Root Exudates Promoted Microbial Diversity in the Sugar Beet Rhizosphere for Organic Nitrogen Mineralization
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Experimental Design
2.2. Soil Sampling and Determination of Soil Nitrogen, Amino Acids, Organic Acids, Soil Microbial Biomass, and Sugar Content
2.3. Soil Bacterial Amplicon Extraction and Sequencing
2.4. Sequencing Data Processing
2.5. Data Analyses
3. Results
3.1. The Structure of the Soil Microbial Community
3.2. Effects of the Sugar Beet Rhizosphere on Bacterial Co-Occurrence Patterns
3.3. Diversity Analysis of Microbial Communities by LEfSe
3.4. Differential Analysis of Sugar Beet Root Exudates and Soil Nitrogen Content
3.5. Correlation between Sugar Beet Root Exudates, Soil Nitrogen Content, and Bacterial Communities
4. Discussion
4.1. Complexity of Microbial Diversity and Networks in the Sugar Beet Rhizosphere Environment
4.2. The Role of the Root Exudates in Regulating Microbial Community Structure and Soil Nitrogen Mineralization
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, Y.C.; Chen, Y.; Tang, M.D.; Li, L.F.; Lin, X.Y.; Wang, Y.H.; Xu, D.H.; Ai, S.Y. Effects of ferrous sulfate and ferric nitrate on cadmium transportation in the rhizosphere soil-rice system. Huan Jing Ke Xue 2020, 41, 5143–5150. (In Chinese) [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Hu, B.; Chu, C. Nitrogen assimilation in plants: Current status and future prospects. J. Genet. Genomics 2022, 49, 394–404. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Ma, X.; Liu, J.; Wang, C.; Qin, J. Soil net organic nitrogen mineralization kinetics in response to short- and long-term land-use conversion of woodland to tea fields. Pol. J. Environ. Stud. 2021, 30, 4475–4483. [Google Scholar] [CrossRef]
- Keuper, F.; Dorrepaal, E.; Van Bodegom, P.M.; Logtestijn, R.; Venhuizen, G.; Hal, J.; Aerts, R. Experimentally increased nutrient availability at the permafrost thaw front selectively enhances biomass production of deep-rooting subarctic peatland species. Global Chang. Biol. 2017, 23, 4257–4266. [Google Scholar] [CrossRef]
- Liu, Y.; Evans, S.E.; Friesen, M.L.; Tiemann, L.K. Root exudates shift how N mineralization and N fixation contribute to the plant-available N supply in low fertility soils. Soil. Biol. Biochem. 2022, 165, 108541. [Google Scholar] [CrossRef]
- Cheng, Y.; Wang, J.; Chang, S.X.; Cai, Z.; Müller, C.; Zhang, J. Nitrogen deposition affects both net and gross soil nitrogen transformations in forest ecosystems: A review. Environ. Pollut. 2019, 244, 608–616. [Google Scholar] [CrossRef]
- Hui, C.; Sun, P.; Guo, X.; Jiang, H.; Zhao, Y.; Xu, L. Shifts in microbial community structure and soil nitrogen mineralization following short-term soil amendment with the ammonifier Bacillus amyloliquefaciens DT. Int. Biodeter. Biodegr. 2018, 132, 40–48. [Google Scholar] [CrossRef]
- Zecchin, S.; Mueller, R.C.; Seifert, J.; Stingl, U.; Anantharaman, K.; Bergen, M.; Cavalca, L.; Pester, M. Rice paddy Nitrospirae carry and express genes related to sulfate respiration: Proposal of the new genus “Candidatus Sulfobium”. Appl. Environ. Microbiol. 2018, 84, e02224-17. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Han, J.; Li, J.; Xu, Y.; Wang, X. Correction: Effects of long-term fertilization on soil organic carbon mineralization and microbial community structure. PLoS ONE 2019, 14, e0216006. [Google Scholar] [CrossRef]
- Mastný, J.; Bárta, J.; Kaštovská, E.; Picek, T. Decomposition of peatland DOC affected by root exudates is driven by specific r and K strategic bacterial taxa. Sci. Rep. 2021, 11, 18612–18677. [Google Scholar] [CrossRef]
- Badri, D.V.; Vivanco, J.M. Regulation and function of root exudates. Plant Cell Environ. 2009, 32, 666–681. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Zhang, W.; Hu, F.; Zhao, Q.-Q.; Lu, Y.; Kong, X.; Lü, J.-G. Influence of artificial root exudates and actual root exudates on the microbial community in pyrene-contaminated soil. Huan Jing Ke Xue 2022, 43, 1077–1088. (In Chinese) [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.J.; Wang, T.Q.; Hou, Z.J.; Wang, X.H.; Su, G.J.; Liu, Y.Q.; Zhou, Q. Responses of root exudates to intercropping of Chinese milk vetch with rape. Ying Yong Sheng Tai Xue Bao 2021, 32, 1783–1790. (In Chinese) [Google Scholar] [CrossRef] [PubMed]
- Meier, I.C.; Finzi, A.C.; Phillips, R.P. Root exudates increase N availability by stimulating microbial turnover of fast-cycling N pools. Soil. Biol. Biochem. 2016, 106, 119–128. [Google Scholar] [CrossRef]
- Zhao, M.; Zhao, J.; Yuan, J.; Hale, L.; Wen, T.; Huang, Q.; Vivanco, J.M.; Zhou, J.; Kowalchuk, G.A.; Shen, Q. Root exudates drive soil-microbe-nutrient feedbacks in response to plant growth. Plant Cell Environ. 2021, 44, 613–628. [Google Scholar] [CrossRef] [PubMed]
- Dessaux, Y.; Grandclement, C.; Faure, D. Engineering the rhizosphere. Trends Plant Sci. 2016, 21, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ruyter-Spira, C.; Bouwmeester, H.J. Engineering the plant rhizosphere. Curr. Opin. Biotechnol. 2015, 32, 136–142. [Google Scholar] [CrossRef]
- Li, J.; Liu, X.; Xu, L.; Li, W.; Yao, Q.; Yin, X.; Wang, Q.; Tan, W.; Xing, W.; Liu, D. Low nitrogen stress-induced transcriptome changes revealed the molecular response and tolerance characteristics in maintaining the C/N balance of sugar beet (Beta vulgaris L.). Front. Plant Sci. 2023, 14, 1164151. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.; Zhang, Y.; Xu, L.; Li, J.; Xing, W.; Liu, D. Effects of Different Nitrogen form Ratio on Key Enzymes of Nitrogen Assimilation in Sugar Beet. Crops 2024, 1–9. Available online: http://hfffg5fce84748f1d4cc2h9v90opkpcnww6fqq.fgfy.hlju.cwkeji.cn/kcms/detail/11.1808.S.20240318.1817.002.html (accessed on 13 June 2024).
- Wang, Q.H.; Guo, Y.N.; Hu, X.H.; Wang, X.C.; Deng, Y.H.; Zhou, J.C. Screening and variation analysis of soil organic nitrogen efficient in different Beta vulgaris genotypes. Bull. Bot. Res. 2017, 37, 563–571. (In Chinese) [Google Scholar] [CrossRef]
- Liu, S.; Song, B.; Sehar, S.; Adil, M.F.; Lin, X.; Huo, J.; Zhao, X.; Riaz, M. Variations in rhizosphere soil dominant and pathogenic flora improve boron-efficient Beta vulgaris L. yield under boron deficit. J. Clean. Prod. 2024, 444, 141241. [Google Scholar] [CrossRef]
- Lu, R.K. Analysis Methods of Soil Agricultural Chemistry; China Agricultural Science Technology Press: Beijing, China, 2000. (In Chinese) [Google Scholar]
- Devi, N.S.; Saha, D. Influence of organic matter vis-à-vis humic acid on the transformation of inorganic and organic forms of nitrogen in a Typic Haplustept soil. Commun. Soil. Sci. Plant Anal. 2017, 48, 1042–1051. [Google Scholar] [CrossRef]
- Díaz, J.; Lliberia, J.L.; Comellas, L.; Broto-Puig, F. Amino acid and amino sugar determination by derivatization with 6-aminoquinolyl-n-hydroxysuccinimidyl carbamate followed by high-performance liquid chromatography and fluorescence detection. J. Chromatogr. A 1996, 719, 171–179. [Google Scholar] [CrossRef]
- Edgar, R.C. Uparse: Highly accurate out sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microb. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microb. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Cui, R.; Geng, G.; Wang, G.; Stevanato, P.; Dong, Y.; Li, T.; Yu, L.; Wang, Y. The response of sugar beet rhizosphere micro-ecological environment to continuous cropping. Front. Microb. 2022, 13, 956785. [Google Scholar] [CrossRef]
- Hernández-Álvarez, C.; García-Oliva, F.; Cruz-Ortega, R.; Romero, M.F.; Barajas, H.R.; Piñero, D.; Alcaraz, L.D. Squash root microbiome transplants and metagenomic inspection for in situ arid adaptations. Sci. Total Environ. 2022, 805, 150136. [Google Scholar] [CrossRef]
- Bertoldo, G.; Della Lucia, M.C.; Squartini, A.; Concheri, G.; Broccanello, C.; Romano, A.; Ravi, S.; Cagnin, M.; Baglieri, A.; Stevanato, P. Endophytic microbiome responses to sulfur availability in Beta vulgaris (L.). Int. J. Mol. Sci. 2021, 22, 7184. [Google Scholar] [CrossRef]
- Kim, H.-S.; Lee, S.-H.; Jo, H.Y.; Finneran, K.T.; Kwon, M.J. Diversity and composition of soil Acidobacteria and Proteobacteria communities as a bacterial indicator of past land-use change from forest to farmland. Sci. Total Environ. 2021, 797, 148944. [Google Scholar] [CrossRef]
- Chen, L.; Jiang, Y.; Liang, C.; Luo, Y.; Xu, Q.; Han, C.; Zhao, Q.; Sun, B. Competitive interaction with keystone taxa induced negative priming under biochar amendments. Microbiome 2019, 7, 77. [Google Scholar] [CrossRef] [PubMed]
- Daims, H.; Nielsen, J.L.; Nielsen, P.H.; Schleifer, K.-H.; Wagner, M. In situ characterization of nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl. Environ. Microb. 2001, 67, 5273–5284. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.; Tong, D.; Li, Z.; Nie, X.; Liu, Y.; Ran, F.; Liao, S. Characteristics of microbial community composition and its relationship with carbon, nitrogen and sulfur in sediments. Sci. Total Environ. 2021, 795, 148848. [Google Scholar] [CrossRef] [PubMed]
- Gan, D.; Zeng, H.; Zhu, B. The rhizosphere effect on soil gross nitrogen mineralization: A meta-analysis. Soil. Ecol. Lett. 2022, 4, 144–154. [Google Scholar] [CrossRef]
- Kuypers, M.M.M.; Marchant, H.K.; Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microb. 2018, 16, 263–276. [Google Scholar] [CrossRef]
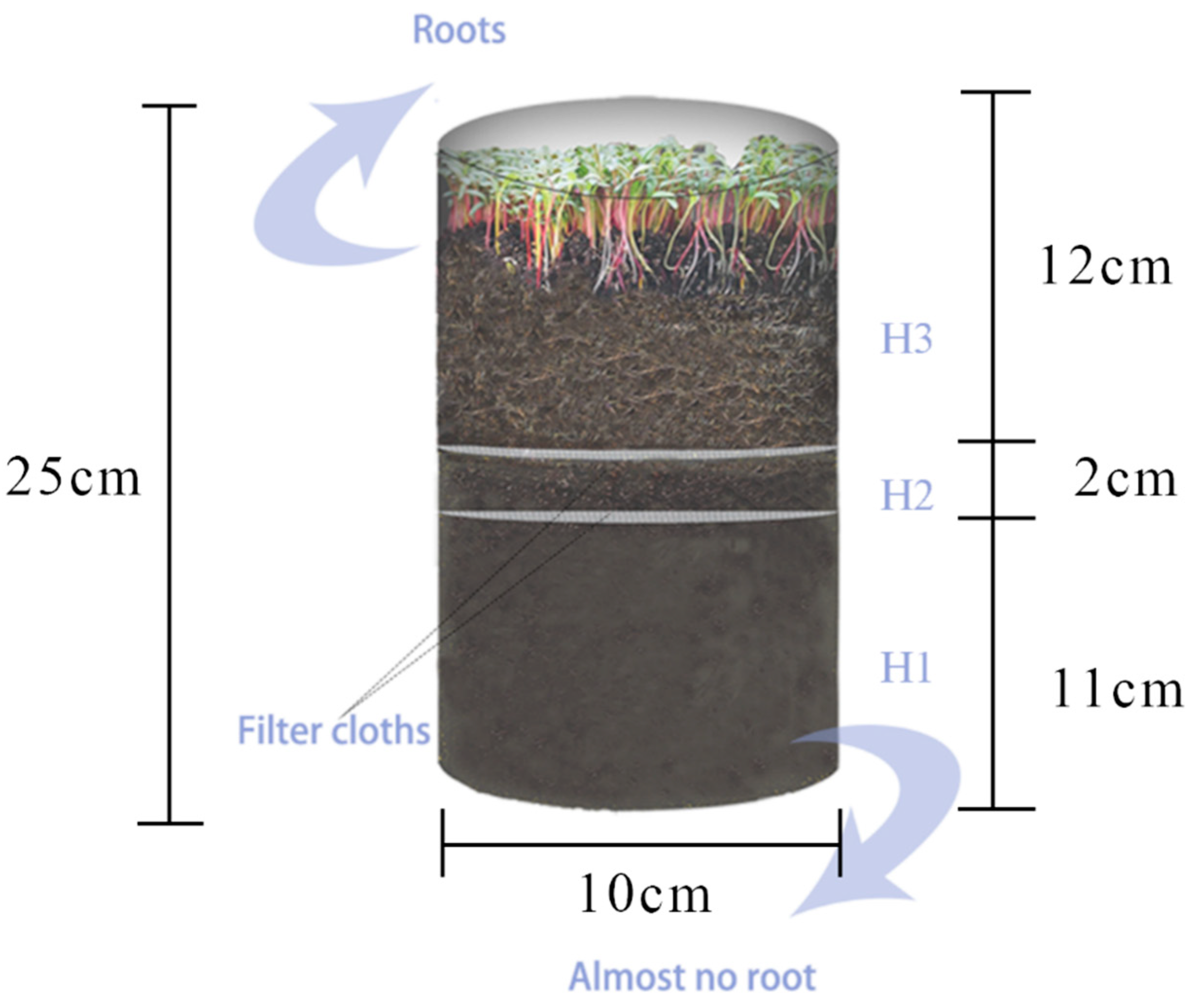

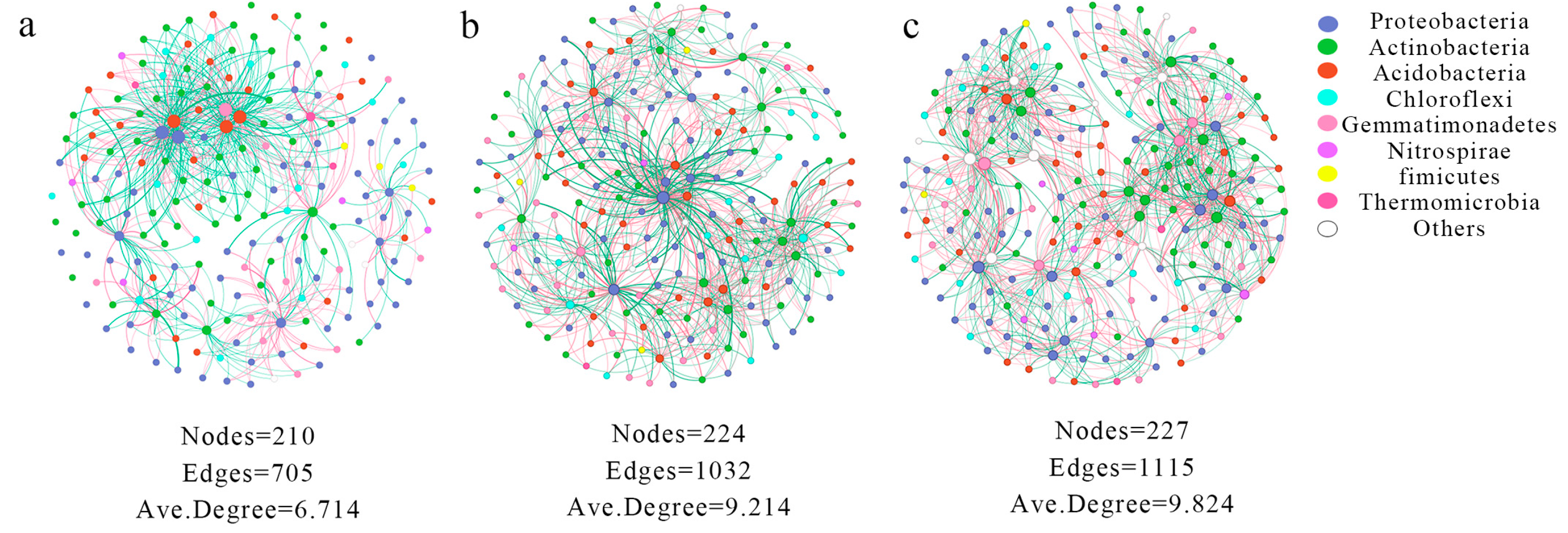
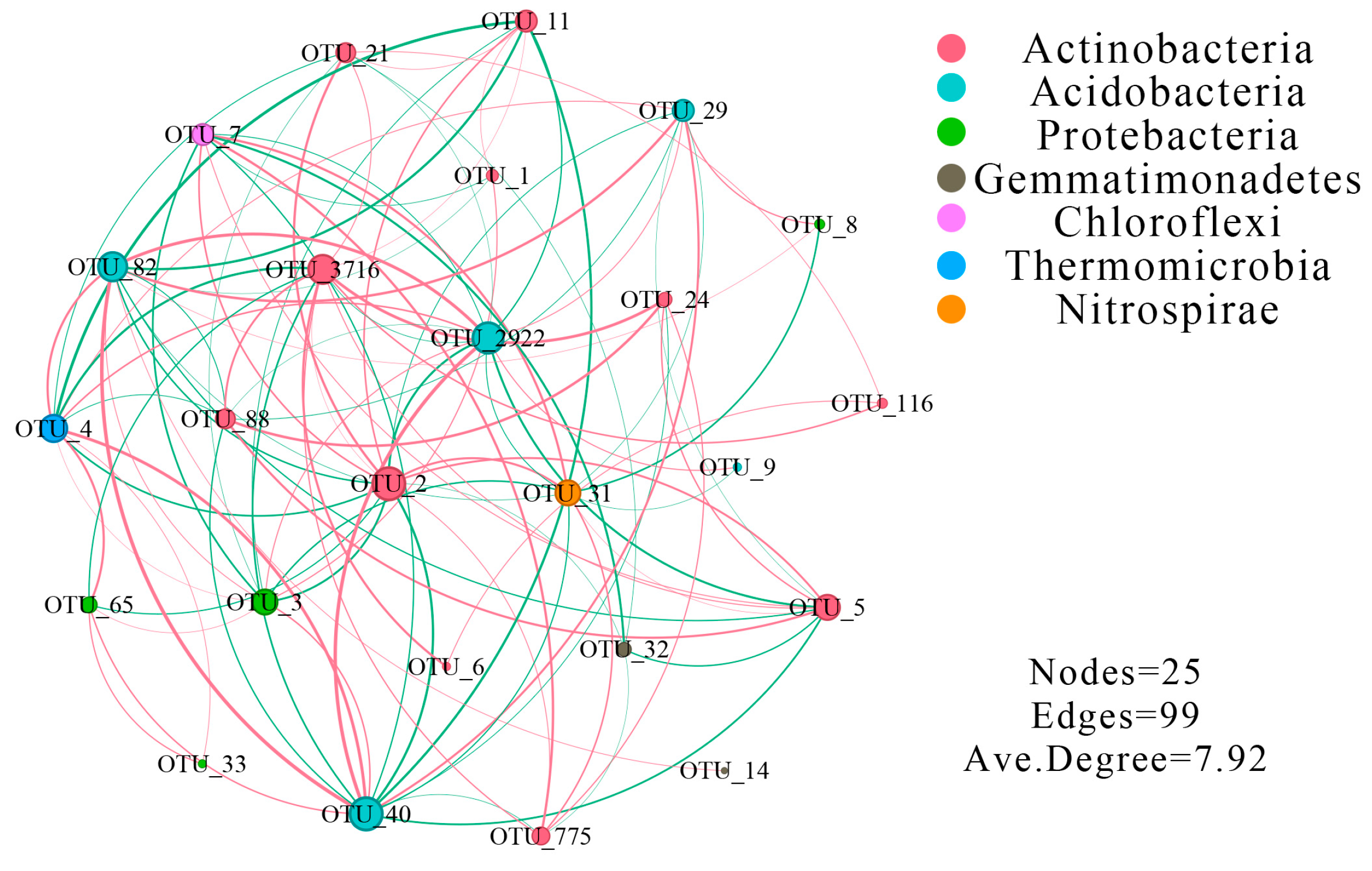
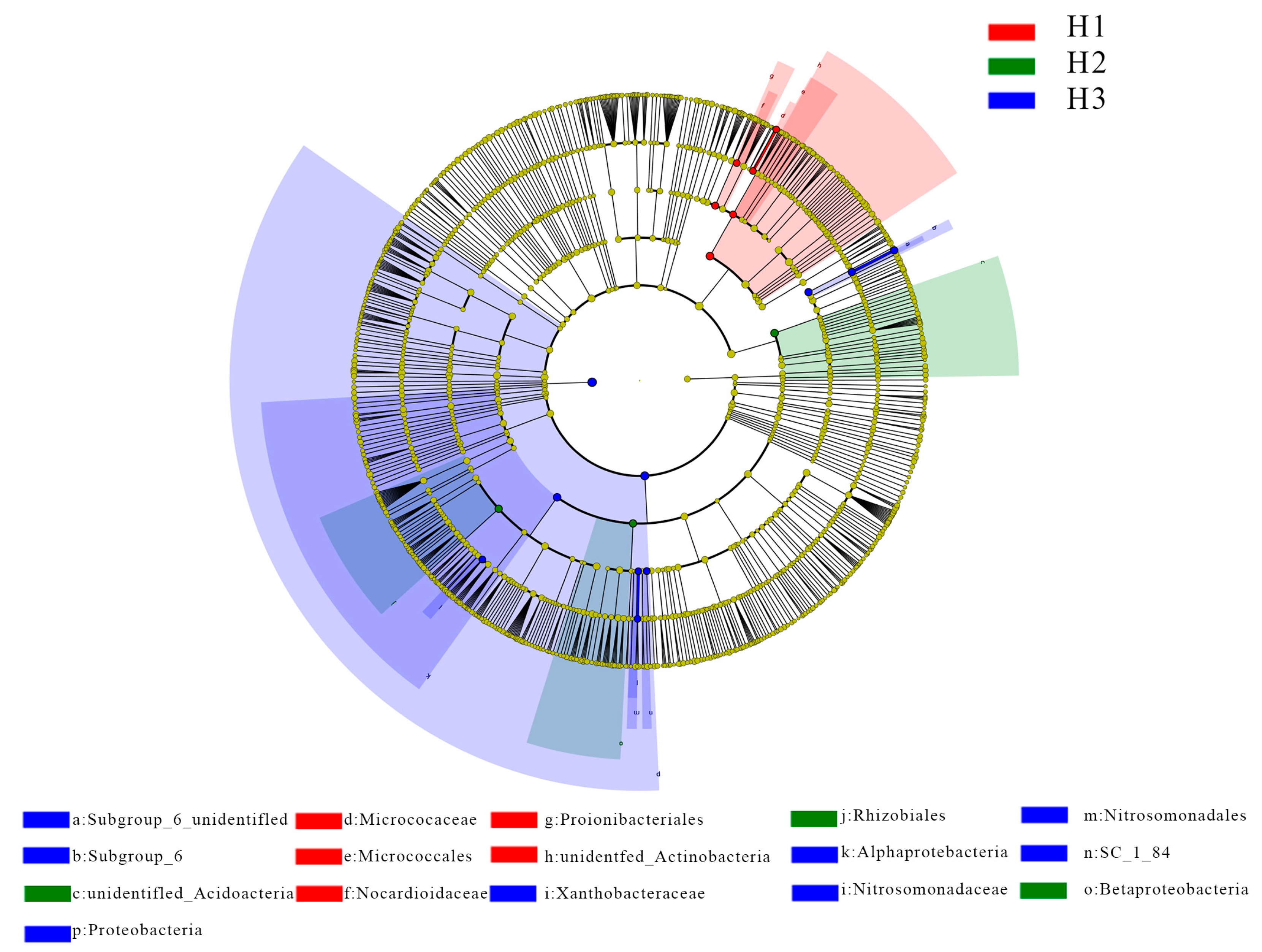



| Distance from the Root Surface (cm) | Bacteria (×104 cfu/g) | ||
|---|---|---|---|
| H3 | H2 | H1 | |
| KWS8138 | 60.3 ± 1.45a | 54.0 ± 1.20b | 47.0 ± 1.14c |
| Substance (µg/L) | H1 | H2 | H3 |
|---|---|---|---|
| Ser | 23.45 ± 0.04c | 26.12 ± 0.25b | 28.8 ± 0.28a |
| Gly | 10.35 ± 0.04a | 6.22 ± 0.12b | 3.95 ± 0.98c |
| Ala | 36.65 ± 0.04a | 33.46 ± 0.12b | 31 ± 0.21c |
| Cys | 61.5 ± 0.70b | 60.13 ± 0.59c | 59.45 ± 0.01c |
| Met | 31.6 ± 0.84c | 50.32 ± 1.81b | 54.85 ± 2.54b |
| Ile | 23.55 ± 0.53c | 26.21 ± 0.11b | 28.9 ± 0.16a |
| Leu | 38.25 ± 0.83c | 43.11 ± 0.57b | 46.4 ± 0.14a |
| Tyr | 22.3 ± 0.45a | 19.67 ± 0.55c | 18.95 ± 0.38c |
| Phe | 16.6 ± 0.42c | 18.05 ± 0.09b | 18.6 ± 0.28b |
| Lys | 28.25 ± 1.32c | 29.07 ± 0.19c | 29.4 ± 0.10c |
| His | 5.25 ± 0.02b | 5.41 ± 0.11c | 5.45 ± 0.07c |
| Arg | 22.7 ± 0.31c | 23.98 ± 0.014b | 24.3 ± 0.56b |
| Oxalic acid | 41.1 ± 0.56c | 130 ± 2.26b | 161.6 ± 1.66a |
| Formic acid | 5.3 ± 0.07c | 12.4 ± 0.14b | 16.5 ± 0.25a |
| Glucose | 0.45 ± 0.04c | 1.22 ± 0.01b | 1.52 ± 0.002a |
| N (g/kg) | H1 | H2 | H3 |
|---|---|---|---|
| AHON | 1.444 ± 0.075ab | 1.284 ± 0.018b | 1.576 ± 0.094a |
| AN | 0.466 ± 0.060b | 0.432 ± 0.001b | 0.597 ± 0.047a |
| ASN | 0.083 ± 0.012b | 0.077 ± 0.298a | 0.0436 ± 0.034b |
| AAN | 0.463 ± 0.089ab | 0.375 ± 0.018b | 0.531 ± 0.077a |
| TN | 2.186 ± 0.068b | 2.213 ± 0.350b | 2.48 ± 0.181a |
| TIN | 0.0197 ± 0.005b | 0.0206 ± 0.004b | 0.480 ± 0.069a |
| TON | 2.166 ± 0.069b | 2.213 ± 0.328a | 2.008 ± 0.130b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, D.; Xu, L.; Wang, H.; Xing, W.; Song, B.; Wang, Q. Root Exudates Promoted Microbial Diversity in the Sugar Beet Rhizosphere for Organic Nitrogen Mineralization. Agriculture 2024, 14, 1094. https://doi.org/10.3390/agriculture14071094
Liu D, Xu L, Wang H, Xing W, Song B, Wang Q. Root Exudates Promoted Microbial Diversity in the Sugar Beet Rhizosphere for Organic Nitrogen Mineralization. Agriculture. 2024; 14(7):1094. https://doi.org/10.3390/agriculture14071094
Chicago/Turabian StyleLiu, Dali, Lingqing Xu, Hao Wang, Wang Xing, Baiquan Song, and Qiuhong Wang. 2024. "Root Exudates Promoted Microbial Diversity in the Sugar Beet Rhizosphere for Organic Nitrogen Mineralization" Agriculture 14, no. 7: 1094. https://doi.org/10.3390/agriculture14071094
APA StyleLiu, D., Xu, L., Wang, H., Xing, W., Song, B., & Wang, Q. (2024). Root Exudates Promoted Microbial Diversity in the Sugar Beet Rhizosphere for Organic Nitrogen Mineralization. Agriculture, 14(7), 1094. https://doi.org/10.3390/agriculture14071094





