An Evaluation of Feature Selection Robustness on Class Noisy Data
Abstract
:1. Introduction
2. Background and Related Work
3. Methodology
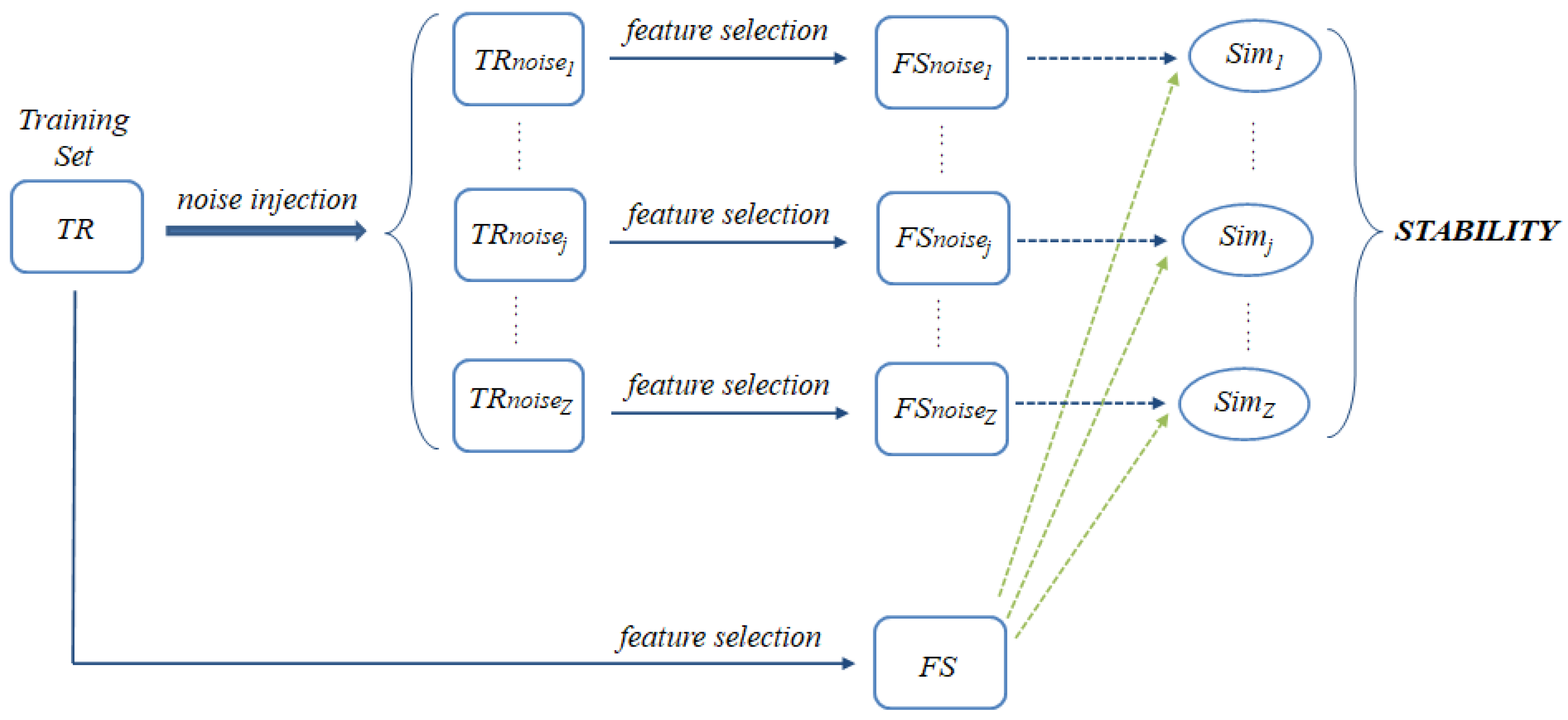
3.1. Noise Injection
3.2. Evaluating the Impact of Noise on Feature Selection
- The original training data () are perturbed according to the noise injection mechanism described in Section 3.1. Being such a mechanism completely random, the noise injection procedure is repeated several times (Z iterations), resulting in different perturbed training sets . The considered feature selection method is then applied to the original training set as well as to each perturbed training set , as shown in Figure 1. The feature subset selected from the original and the perturbed data are denoted, respectively, as and .
- To evaluate the impact of noise on the composition of the selected subsets, a proper consistency index is applied to compute the similarity [34] between each and , resulting in Z similarity scores , which are finally averaged to obtain an overall stability measure: the more similar the selection outcome obtained with and without noise injection, the more stable (robust) the selection process.
- Finally, to also evaluate the impact of noise on the final classification performance, a suitable learning algorithm is applied to the original training data , filtered to retain only the features in , as well as to each perturbed training set , in turn, filtered to retain only the features in . The induced models are evaluated on the same noise-free test set in order to compare the resulting performance (see Figure 2). Specifically, the average performance over the Z noise injection iterations is measured: the more similar it is to the performance without noise, the lower the overall impact of noise on the learning process.
4. Materials and Methods
4.1. Stability and Performance Metrics
4.2. Selection and Classification Methods
- Pearson’s Correlation (Correl): evaluates the importance of each feature by measuring its linear correlation with the target class [38]. The stronger the correlation, the more relevant the feature is for prediction. More in detail, it is defined as:where X is a generic feature, Y is the class label, is the covariance of X and Y while and are, respectively, the standard deviations of X and Y.
- Information Gain (InfoG): assesses the extent to which we can reduce the entropy of the class (i.e., the degree of uncertainty about its prediction) by observing the value of a given feature [39]:where and represent the entropy of the class Y before and after observing the feature X, respectively.
- Gain Ratio (GainR): basically, this is a variant of InfoG that attempts to compensate for its inherent tendency to favor features with more values [39]. Specifically, the InfoG definition is changed as follows:i.e., through a normalization factor expressing how broadly X splits the data:where is the number of instances in which X assumes the value , r is the number of distinct values of X, and R is the total number of training instances.
- One Rule (OneR): is a representative of embedded feature selection methods [40], which exploit a classifier to derive a relevance score for the features. Basically, for each feature in the training data, a one-level decision tree is generated based on that feature: this involves creating a simple classification rule by determining the majority class for each feature value. The accuracy of each rule is then computed, and the features are ranked based on the quality of the corresponding rules.Furthermore, among the multivariate approaches, we considered the following:
- ReliefF: evaluates the relevance of the features based on their ability to distinguish between data instances that are close to each other [41]. More in detail, the algorithm iteratively draws a sample instance from the training set in a repeated process, as per its original two-class formulation. Then, its nearest neighbors are considered, one from the same class (nearest hit H) and one from the opposite class (nearest miss M). For each feature X, a weight is then computed as follows:where m is the number of sample instances considered (which may coincide with the size of the training set), while represents the difference between the values of X in and M, and is the difference computed for and H. The rationale is that “good” features should have the same value for instances that belong to the same class and different values for instances of different classes.
- SVM-AW: exploits a linear SVM classifier to assign a weight to each feature, thus relying on the embedded feature selection paradigm [42]. In particular, a feature is ranked based on the weight given to the feature in the hyperplane function induced by the classifier:where is the N-dimensional feature vector, b is a bias constant, and is the weight vector (note that the absolute value of each weight, AW, is considered for feature ranking).
- SVM-RFE: also uses a linear SVM classifier to assign a weight to the features but adopts a recursive feature elimination (RFE) strategy that consists of removing the features with the lowest weights and repeating the evaluation on the remaining features, as originally proposed in [43]. The ranking process involves multiple iterations, in each of which a fixed percentage p of features is removed: the lower p, the higher the computational cost of the method (since more iterations occur). Given the high dimensionality of the datasets involved in our analysis, we set to keep the computational cost contained.
4.3. Datasets
5. Experimental Analysis
5.1. Methodological Implementation
- The implementation of an algorithm to introduce perturbations in the training set;
- The creation of procedures for applying iterative protocols such as simple holdout, repeated holdout, and cross-validation;
- The implementation of methods to calculate and generate output for stability and performance measures.
5.2. Settings
5.3. Results on Text Categorization Datasets
5.4. Results on Microarray Datasets
5.5. Results on Others Dataset
5.6. Discussion
6. Conclusions and Future Work
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bolón-Canedo, V.; Alonso-Betanzos, A.; Morán-Fernández, L.; Cancela, B. Feature Selection: From the Past to the Future. In Advances in Selected Artificial Intelligence Areas: World Outstanding Women in Artificial Intelligence; Springer International Publishing: Cham, Switerland, 2022; pp. 11–34. [Google Scholar] [CrossRef]
- Gupta, S.; Gupta, A. Dealing with Noise Problem in Machine Learning Data-sets: A Systematic Review. Procedia Comput. Sci. 2019, 161, 466–474. [Google Scholar] [CrossRef]
- García, S.; Luengo, J.; Herrera, F. Dealing with Noisy Data. In Data Preprocessing in Data Mining; Springer International Publishing: Cham, Switerland, 2015; pp. 107–145. [Google Scholar] [CrossRef]
- Frénay, B.; Doquire, G.; Verleysen, M. Estimating mutual information for feature selection in the presence of label noise. Comput. Stat. Data Anal. 2014, 71, 832–848. [Google Scholar] [CrossRef]
- Wald, R.; Khoshgoftaar, T.M.; Shanab, A.A. The effect of measurement approach and noise level on gene selection stability. In Proceedings of the 2012 IEEE International Conference on Bioinformatics and Biomedicine, Philadelphia, PA, USA, 4–7 October 2012; pp. 1–5. [Google Scholar] [CrossRef]
- Saseendran, A.; Setia, L.; Chhabria, V.; Chakraborty, D.; Barman Roy, A. Impact of Noise in Dataset on Machine Learning Algorithms. Mach. Learn. Res. 2019; early-review. [Google Scholar] [CrossRef]
- Shanthini, A.; Vinodhini, G.; Chandrasekaran, R.M.; Supraja, P. A taxonomy on impact of label noise and feature noise using machine learning techniques. Soft Comput. 2019, 23, 8597–8607. [Google Scholar] [CrossRef]
- Fawzi, A.; Moosavi-Dezfooli, S.M.; Frossard, P. Robustness of Classifiers: From Adversarial to Random Noise. Adv. Neural Inf. Process. Syst. 2016, 29, 1632–1640. [Google Scholar]
- Anyfantis, D.; Karagiannopoulos, M.; Kotsiantis, S.; Pintelas, P. Robustness of learning techniques in handling class noise in imbalanced datasets. In Artificial Intelligence and Innovations 2007: From Theory to Applications; Springer: Boston, MA, USA, 2007; Volume 247, pp. 21–28. [Google Scholar] [CrossRef] [Green Version]
- Frenay, B.; Verleysen, M. Classification in the Presence of Label Noise: A Survey. IEEE Trans. Neural Netw. Learn. Syst. 2014, 25, 845–869. [Google Scholar] [CrossRef]
- Bolón-Canedo, V.; Sánchez-Maroño, N.; Alonso-Betanzos, A. A review of feature selection methods on synthetic data. Knowl. Inf. Syst. 2013, 34, 483–519. [Google Scholar] [CrossRef]
- Nogueira, S.; Sechidis, K.; Brown, G. On the Stability of Feature Selection Algorithms. J. Mach. Learn. Res. 2018, 18, 1–54. [Google Scholar]
- Altidor, W.; Khoshgoftaar, T.M.; Napolitano, A. A noise-based stability evaluation of threshold-based feature selection techniques. In Proceedings of the 2011 IEEE International Conference on Information Reuse & Integration, Las Vegas, NV, USA, 3–5 August 2011; pp. 240–245. [Google Scholar] [CrossRef]
- Pes, B. Evaluating Feature Selection Robustness on High-Dimensional Data. In Hybrid Artificial Intelligence Systems; Springer: Boston, MA, USA, 2018. [Google Scholar]
- Gamberger, D.; Lavrac, N.; Džeroski, S. Noise Detection and Elimination in data Preprocessing: Experiments in Medical Domains. Appl. Artif. Intell. 2000, 14, 205–223. [Google Scholar] [CrossRef]
- Sánchez, J.; Barandela, R.; Marqués, A.; Alejo, R.; Badenas, J. Analysis of new techniques to obtain quality training sets. Pattern Recognit. Lett. 2003, 24, 1015–1022. [Google Scholar] [CrossRef]
- Zhu, X.; Wu, X.; Yang, Y. Error Detection and Impact-Sensitive Instance Ranking in Noisy Datasets. Proc. Natl. Conf. Artif. Intell. 2004, 1, 378–384. [Google Scholar]
- Kim, S.; Zhang, H.; Wu, R.; Gong, L. Dealing with noise in defect prediction. In Proceedings of the 2011 33rd International Conference on Software Engineering (ICSE), Honolulu, HI, USA, 21–28 May 2011; pp. 481–490. [Google Scholar] [CrossRef]
- Van Hulse, J.; Khoshgoftaar, T. Knowledge discovery from imbalanced and noisy data. Data & Knowl. Eng. 2009, 68, 1513–1542. [Google Scholar] [CrossRef]
- Nettleton, D.F.; Orriols-Puig, A.; Fornells, A. A study of the effect of different types of noise on the precision of supervised learning techniques. Artif. Intell. Rev. 2010, 33, 275–306. [Google Scholar] [CrossRef]
- Johnson, J.M.; Khoshgoftaar, T.M. A Survey on Classifying Big Data with Label Noise. J. Data Inf. Qual. 2022, 14, 1–43. [Google Scholar] [CrossRef]
- He, S.; Chen, H.; Zhu, Z.; Ward, D.G.; Cooper, H.J.; Viant, M.R.; Heath, J.K.; Yao, X. Robust twin boosting for feature selection from high-dimensional omics data with label noise. Inf. Sci. 2015, 291, 1–18. [Google Scholar] [CrossRef]
- Zhang, W.; Rekaya, R.; Bertrand, K. A method for predicting disease subtypes in presence of misclassification among training samples using gene expression: Application to human breast cancer. Bioinformatics 2006, 22, 317–325. [Google Scholar] [CrossRef] [Green Version]
- Abu Shanab, A.; Khoshgoftaar, T. Filter-Based Subset Selection for Easy, Moderate, and Hard Bioinformatics Data. In Proceedings of the 2018 IEEE International Conference on Information Reuse and Integration (IRI), Salt Lake City, UT, USA, 6–9 July 2018; pp. 372–377. [Google Scholar] [CrossRef]
- Pes, B. Feature Selection for High-Dimensional Data: The Issue of Stability. In Proceedings of the 2017 IEEE 26th International Conference on Enabling Technologies: Infrastructure for Collaborative Enterprises (WETICE), Poznan, Poland, 21–23 June 2017; pp. 170–175. [Google Scholar] [CrossRef]
- Khaire, U.M.; Dhanalakshmi, R. Stability of feature selection algorithm: A review. J. King Saud Univ.-Comput. Inf. Sci. 2022, 34, 1060–1073. [Google Scholar] [CrossRef]
- Li, F.; Mi, H.; Yang, F. Exploring the stability of feature selection for imbalanced intrusion detection data. In Proceedings of the 2011 9th IEEE International Conference on Control and Automation (ICCA), Santiago, Chile, 19–21 December 2011; pp. 750–754. [Google Scholar] [CrossRef]
- Dessì, N.; Pes, B. Stability in Biomarker Discovery: Does Ensemble Feature Selection Really Help? In Proceedings of the Current Approaches in Applied Artificial Intelligence; Springer International Publishing: Cham, Switerland, 2015; pp. 191–200. [Google Scholar]
- Dessì, N.; Pes, B.; Angioni, M. On Stability of Ensemble Gene Selection. In Proceedings of the Intelligent Data Engineering and Automated Learning—IDEAL 2015; Springer International Publishing: Cham, Switerland, 2015; pp. 416–423. [Google Scholar]
- Wang, H.; Khoshgoftaar, T.M.; Seliya, N. On the Stability of Feature Selection Methods in Software Quality Prediction: An Empirical Investigation. Int. J. Softw. Eng. Knowl. Eng. 2015, 25, 1467–1490. [Google Scholar] [CrossRef]
- Jiang, L.; Haiminen, N.; Carrieri, A.P.; Huang, S.; Vázquez-Baeza, Y.; Parida, L.; Kim, H.C.; Swafford, A.D.; Knight, R.; Natarajan, L. Utilizing stability criteria in choosing feature selection methods yields reproducible results in microbiome data. Biometrics 2022, 78, 1155–1167. [Google Scholar] [CrossRef]
- Manca, M.M.; Pes, B.; Riboni, D. Exploiting Feature Selection in Human Activity Recognition: Methodological Insights and Empirical Results Using Mobile Sensor Data. IEEE Access 2022, 10, 64043–64058. [Google Scholar] [CrossRef]
- Zhu, X.; Wu, X. Cost-guided class noise handling for effective cost-sensitive learning. In Proceedings of the Fourth IEEE International Conference on Data Mining (ICDM’04), Brighton, UK, 1–4 November 2004; pp. 297–304. [Google Scholar] [CrossRef]
- Cannas, L.M.; Dessì, N.; Pes, B. Assessing similarity of feature selection techniques in high-dimensional domains. Pattern Recognit. Lett. 2013, 34, 1446–1453. [Google Scholar] [CrossRef] [Green Version]
- Kuncheva, L. A stability index for feature selection. In Proceedings of the 25th IASTED International Multi-Conference: Artificial Intelligence and Applications, Innsbruck, Austria, 12–14 February 2007; ACTA Press: Anaheim, CA, USA, 2007; pp. 390–395. [Google Scholar]
- Almugren, N.; Alshamlan, H. A survey on hybrid feature selection methods in microarray gene expression data for cancer classification. IEEE Access 2019, 7, 78533–78548. [Google Scholar] [CrossRef]
- Cannas, L.M.; Dessì, N.; Pes, B. A filter-based evolutionary approach for selecting features in high-dimensional micro-array data. In Intelligent Information Processing V, Proceedings of the 6th IFIP TC 12 International Conference, IIP 2010, Manchester, UK, 13–16 October 2010; Proceedings 6; Springer: Berlin/Heidelberg, Germany, 2010; pp. 297–307. [Google Scholar]
- Tan, P.N.; Steinbach, M.; Karpatne, A.; Kumar, V. Introduction to Data Mining; Pearson: London, UK, 2019. [Google Scholar]
- Witten, I.H.; Frank, E.; Hall, M.A.; Pal, C.J. Data Mining: Practical Machine Learning Tools and Techniques, 4th ed.; Morgan Kaufmann: San Francisco, CA, USA, 2016. [Google Scholar]
- Dessì, N.; Pes, B. Similarity of feature selection methods: An empirical study across data intensive classification tasks. Expert Syst. Appl. 2015, 42, 4632–4642. [Google Scholar] [CrossRef]
- Urbanowicz, R.J.; Meeker, M.; La Cava, W.; Olson, R.S.; Moore, J.H. Relief-based feature selection: Introduction and review. J. Biomed. Inform. 2018, 85, 189–203. [Google Scholar] [CrossRef]
- Bolón-Canedo, V.; Sánchez-Maroño, N.; Alonso-Betanzos, A. Feature Selection for High-Dimensional Data; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Guyon, I.; Weston, J.; Barnhill, S.; Vapnik, V. Gene Selection for Cancer Classification using Support Vector Machines. Mach. Learn. 2002, 46, 389–422. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Golub, T.R.; Slonim, D.K.; Tamayo, P.; Huard, C.; Gaasenbeek, M.; Mesirov, J.P.; Coller, H.A.; Loh, M.L.; Downing, J.R.; Caligiuri, M.A.; et al. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science 1999, 286, 531–537. [Google Scholar] [CrossRef] [Green Version]
- Shipp, M.; Ross, K.; Tamayo, P.; Weng, A.; Kutok, J.; Aguiar, T.; Gaasenbeek, M.; Angelo, M.; Reich, M.; Pinkus, G.; et al. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat. Med. 2002, 8, 68–74. [Google Scholar] [CrossRef]
- Beer, D.; Kardia, S.; Huang, C.; Giordano, T.; Levin, A.; Misek, D.; Lin, L.; Chen, G.; Gharib, T.; Thomas, D.; et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat. Med. 2002, 8, 816–824. [Google Scholar] [CrossRef]
- Petricoin, E.F.; Ardekani, A.M.; Hitt, B.A.; Levine, P.J.; Fusaro, V.A.; Steinberg, S.M.; Mills, G.B.; Simone, C.; Fishman, D.A.; Kohn, E.C.; et al. Use of proteomic patterns in serum to identify ovarian cancer. Lancet 2002, 359, 572–577. [Google Scholar] [CrossRef]
- Tsanas, A.; Little, M.A.; Fox, C.; Ramig, L.O. Objective Automatic Assessment of Rehabilitative Speech Treatment in Parkinson’s Disease. IEEE Trans. Neural Syst. Rehabil. Eng. 2014, 22, 181–190. [Google Scholar] [CrossRef] [Green Version]
- Witten, I.H.; Frank, E.; Hall, M.A.; Pal, C.J. (Eds.) Appendix B—The WEKA workbench. In Data Mining, 4th ed.; Morgan Kaufmann: San Francisco, CA, USA, 2017; pp. 553–571. [Google Scholar] [CrossRef]
- Forman, G. An Extensive Empirical Study of Feature Selection Metrics for Text Classification. J. Mach. Learn. Res. 2003, 3, 1289–1305. [Google Scholar]
- Debole, F.; Sebastiani, F. An Analysis of the Relative Hardness of Reuters-21578 Subsets: Research Articles. J. Am. Soc. Inf. Sci. Technol. 2005, 56, 584–596. [Google Scholar] [CrossRef] [Green Version]




| Datasets | Number of Features | Number of Instances | Type of Datasets |
|---|---|---|---|
| Earn | 9499 | 12,897 | text categorization |
| Acq | 7494 | 12,897 | text categorization |
| Money | 7756 | 12,897 | text categorization |
| Leukemia | 7129 | 72 | microarray |
| Lymphoma | 7129 | 77 | microarray |
| Lung | 7129 | 96 | microarray |
| Ovarian | 15,155 | 253 | proteomics |
| Lsvt | 310 | 126 | biomedical |
| Datasets | Number of Total Instances | Number of Training Instances | Number of Test Instances |
|---|---|---|---|
| Earn | 12,897 | 9598 | 3299 |
| Acq | 12,897 | 9598 | 3299 |
| Money | 12,897 | 9598 | 3299 |
| Datasets | Number of Instances (Training Set) | Number of Positive Instances (Training Set) | noiseP | noiseT |
|---|---|---|---|---|
| Earn | 9598 | 2877 | 10% 20% | 6% 12% |
| Acq | 9598 | 1650 | 10% 20% | 3.5% 7% |
| Money | 9598 | 538 | 10% 20% | 1% 2% |
| Datasets | Number of Instances (Training Set) | Number of Positive Instances (Training Set) | noiseP | noiseT |
|---|---|---|---|---|
| Leukemia | 57 | 20 | 10% 20% | 7% 14% |
| Lymphoma | 61 | 15 | 10% 20% | 6% 10% |
| Lung | 76 | 8 | 10% 20% | 2.5% 5% |
| Datasets | Number of Total Instances | Number of Training Instances | Number of Test Instances |
|---|---|---|---|
| Leukemia | 72 | 57 | 15 |
| Lymphoma | 77 | 61 | 16 |
| Lung | 96 | 76 | 20 |
| Datasets | Number of Instances (Training Set) | Number of Positive Instances (Training Set) | noiseP | noiseT |
|---|---|---|---|---|
| Ovarian | 201 | 72 | 10% 20% | 7% 14% |
| LSVT | 100 | 33 | 10% 20% | 6% 13% |
| Datasets | Number of Total Instances | Number of Training Instances | Number of Test Instances |
|---|---|---|---|
| Ovarian | 253 | 201 | 52 |
| LSVT | 126 | 100 | 26 |
| Dataset | 1% of Features | NO FS | Correl | InfoG | GainR | OneR |
|---|---|---|---|---|---|---|
| Earn | clean | 0.96 | 0.98 | 0.97 | 0.96 | 0.97 |
| noiseP = 10% (noiseT = 6%) | 0.96 | 0.97 | 0.97 | 0.95 | 0.96 | |
| noiseP = 20% (noiseT = 12%) | 0.95 | 0.94 | 0.95 | 0.94 | 0.93 | |
| Acq | clean | 0.84 | 0.86 | 0.86 | 0.77 | 0.80 |
| noiseP = 10% (noiseT = 3.5%) | 0.74 | 0.83 | 0.83 | 0.51 | 0.77 | |
| noiseP = 20% (noiseT = 7%) | 0.58 | 0.78 | 0.79 | 0.17 | 0.70 | |
| Money | clean | 0.36 | 0.62 | 0.66 | 0.33 | 0.49 |
| noiseP = 10% (noiseT = 1%) | 0.33 | 0.56 | 0.60 | 0.17 | 0.41 | |
| noiseP = 20% (noiseT = 2%) | 0.28 | 0.49 | 0.54 | 0.13 | 0.32 |
| Dataset | 1% of Features | NO FS | Correl | InfoG | GainR | OneR | SVM-AW | SVM-RFE | ReliefF |
|---|---|---|---|---|---|---|---|---|---|
| Leukemia | clean | 0.78 | 0.94 | 0.91 | 0.96 | 0.91 | 0.89 | 0.95 | 0.96 |
| noiseP = 10% (noiseT = 7%) | 0.70 | 0.91 | 0.92 | 0.90 | 0.93 | 0.86 | 0.87 | 0.90 | |
| noiseP = 20% (noiseT = 14%) | 0.63 | 0.86 | 0.87 | 0.82 | 0.81 | 0.59 | 0.78 | 0.85 | |
| Lymphoma | clean | 0.47 | 0.83 | 0.70 | 0.75 | 0.74 | 0.84 | 0.91 | 0.78 |
| noiseP = 10% (noiseT = 6%) | 0.34 | 0.75 | 0.63 | 0.56 | 0.66 | 0.55 | 0.66 | 0.73 | |
| noiseP = 20% (noiseT = 10%) | 0.25 | 0.71 | 0.62 | 0.49 | 0.46 | 0.44 | 0.42 | 0.72 | |
| Lung-cancer | clean | 0.67 | 1.00 | 1.00 | 1.00 | 1.00 | 0.93 | 0.93 | 1.00 |
| noiseP = 10% (noiseT = 2.5%) | 0.51 | 0.93 | 0.91 | 0.84 | 0.88 | 0.74 | 0.80 | 0.94 | |
| noiseP = 20% (noiseT = 5%) | 0.13 | 0.81 | 0.87 | 0.74 | 0.85 | 0.48 | 0.59 | 0.85 |
| Dataset | Setting | NO FS | Correl | InfoG | GainR | OneR | SVM-AW | SVM-RFE | ReliefF |
|---|---|---|---|---|---|---|---|---|---|
| Ovarian (1% of features) | clean | 0.93 | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 | 0.99 | 0.98 |
| noiseP = 10% (noiseT = 7%) | 0.89 | 0.97 | 0.98 | 0.98 | 0.97 | 0.93 | 0.97 | 0.97 | |
| noiseP = 20% (noiseT = 14%) | 0.88 | 0.94 | 0.95 | 0.96 | 0.93 | 0.81 | 0.92 | 0.95 | |
| LSVT (2% of features) | clean | 0.77 | 0.70 | 0.64 | 0.60 | 0.77 | 0.63 | 0.71 | 0.75 |
| noiseP = 10% (noiseT = 6%) | 0.72 | 0.70 | 0.66 | 0.58 | 0.66 | 0.59 | 0.68 | 0.66 | |
| noiseP = 20% (noiseT = 13%) | 0.63 | 0.64 | 0.60 | 0.55 | 0.61 | 0.52 | 0.58 | 0.66 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pau, S.; Perniciano, A.; Pes, B.; Rubattu, D. An Evaluation of Feature Selection Robustness on Class Noisy Data. Information 2023, 14, 438. https://doi.org/10.3390/info14080438
Pau S, Perniciano A, Pes B, Rubattu D. An Evaluation of Feature Selection Robustness on Class Noisy Data. Information. 2023; 14(8):438. https://doi.org/10.3390/info14080438
Chicago/Turabian StylePau, Simone, Alessandra Perniciano, Barbara Pes, and Dario Rubattu. 2023. "An Evaluation of Feature Selection Robustness on Class Noisy Data" Information 14, no. 8: 438. https://doi.org/10.3390/info14080438
APA StylePau, S., Perniciano, A., Pes, B., & Rubattu, D. (2023). An Evaluation of Feature Selection Robustness on Class Noisy Data. Information, 14(8), 438. https://doi.org/10.3390/info14080438







