive Tree. In Olives and Olive Oil in Health and Disease Prevention; Academic Press: Cambridge, MA, USA, 2010; pp. 25–32. [Google Scholar] [CrossRef]
- Ghosh, D.; Xu, J. Abiotic Stress Responses in Plant Roots: A Proteomics Perspective. Front. Plant Sci. 2014, 5, 66977. [Google Scholar] [CrossRef]
- Cao, Y.; Luo, Q.; Tian, Y.; Meng, F. Physiological and Proteomic Analyses of the Drought Stress Response in Amygdalus Mira (Koehne) Yü et Lu Roots. BMC Plant Biol. 2017, 17, 53. [Google Scholar] [CrossRef]
- Habibpourmehraban, F.; Wu, Y.; Wu, J.X.; Hamzelou, S.; Masoomi-Aladizgeh, F.; Kamath, K.S.; Amirkhani, A.; Atwell, B.J.; Haynes, P.A. Multiple Abiotic Stresses Applied Simultaneously Elicit Distinct Responses in Two Contrasting Rice Cultivars. Int. J. Mol. Sci. 2022, 23, 1739. [Google Scholar] [CrossRef]
- Zeng, W.; Peng, Y.; Zhao, X.; Wu, B.; Chen, F.; Ren, B.; Zhuang, Z.; Gao, Q.; Ding, Y. Comparative Proteomics Analysis of the Seedling Root Response of Drought-Sensitive and Drought-Tolerant Maize Varieties to Drought Stress. Int. J. Mol. Sci. 2019, 20, 2793. [Google Scholar] [CrossRef]
- Halder, T.; Choudhary, M.; Liu, H.; Chen, Y.; Yan, G.; Siddique, K.H.M. Wheat Proteomics for Abiotic Stress Tolerance and Root System Architecture: Current Status and Future Prospects. Proteomes 2022, 10, 17. [Google Scholar] [CrossRef]
- Kosová, K.; Prášil, I.T.; Klíma, M.; Nesvadba, Z.; Vítámvás, P.; Ovesná, J. Proteomics of Wheat and Barley Cereals in Response to Environmental Stresses: Current State and Future Challenges. J. Proteom. 2023, 282, 104923. [Google Scholar] [CrossRef]
- Li, H.; Li, Y.; Ke, Q.; Kwak, S.S.; Zhang, S.; Deng, X. Physiological and Differential Proteomic Analyses of Imitation Drought Stress Response in Sorghum bicolor Root at the Seedling Stage. Int. J. Mol. Sci. 2020, 21, 9174. [Google Scholar] [CrossRef]
- Yahoueian, S.H.; Bihamta, M.R.; Babaei, H.R.; Bazargani, M.M. Proteomic Analysis of Drought Stress Response Mechanism in Soybean (Glycine max L.) Leaves. Food Sci. Nutr. 2021, 9, 2010–2020. [Google Scholar] [CrossRef]
- Idle, J.R.; Gonzalez, F.J. Metabolomics. Cell Metab. 2007, 6, 348–351. [Google Scholar] [CrossRef]
- Junaid, M.D.; Öztürk, Z.N.; Gökçe, A.F. Exploitation of Tolerance to Drought Stress in Carrot (Daucus carota L.): An Overview. Stress Biol. 2023, 3, 55. [Google Scholar] [CrossRef]
- Razavi, F.; De Keyser, E.; De Riek, J.; Van Labeke, M.C. A Method for Testing Drought Tolerance in Fragaria Based on Fast Screening for Water Deficit Response and Use of Associated AFLP and EST Candidate Gene Markers. Euphytica 2011, 180, 385–409. [Google Scholar] [CrossRef]
- Dias, M.C.; Mariz-Ponte, N.; Santos, C. Lead Induces Oxidative Stress in Pisum Sativum Plants and Changes the Levels of Phytohormones with Antioxidant Role. Plant Physiol. Biochem. 2019, 137, 121–129. [Google Scholar] [CrossRef]
- Talhaoui, N.; Taamalli, A.; Gómez-Caravaca, A.M.; Fernández-Gutiérrez, A.; Segura-Carretero, A. Phenolic Compounds in Olive Leaves: Analytical Determination, Biotic and Abiotic Influence, and Health Benefits. Food Res. Int. 2015, 77, 92–108. [Google Scholar] [CrossRef]
- Sharma, M.; Kumar, P.; Verma, V.; Sharma, R.; Bhargava, B.; Irfan, M. Understanding Plant Stress Memory Response for Abiotic Stress Resilience: Molecular Insights and Prospects. Plant Physiol. Biochem. 2022, 179, 10–24. [Google Scholar] [CrossRef]
- Fàbregas, N.; Fernie, A.R. The Metabolic Response to Drought. J. Exp. Bot. 2019, 70, 1077–1085. [Google Scholar] [CrossRef]
- Noleto-Dias, C.; Picoli, E.A.d.T.; Porzel, A.; Wessjohann, L.A.; Tavares, J.F.; Farag, M.A. Metabolomics Characterizes Early Metabolic Changes and Markers of Tolerant Eucalyptus ssp. Clones against Drought Stress. Phytochemistry 2023, 212, 113715. [Google Scholar] [CrossRef]
- Jia, H.; Wang, L.; Li, J.; Sun, P.; Lu, M.; Hu, J. Comparative Metabolomics Analysis Reveals Different Metabolic Responses to Drought in Tolerant and Susceptible Poplar Species. Physiol. Plant. 2020, 168, 531–546. [Google Scholar] [CrossRef]
- You, J.; Zhang, Y.; Liu, A.; Li, D.; Wang, X.; Dossa, K.; Zhou, R.; Yu, J.; Zhang, Y.; Wang, L.; et al. Transcriptomic and Metabolomic Profiling of Drought-Tolerant and Susceptible Sesame Genotypes in Response to Drought Stress. BMC Plant Biol. 2019, 19, 267. [Google Scholar] [CrossRef]
- Soliman, S.S.M.; Abouleish, M.; Abou-Hashem, M.M.M.; Hamoda, A.M.; El-Keblawy, A.A. Lipophilic Metabolites and Anatomical Acclimatization of Cleome amblyocarpa in the Drought and Extra-Water Areas of the AridDesert of UAE. Plants 2019, 8, 132. [Google Scholar] [CrossRef]
- Saeed, F.; Chaudhry, U.K.; Bakhsh, A.; Raza, A.; Saeed, Y.; Bohra, A.; Varshney, R.K. Moving Beyond DNA Sequence to Improve Plant Stress Responses. Front. Genet. 2022, 13, 874648. [Google Scholar] [CrossRef]
- Sun, C.; Ali, K.; Yan, K.; Fiaz, S.; Dormatey, R.; Bi, Z.; Bai, J. Exploration of Epigenetics for Improvement of Drought and Other Stress Resistance in Crops: A Review. Plants 2021, 10, 1226. [Google Scholar] [CrossRef]
- Pikaard, C.S.; Scheid, O.M. Epigenetic Regulation in Plants. Cold Spring Harb. Perspect. Biol. 2014, 6, a019315. [Google Scholar] [CrossRef]
- Akhter, Z.; Bi, Z.; Ali, K.; Sun, C.; Fiaz, S.; Haider, F.U.; Bai, J. In Response to Abiotic Stress, Dna Methylation Confers Epigenetic Changes in Plants. Plants 2021, 10, 1096. [Google Scholar] [CrossRef]
- Badad, O.; Lakhssassi, N.; Zaid, N.; El Baze, A.; Zaid, Y.; Meksem, J.; Lightfoot, D.A.; Tombuloglu, H.; Zaid, E.H.; Unver, T.; et al. Genome Wide Medip-Seq Profiling of Wild and Cultivated Olives Trees Suggests Dna Methylation Fingerprint on the Sensory Quality of Olive Oil. Plants 2021, 10, 1405. [Google Scholar] [CrossRef]
- Zhao, L.; Zhou, Q.; He, L.; Deng, L.; Lozano-Duran, R.; Li, G.; Zhu, J.K. DNA Methylation Underpins the Epigenomic Landscape Regulating Genome Transcription in Arabidopsis. Genome Biol. 2022, 23, 197. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; Zhao, L.; Zheng, X.; Gautam, M.; Yue, M.; Hou, J.; Chen, Z.; Wang, P.; Li, L. Dynamic Changes in Histone Modification Are Associated with Upregulation of Hsf and RRNA Genes during Heat Stress in Maize Seedlings. Protoplasma 2019, 256, 1245–1256. [Google Scholar] [CrossRef] [PubMed]
- Gayacharan; Joel, A.J. Epigenetic Responses to Drought Stress in Rice (Oryza sativa L.). Physiol. Mol. Biol. Plants 2013, 19, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Cheng, S.; Liu, X.; Du, H.; Dai, M.; Zhou, D.X.; Yang, W.; Zhao, Y. The R2R3-Type MYB Gene OsMYB91 Has a Function in Coordinating Plant Growth and Salt Stress Tolerance in Rice. Plant Sci. 2015, 236, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Abid, G.; Mingeot, D.; Muhovski, Y.; Mergeai, G.; Aouida, M.; Abdelkarim, S.; Aroua, I.; El Ayed, M.; M’hamdi, M.; Sassi, K.; et al. Analysis of DNA Methylation Patterns Associated with Drought Stress Response in Faba Bean (Vicia faba L.) Using Methylation-Sensitive Amplification Polymorphism (MSAP). Environ. Exp. Bot. 2017, 142, 34–44. [Google Scholar] [CrossRef]
- Benoit, M.; Drost, H.G.; Catoni, M.; Gouil, Q.; Lopez-Gomollon, S.; Baulcombe, D.; Paszkowski, J. Environmental and Epigenetic Regulation of Rider Retrotransposons in Tomato. PLoS Genet. 2019, 15, e1008370. [Google Scholar] [CrossRef]
- Sow, M.D.; Le Gac, A.L.; Fichot, R.; Lanciano, S.; Delaunay, A.; Le Jan, I.; Lesage-Descauses, M.C.; Citerne, S.; Caius, J.; Brunaud, V.; et al. RNAi Suppression of DNA Methylation Affects the Drought Stress Response and Genome Integrity in Transgenic Poplar. New Phytol. 2021, 232, 80–97. [Google Scholar] [CrossRef] [PubMed]
- Neves, D.M.; Almeida, L.A.D.H.; Santana-Vieira, D.D.S.; Freschi, L.; Ferreira, C.F.; Soares Filho, W.D.S.; Costa, M.G.C.; Micheli, F.; Coelho Filho, M.A.; Gesteira, A.D.S. Recurrent Water Deficit Causes Epigenetic and Hormonal Changes in Citrus Plants. Sci. Rep. 2017, 7, 13684. [Google Scholar] [CrossRef] [PubMed]
- Perrin, A.; Daccord, N.; Roquis, D.; Celton, J.M.; Vergne, E.; Bucher, E. Divergent DNA Methylation Signatures of Juvenile Seedlings, Grafts and Adult Apple Trees. Epigenomes 2020, 4, 4. [Google Scholar] [CrossRef]
- Santos, A.S.; Neves, D.M.; Santana-Vieira, D.D.S.; Almeida, L.A.H.; Costa, M.G.C.; Soares Filho, W.S.; Pirovani, C.P.; Coelho Filho, M.A.; Ferreira, C.F.; Gesteira, A.S. Citrus Scion and Rootstock Combinations Show Changes in DNA Methylation Profiles and ABA Insensitivity under Recurrent Drought Conditions. Sci. Hortic. 2020, 267, 109313. [Google Scholar] [CrossRef]
- Zhou, R.; Jiang, F.; Niu, L.; Song, X.; Yu, L.; Yang, Y.; Wu, Z. Increase Crop Resilience to Heat Stress Using Omic Strategies. Front. Plant Sci. 2022, 13, 891861. [Google Scholar] [CrossRef] [PubMed]
- Roychowdhury, R.; Das, S.P.; Gupta, A.; Parihar, P.; Chandrasekhar, K.; Sarker, U.; Kumar, A.; Ramrao, D.P.; Sudhakar, C. Multi-Omics Pipeline and Omics-Integration Approach To Decipher Plant’s Abiotic Stress Tolerance Responses. Genes 2023, 14, 1281. [Google Scholar] [CrossRef] [PubMed]
- Yadav, V.; Zhong, H.; Patel, M.K.; Zhang, S.; Zhou, X.; Zhang, C.; Zhang, J.; Su, J.; Zhang, F.; Wu, X. Integrated Omics-Based Exploration for Temperature Stress Resilience: An Approach to Smart Grape Breeding Strategies. Plant Stress 2024, 11, 100356. [Google Scholar] [CrossRef]
- Weighill, D.; Tschaplinski, T.J.; Tuskan, G.A.; Jacobson, D. Data Integration in Poplar: ‘omics Layers and Integration Strategies. Front. Genet. 2019, 10, 461699. [Google Scholar] [CrossRef] [PubMed]
- Leão, A.P.; Bittencourt, C.B.; Carvalho da Silva, T.L.; Rodrigues Neto, J.C.; Braga, Í.D.O.; Vieira, L.R.; de Aquino Ribeiro, J.A.; Abdelnur, P.V.; de Sousa, C.A.F.; Souza Júnior, M.T. Insights from a Multi-Omics Integration (MOI) Study in Oil Palm (Elaeis guineensis Jacq.) Response to Abiotic Stresses: Part Two—Drought. Plants 2022, 11, 2786. [Google Scholar] [CrossRef] [PubMed]
- Bittencourt, C.B.; Carvalho da Silva, T.L.; Rodrigues Neto, J.C.; Vieira, L.R.; Leão, A.P.; de Aquino Ribeiro, J.A.; Abdelnur, P.V.; de Sousa, C.A.F.; Souza, M.T. Insights from a Multi-Omics Integration (MOI) Study in Oil Palm (Elaeis guineensis Jacq.) Response to Abiotic Stresses: Part One—Salinity. Plants 2022, 11, 1755. [Google Scholar] [CrossRef]
- Guerrero-Sánchez, V.M.; López-Hidalgo, C.; Rey, M.-D.; Castillejo, M.Á.; Jorrín-Novo, J.V.; Escandón, M. Multiomic Data Integration in the Analysis of Drought-Responsive Mechanisms in Quercus ilex Seedlings. Plants 2022, 11, 3067. [Google Scholar] [CrossRef] [PubMed]
- Quiles, C.; Viera, I.; Roca, M. Multiomics Approach To Decipher the Origin of Chlorophyll Content in Virgin Olive Oil. J. Agric. Food Chem. 2022, 70, 3807–3817. [Google Scholar] [CrossRef]
- Sirangelo, T.M.; Forgione, I.; Zelasco, S.; Benincasa, C.; Perri, E.; Vendramin, E.; Angilè, F.; Fanizzi, F.P.; Sunseri, F.; Salimonti, A.; et al. Combined Transcriptomic and Metabolomic Approach Revealed a Relationship between Light Control, Photoprotective Pigments, and Lipid Biosynthesis in Olives. Int. J. Mol. Sci. 2023, 24, 14448. [Google Scholar] [CrossRef]
- Miladinov, G. Impacts of Population Growth and Economic Development on Food Security in Low-Income and Middle-Income Countries. Front. Hum. Dyn. 2023, 5, 1121662. [Google Scholar] [CrossRef]
| Species/Variety | Genome Size (GB) | Sequencing Technology | Year | References |
|---|---|---|---|---|
| Olea europaea L. cv. Farga | 1.38 | Illumina MiSeq—HiSeq | 2016 | [68] |
| Olea europaea L. subsp. Sylvestris | 1.46 | Illumina HiSEq 2000 | 2017 | [69] |
| Olea europaea L. cv. Picual | 1.63 to 1.81 | Illumina short reads, PacBio long reads | 2020 | [79] |
| Olea europaea L. cv. Farga | 1.31 | Illumina HiSeq 2000 | 2020 | [80] |
| Olea europaea L. cv. Arbequina | 1.30 | Oxford Nanopore third-generation sequencing, Hi-C technology | 2021 | [71] |
| Olea europaea L. cv. Farga | 1.31 | Illumina HiSeq 2000, Roche 454 | 2022 | [81] |
| Olea europaea L. cv. Leccino | 0.54 | |||
| Olea europaea L. cv. Ayvalik | 0.93 |
| Approach | Type of Abiotic Stress | Varieties | Tissue | References |
|---|---|---|---|---|
| Transcriptomics | Salinity | Kalamon Chondrolia Chalkidikis | roots leaves | [96,97] |
| Koroneiki Picual Royal de Cazorla Fadak86 | leaves | [98] | ||
| Leccino Frantoio | roots stems leaves | [99] | ||
| Arbequina | leaves | [100] | ||
| Kalamon | roots | [104] | ||
| Cold | Leccino | leaves | [101] | |
| Picual | leaves | [102] | ||
| Frantoio | seed coats embryos | [103] | ||
| Drought | Souri | Branches roots | [105] | |
| Souri | branches | [106] | ||
| Proteomics | Salinity | Chondrolia Chalkidikis | roots leaves | [107] |
| Chetoui | leaves | [108] | ||
| Metabolomics | UV-B radiation/heat shock | Cobrançosa | leaves | [109] |
| Koroneiki | leaves | [110] | ||
| Drought | Cobrançosa | leaves | [111] | |
| Epigenomics | Salinity | Koroneiki Royal de Cazorla Arbequina Picual | leaves | [112] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nteve, G.-M.; Kostas, S.; Polidoros, A.N.; Madesis, P.; Nianiou-Obeidat, I. Adaptation Mechanisms of Olive Tree under Drought Stress: The Potential of Modern Omics Approaches. Agriculture 2024, 14, 579. https://doi.org/10.3390/agriculture14040579
Nteve G-M, Kostas S, Polidoros AN, Madesis P, Nianiou-Obeidat I. Adaptation Mechanisms of Olive Tree under Drought Stress: The Potential of Modern Omics Approaches. Agriculture. 2024; 14(4):579. https://doi.org/10.3390/agriculture14040579
Chicago/Turabian StyleNteve, Georgia-Maria, Stefanos Kostas, Alexios N. Polidoros, Panagiotis Madesis, and Irini Nianiou-Obeidat. 2024. "Adaptation Mechanisms of Olive Tree under Drought Stress: The Potential of Modern Omics Approaches" Agriculture 14, no. 4: 579. https://doi.org/10.3390/agriculture14040579








