Early Parkinson’s Disease Diagnosis through Hand-Drawn Spiral and Wave Analysis Using Deep Learning Techniques
Abstract
1. Introduction
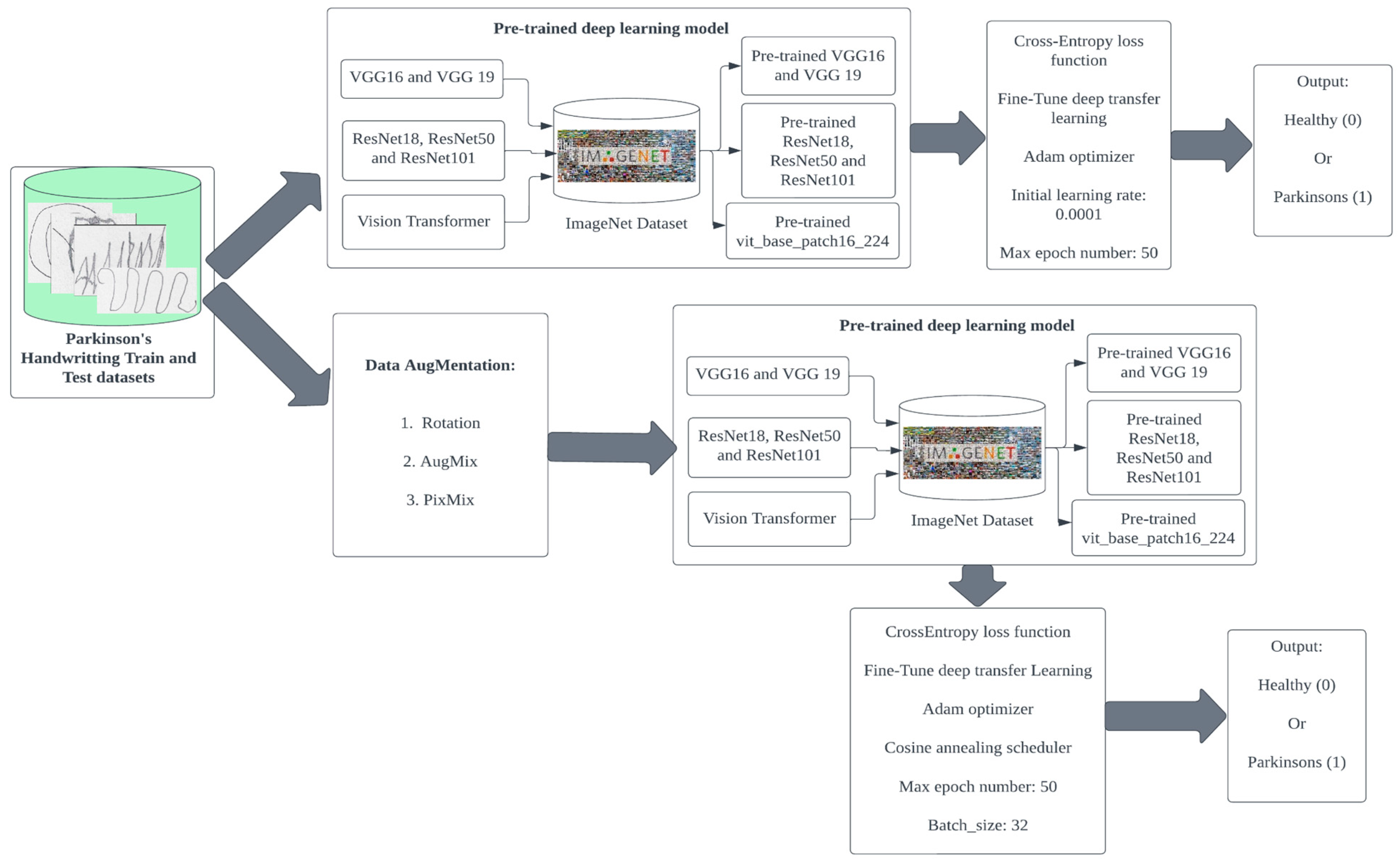
2. Methodology
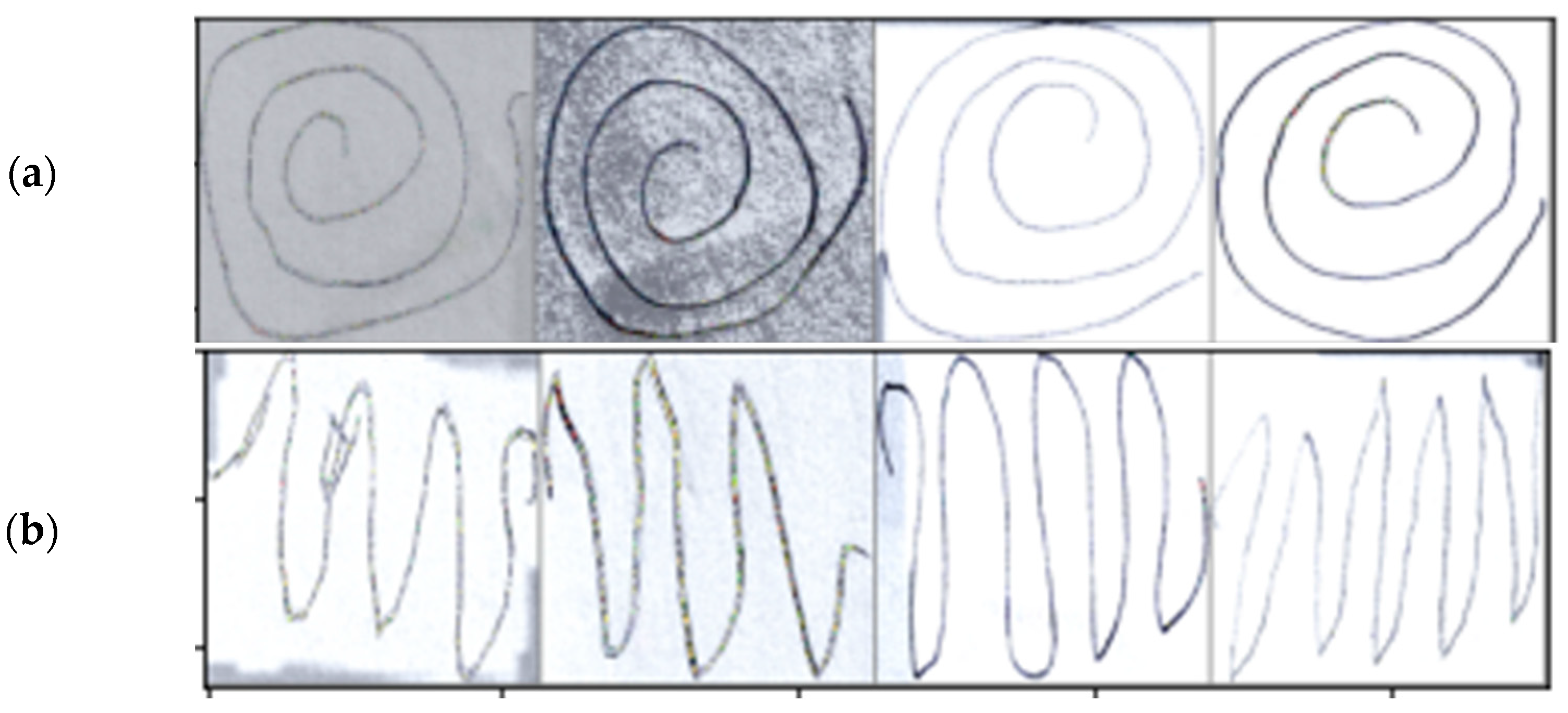
2.1. Dataset Preprocessing
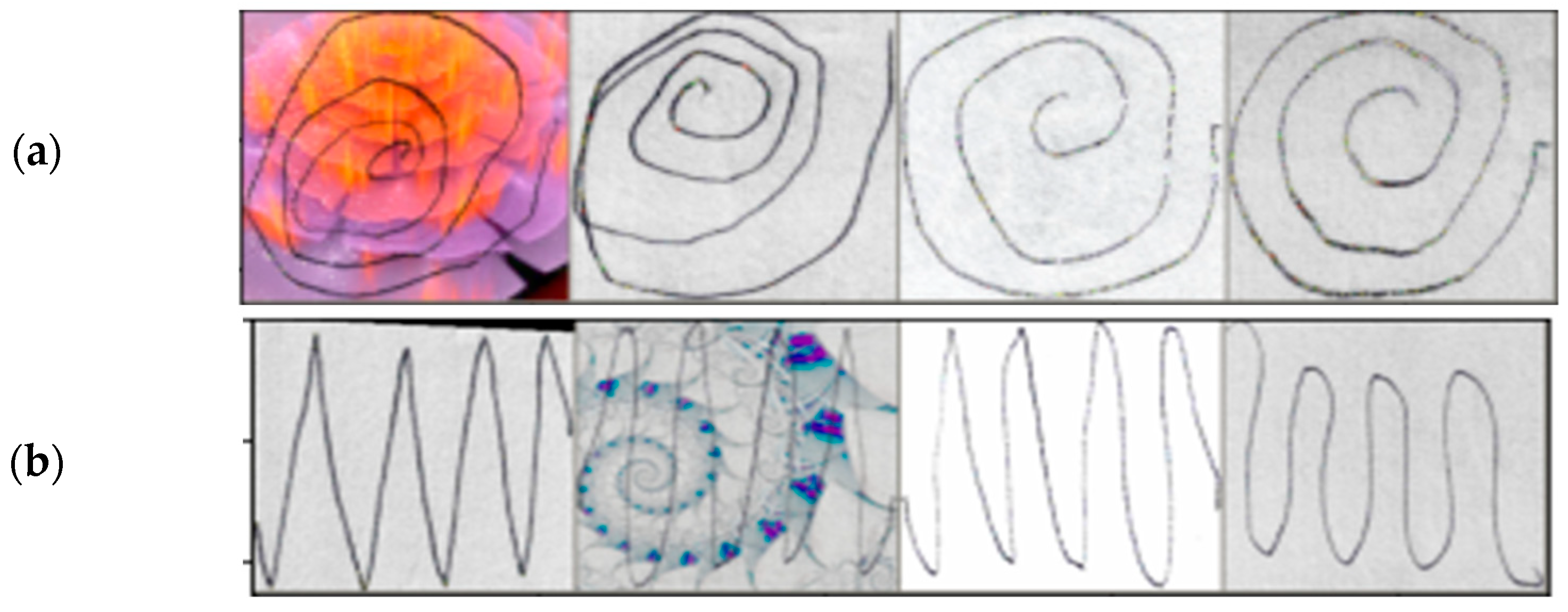
2.2. Data Augmentation
Cosine Annealing Schedule
2.3. Deep Learning Models
2.3.1. Residual Network (ResNet)
2.3.2. Convolutional Neural Networks (CNNs)
2.3.3. Vision Transformers (ViTs)
2.4. Evaluation Metrics
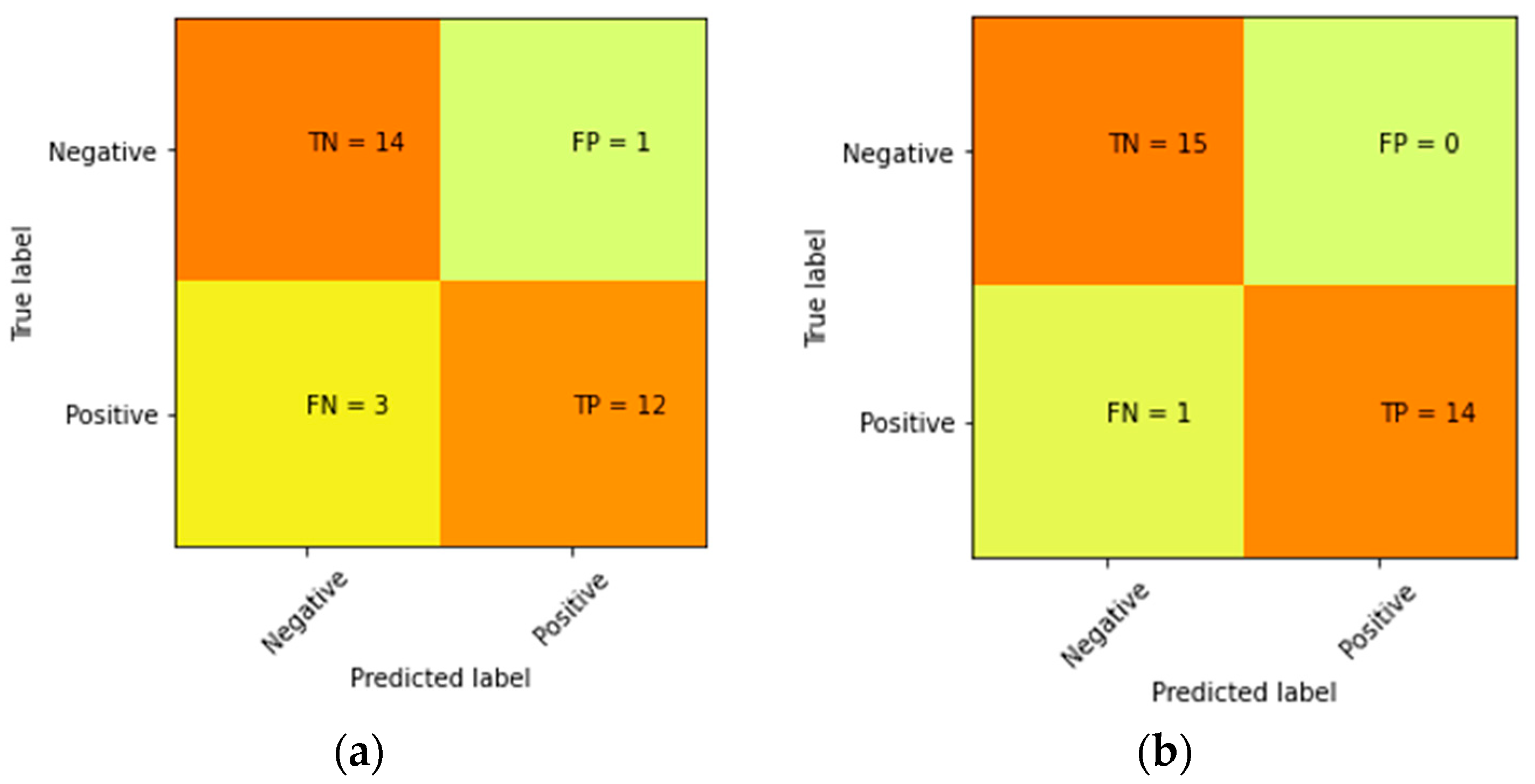
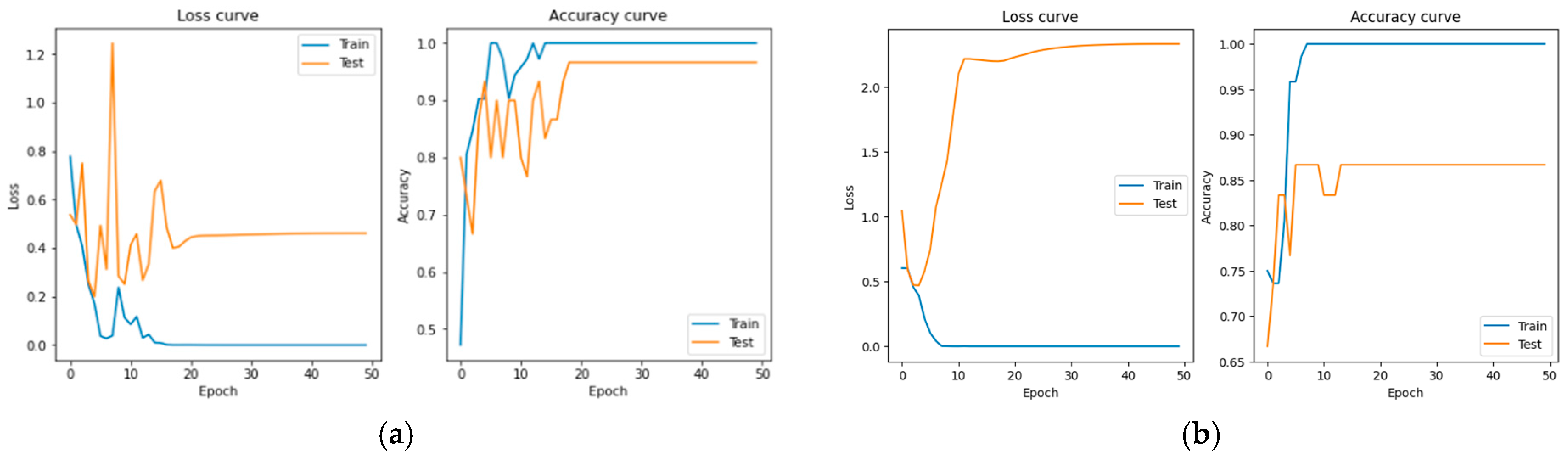
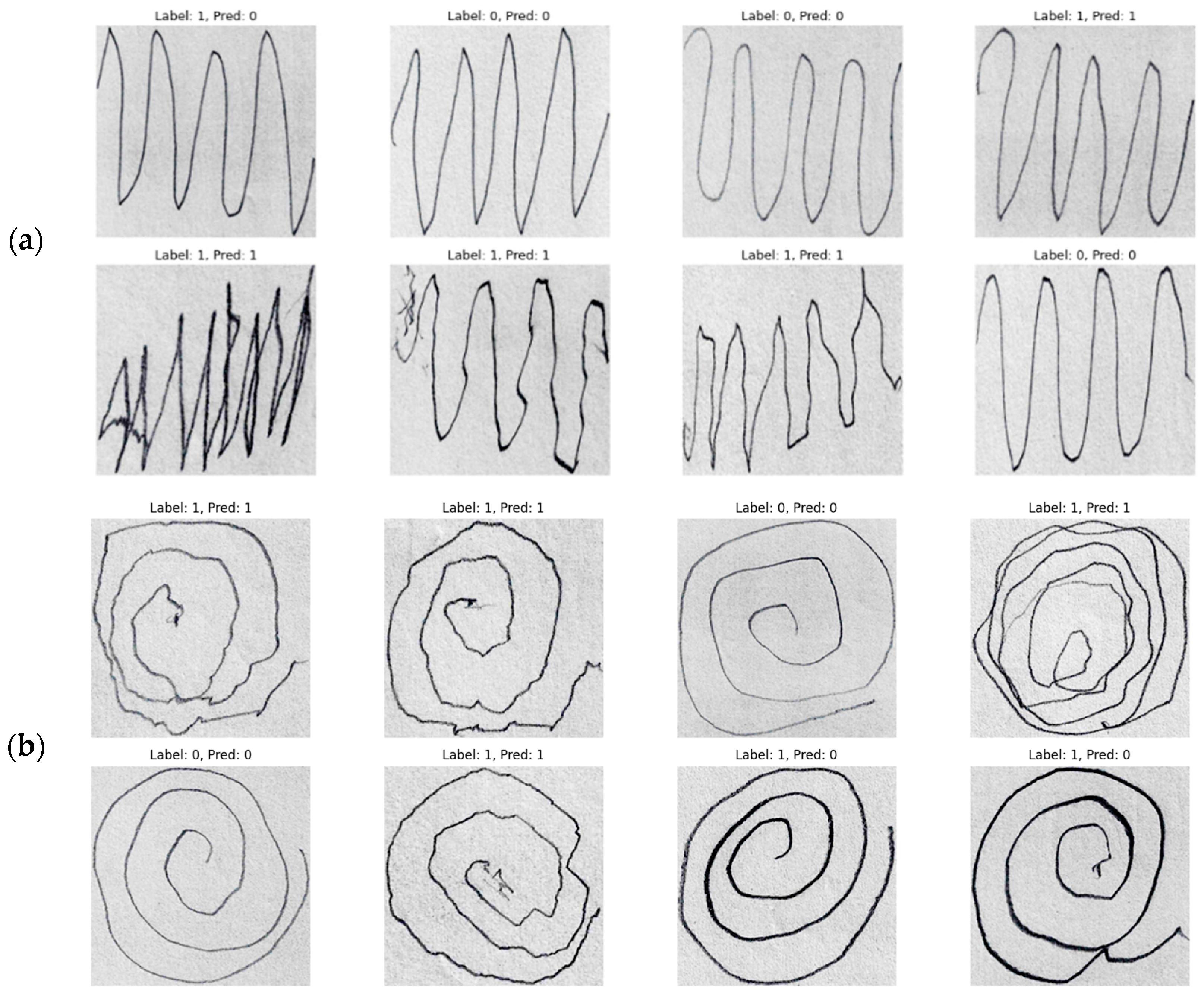
3. Result
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Postuma, R.B.; Berg, D.; Stern, M.; Poewe, W.; Olanow, C.W.; Oertel, W.; Obeso, J.; Marek, K.; Litvan, I.; Lang, A.E.; et al. MDS Clinical Diagnostic Criteria for Parkinson’s Disease. Mov. Disord. 2015, 30, 1591–1601. [Google Scholar] [CrossRef]
- Sveinbjornsdottir, S. The Clinical Symptoms of Parkinson’s Disease. J. Neurochem. 2016, 139, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Parkinson’s Australia. Statistics on Parkinson’s 2022. Available online: https://www.parkinsons.org.au/statistics/ (accessed on 29 February 2024).
- Kalia, L.V.; Lang, A.E. Parkinson’s Disease. Lancet 2015, 386, 896–912. [Google Scholar] [CrossRef] [PubMed]
- Huseyn, E. Deep Learning Based Early Diagnostics of Disease. arXiv 2020, arXiv:2008.01792. [Google Scholar] [CrossRef]
- Shaban, M. Deep Learning for Parkinson’s Disease Diagnosis: A Short Survey. Computer 2023, 12, 58. [Google Scholar] [CrossRef]
- Tanveer, M.; Richhariya, B.; Khan, R.U.; Rashid, A.H.; Khanna, P.; Prasad, M.; Lin, C.T. Machine Learning Techniques for the Diagnosis of Alzheimer’s Disease. ACM Trans. Multimed. Comput. Commun. Appl. 2020, 16, 1–35. [Google Scholar] [CrossRef]
- Zhou, Z.; Kanwal, A.; Chaturvedi, K.; Raza, R.; Prakash, S.; Jan, T.; Prasad, M. Deep Learning-Based Classification of Neurodegenerative Diseases Using Gait Dataset: A Comparative Study. In Proceedings of the 2023 International Conference on Robotics, Control and Vision Engineering, Tokyo, Japan, 21–23 July 2023; ACM: New York, NY, USA, 2023; pp. 59–64. [Google Scholar]
- Chatterjee, K.; Kumar, R.P.; Bandyopadhyay, A.; Swain, S.; Mallik, S.; Li, A.; Ray, K. PDD-ET: Parkinson’s Disease Detection Using ML Ensemble Techniques and Customized Big Dataset. Information 2023, 14, 502. [Google Scholar] [CrossRef]
- Zafeiropoulos, N.; Bitilis, P.; Tsekouras, G.E.; Kotis, K. Evaluating Ontology-Based PD Monitoring and Alerting in Personal Health Knowledge Graphs and Graph Neural Networks. Information 2024, 15, 100. [Google Scholar] [CrossRef]
- Dai, Y.; Song, Y.; Liu, W.; Bai, W.; Gao, Y.; Dong, X.; Lv, W. Multi-Focus Image Fusion Based on Convolution Neural Network for Parkinson’s Disease Image Classification. Diagnostics 2021, 11, 2379. [Google Scholar] [CrossRef] [PubMed]
- Hap, A.U.; Li, J.P.; Agbley, B.L.Y.; Mawuli, C.B.; Ali, Z.; Nazir, S.; Din, S.U. A Survey of Deep Learning Techniques Based Parkinson’s Disease Recognition Methods Employing Clinical Data. Expert Syst. Appl. 2022, 208, 118045. [Google Scholar] [CrossRef]
- Haller, S.; Simon Badoud, S.; Nguyen, D.; Garibotto, V. Individual Detection of Patients with Parkinson Disease Using Support Vector Machine Analysis of Diffusion Tensor Imaging Data: Initial Results. Am. J. Neuroradiol. 2012, 33, 2123–2128. [Google Scholar] [CrossRef] [PubMed]
- Tassew, T.M.; Xuan, N.; Chai, B. PDDS: A Software for the Early Diagnosis of Parkinson’s Disease from MRI and DaT Scan Images Using Detection and Segmentation Algorithms. Biomed. Signal Process. Control 2023, 86, 105140. [Google Scholar] [CrossRef]
- Isaacson, J.R.; Brillman, S.; Chhabria, N.; Isaacson, S.H. Impact of DaTscan Imaging on Clinical Decision Making in Clinically Uncertain Parkinson’s Disease. J. Park. Dis. 2021, 11, 885–889. [Google Scholar] [CrossRef] [PubMed]
- Zham, P.; Kumar, K.D.; Dabnichki, P.; Arjunan, P.S.; Raghav, S. Distinguishing Different Stages of Parkinson’s Disease Using Composite Index of Speed and Pen-Pressure of Sketching a Spiral. Front. Neurol. 2017, 8, 268142. [Google Scholar] [CrossRef] [PubMed]
- Pereira, R.D.; Pereira, D.R.; da Silva, F.A.; Hook, C.; Webber, S.A.; Pereira, L.A.; Papa, J.P. A Step towards the Automated Diagnosis of Parkinson’s Disease: Analyzing Handwriting Movements. In Proceedings of the IEEE International Symposium on Computer-Based Medical Systems 2015, Sao Carlos, Brazil, 22–25 June 2015. [Google Scholar]
- Basnin, N.; Sumi, A.T.; Hossain, S.M.; Andersson, K. Early Detection of Parkinson’s Disease from Micrographic Static Hand Drawings. In Proceedings of the 2021 International Conference on Brain Informatics, Virtual, 17–19 September 2021; Volume 12960, pp. 433–447. [Google Scholar]
- Das, H.S.; Das, A.; Neog, A.; Mallik, S.; Bora, K.; Zhao, Z. Early Detection of Parkinson’s Disease Using Fusion of Discrete Wavelet Transformation and Histograms of Oriented Gradients. Mathematics 2022, 10, 4218. [Google Scholar] [CrossRef]
- Kamran, I.; Naz, S.; Razzak, I.; Imran, M. Handwriting Dynamics Assessment Using Deep Neural Networks for Early Identification of Parkinson’s Disease. Future Gener. Comput. Syst. 2020, 117, 234–244. [Google Scholar] [CrossRef]
- Shaban, M. Deep Convolutional Neural Network for Parkinson’s Disease Based Handwriting Screening. In Proceedings of the IEEE 17th International Symposium on Biomedical Imaging Workshops (ISBI Workshops) 2020, Iowa City, IA, USA, 4 April 2020. [Google Scholar]
- Hendrycks, D.; Mu, N.; Cubuk, E.D.; Zoph, B.; Gilmer, J.; Lakshminarayanan, B. AugMix: A Simple Data Processing Method to Improve Robustness and Uncertainty. arXiv 2019, arXiv:1912.02781. [Google Scholar]
- Hendrycks, D.; Zou, A.; Mazeika, M.; Tang, L.; Song, D.; Steinhardt, J.; Li, B. PIXMIX: Dreamlike Pictures Comprehensively Improve Safety Measures. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022. [Google Scholar]
- Loshchilov, I.; Hutter, F. SGDR: Stochastic Gradient Descent with Warm Restarts. arXiv 2017, arXiv:1608.03983. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- Canziani, A.; Paszke, A.; Culurciello, E. An Analysis of Deep Neural Network Model for Practical Applications. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image Is Worth 16*16 Words: Transformers for Image Recognition at Scale. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020. [Google Scholar]
- Mei, J.; Desrosiers, C.; Frasnelli, J. Machine Learning for the Diagnosis of Parkinson’s Disease: A Review of Literature. Front. Aging Neurosci. 2021, 13, 633752. [Google Scholar] [CrossRef] [PubMed]

| Augmentation Methods | Dataset | VGG16 | VGG19 | ResNet18 | ResNet50 | ResNet101 | Vit_base_patch16_224 |
|---|---|---|---|---|---|---|---|
| No Augmentation | Wave | 92 | 93.33 | 86.67 | 81.32 | 85.33 | 68 |
| Spiral | 77.34 | 66 | 78.7 | 73.34 | 79.33 | 70 | |
| Rotation and flipping | Wave | 86.67 | 96.67 | 92.67 | 87.33 | 94 | 60 |
| Spiral | 80 | 90 | 86.67 | 87.33 | 84.67 | 86.67 | |
| AugMix | Wave | 90 | 86.67 | 90 | 76.67 | 84.67 | 64 |
| Spiral | 83.33 | 80 | 86.67 | 83.33 | 81.33 | 73.33 | |
| PixMix | Wave | 76.67 | 63.33 | 47.33 | 44 | 51.33 | 83.33 |
| Spiral | 46.67 | 53.33 | 52.67 | 50 | 47.33 | 86.67 |
| Methods | Dataset | VGG16 | VGG19 | ResNet18 | ResNet50 | ResNet101 | Vit_base_patch16_224 |
|---|---|---|---|---|---|---|---|
| W/O Cosine Annealing | Wave | 90 | 82 | 92.67 | 87.33 | 79.33 | 82.67 |
| Spiral | 79.33 | 83.33 | 83.33 | 82.67 | 86 | 66 | |
| Cosine Annealing | Wave | 92 | 96.67 | 92.67 | 87.33 | 94 | 83.33 |
| Spiral | 83.33 | 87.66 | 86.67 | 87.33 | 84.67 | 86.67 |
| Model | Dataset | Accuracy | Precision | Recall | MCC |
|---|---|---|---|---|---|
| VGG19 | Wave | 0.97 | 1 | 0.93 | 0.94 |
| Spiral | 0.87 | 0.92 | 0.80 | 0.74 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Chaturvedi, K.; Nayan, A.-A.; Hesamian, M.H.; Braytee, A.; Prasad, M. Early Parkinson’s Disease Diagnosis through Hand-Drawn Spiral and Wave Analysis Using Deep Learning Techniques. Information 2024, 15, 220. https://doi.org/10.3390/info15040220
Huang Y, Chaturvedi K, Nayan A-A, Hesamian MH, Braytee A, Prasad M. Early Parkinson’s Disease Diagnosis through Hand-Drawn Spiral and Wave Analysis Using Deep Learning Techniques. Information. 2024; 15(4):220. https://doi.org/10.3390/info15040220
Chicago/Turabian StyleHuang, Yingcong, Kunal Chaturvedi, Al-Akhir Nayan, Mohammad Hesam Hesamian, Ali Braytee, and Mukesh Prasad. 2024. "Early Parkinson’s Disease Diagnosis through Hand-Drawn Spiral and Wave Analysis Using Deep Learning Techniques" Information 15, no. 4: 220. https://doi.org/10.3390/info15040220
APA StyleHuang, Y., Chaturvedi, K., Nayan, A.-A., Hesamian, M. H., Braytee, A., & Prasad, M. (2024). Early Parkinson’s Disease Diagnosis through Hand-Drawn Spiral and Wave Analysis Using Deep Learning Techniques. Information, 15(4), 220. https://doi.org/10.3390/info15040220










