SMART: On-Site Rapid Detection of Nucleic Acid from Plants, Animals, and Microorganisms in under 25 Minutes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
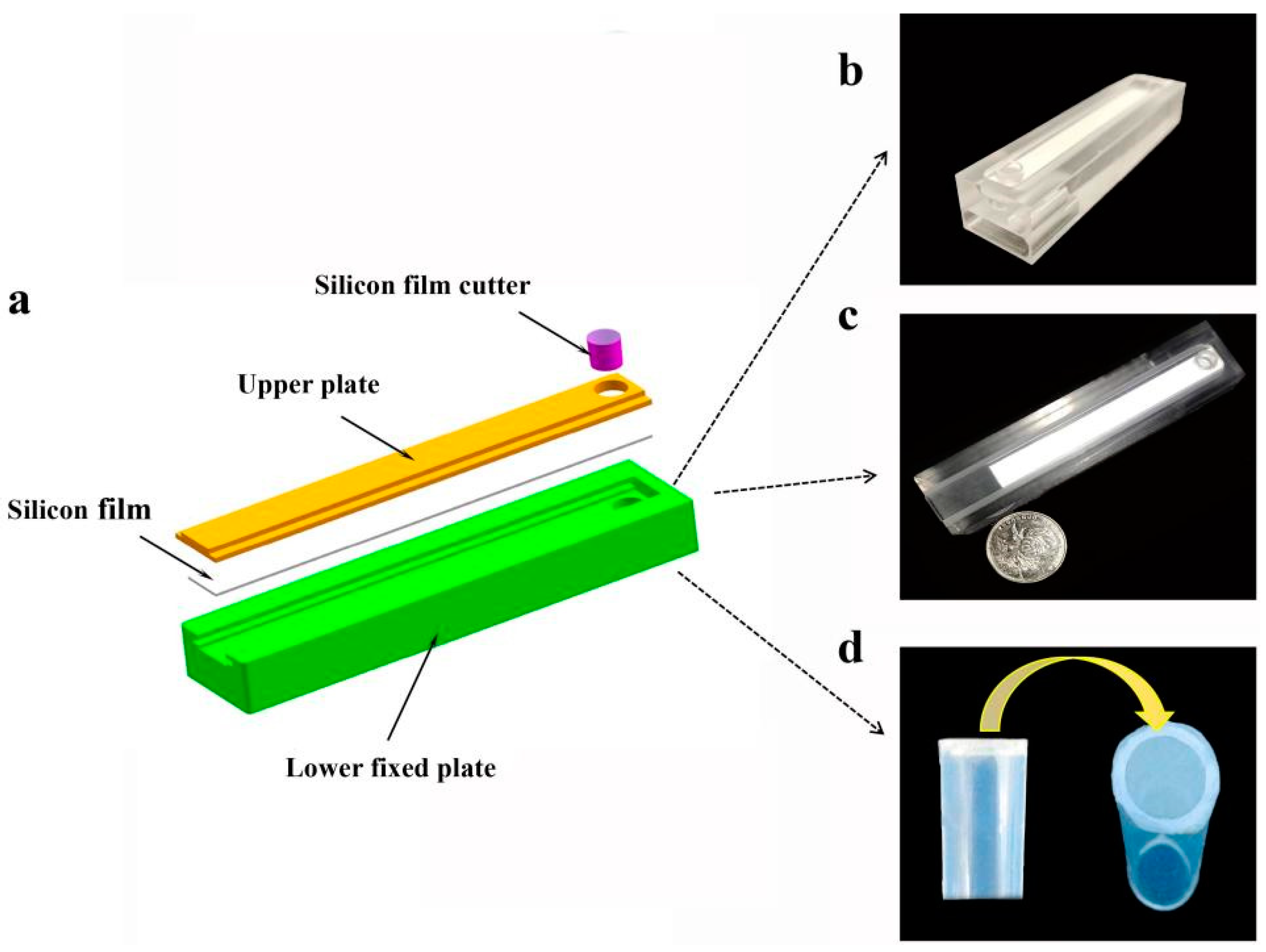
2.2. Design and Manufacturing of SMART
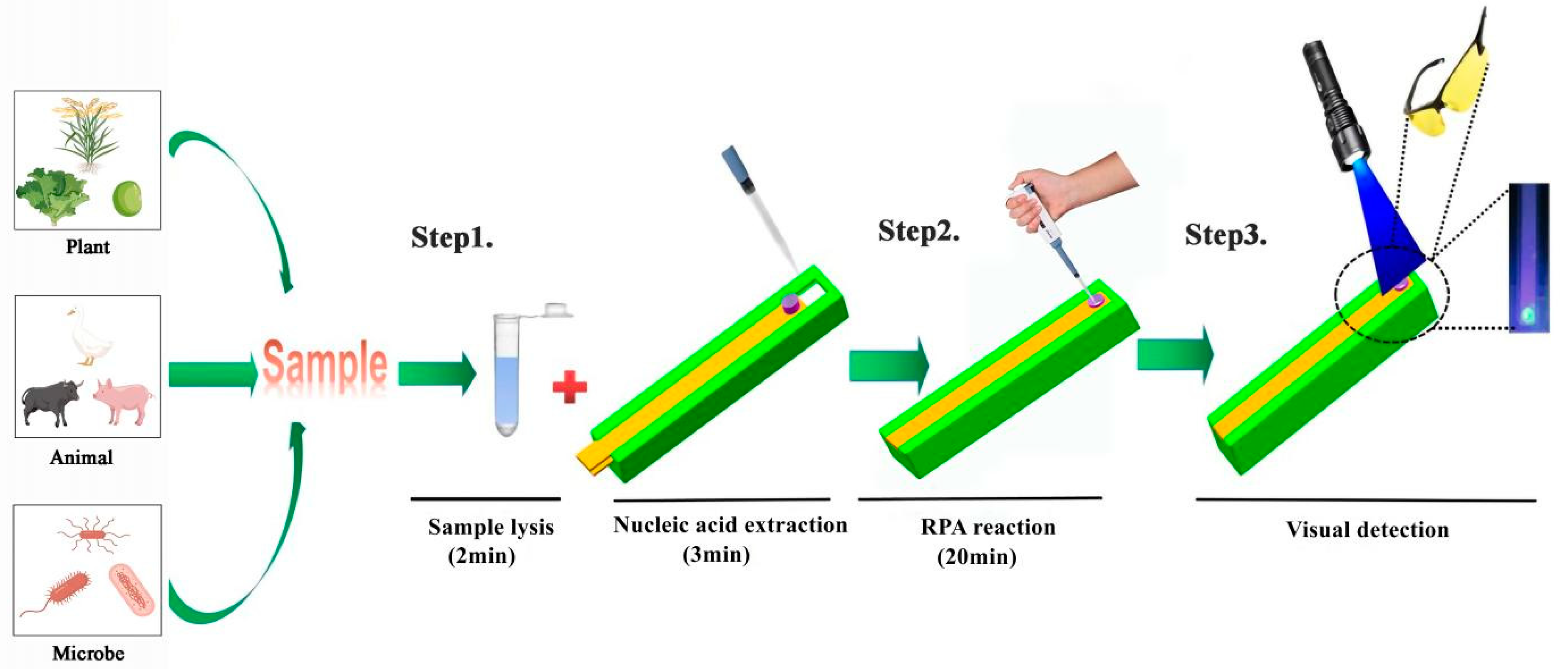
2.3. The Operation of SMART
2.4. Establishment of SMART Ultra-Fast DNA Adsorption System
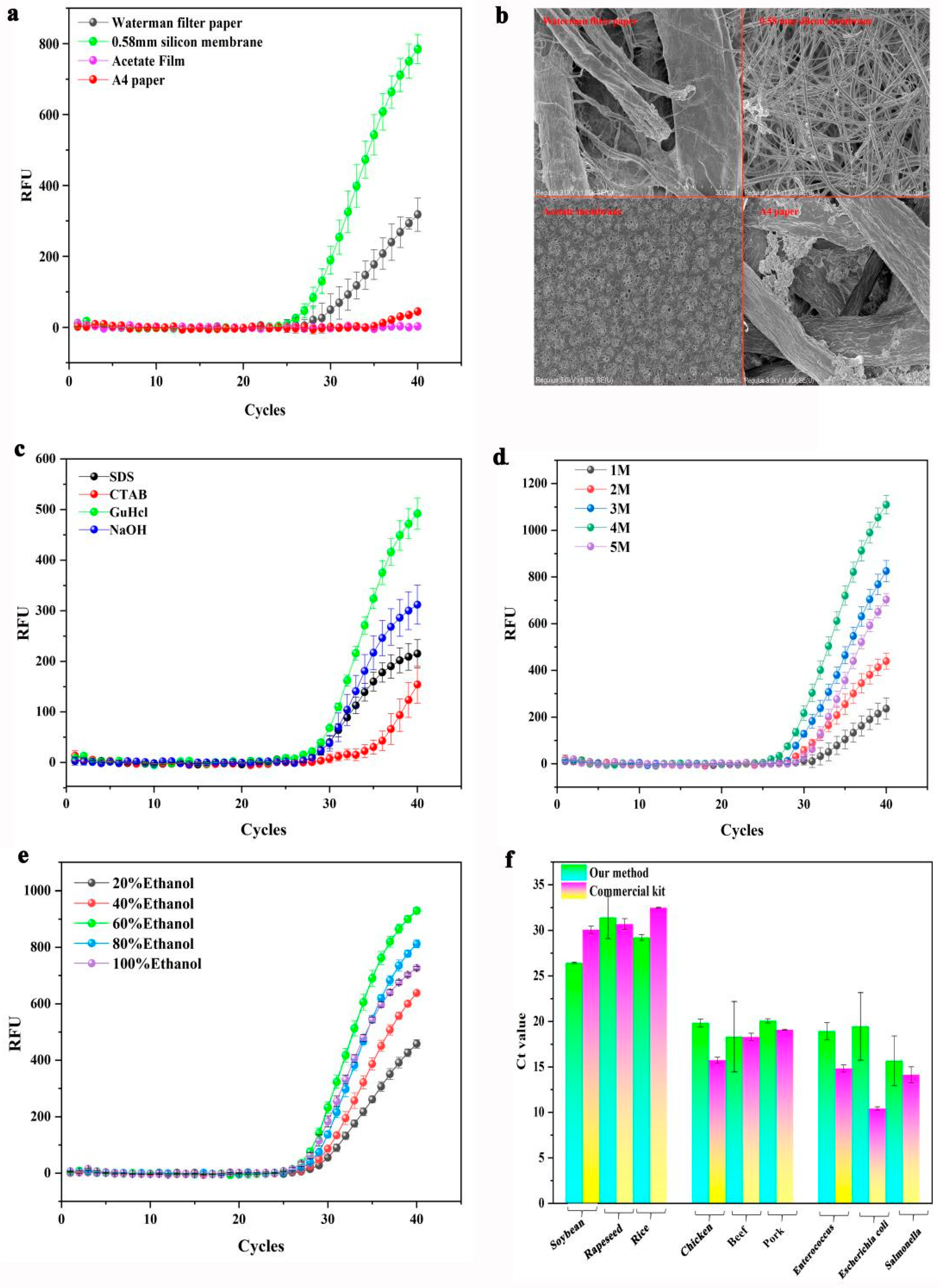
2.4.1. Screening of Adsorption Materials
2.4.2. Screening of Lysis Buffer
2.4.3. Screening of Washing Solution
2.4.4. Ultrafast Adsorption of DNA from Different Samples
2.5. DNA Amplification Reactions and Product Detection
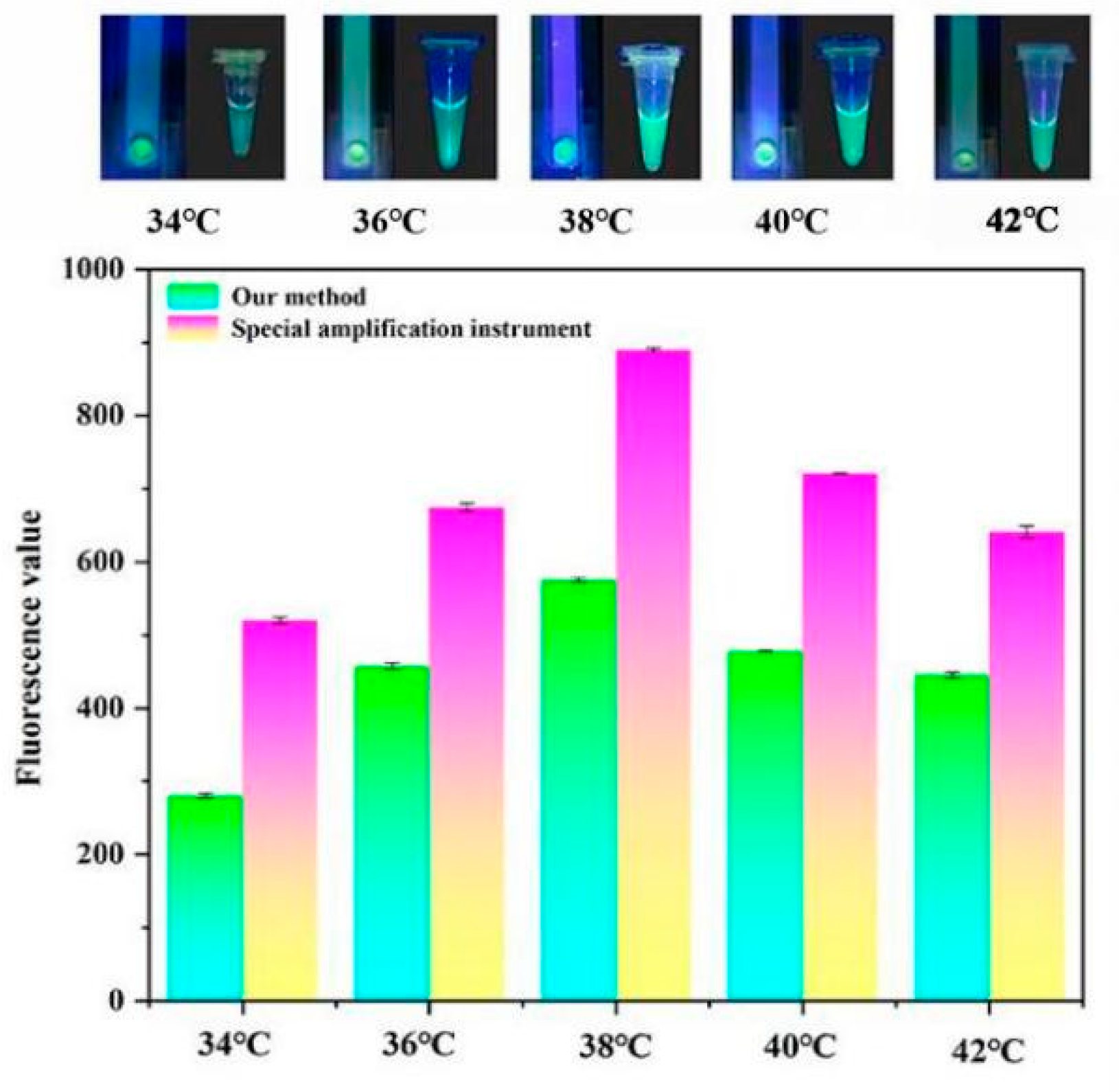
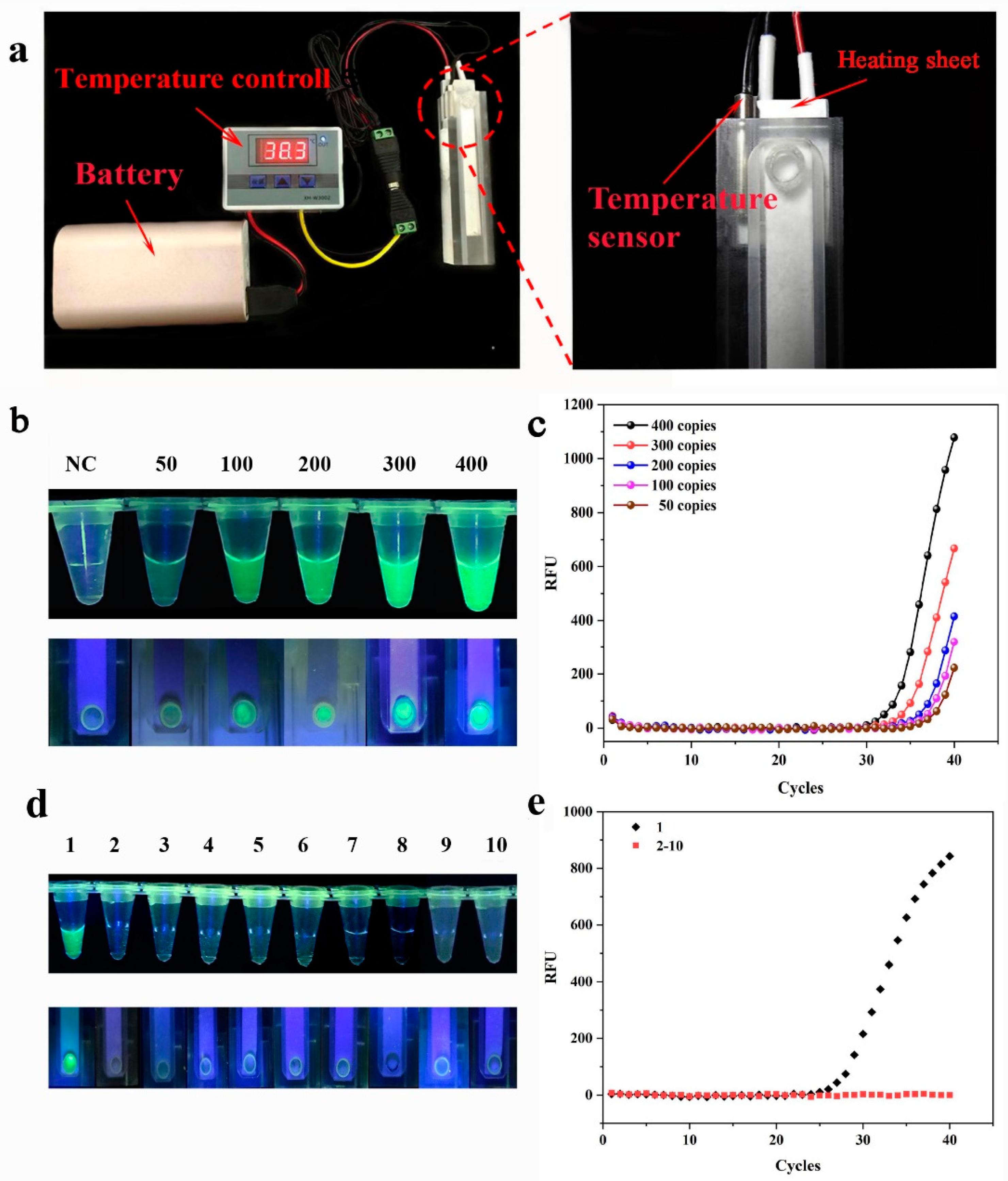
2.6. Temperature Control Experiment
2.7. Sensitivity and Specificity of SMART
2.8. Real Sample On-Site Detection Using SMART
3. Results and Discussions
3.1. DNA Adsorption System
3.1.1. Performance Evaluation of DNA Adsorption Materials
3.1.2. Performance Evaluation of Lysis Buffer
3.1.3. Washing Performance Evaluation of Washing Buffer
3.1.4. Evaluation of Rapid DNA Adsorption Method
3.2. Temperature Control Experiments
3.3. Sensitivity and Specificity of SMART
3.4. On-Site Detection Results of SMART
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Levin, R.E. The application of real-time PCR to food and agricultural systems. A review. Food Biotechnol. 2004, 18, 97–133. [Google Scholar] [CrossRef]
- Millar, B.C.; Xu, J.; Moore, J.E. Risk assessment models, and contamination management: Implications for broad-range ribosomal DNA PCR as a diagnostic tool in medical bacteriology. J. Clin. Microbiol. 2002, 40, 1575–1580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.T.; Jalbani, Y.M.; Zhang, G.L.; Zhao, Z.Y.; Wang, Z.Y.; Zhao, X.Y.; Chen, A.L. Detection of goat meat adulteration by real-time PCR based on a reference primer. Food Chem. 2019, 277, 554–557. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Lu, D.; Zhang, T.; Mao, J.; Shang, S. Application experience of a rapid nucleic acid detection system for COVID-19. Microbes Infect. 2022, 24, 104945. [Google Scholar] [CrossRef]
- Cheng, W.-C.; Horn, T.; Zayats, M.; Rizk, G.; Major, S.; Zhu, H.; Russell, J.; Xu, Z.; Rothman, R.E.; Celedon, A. Ultra-sensitive and rapid detection of nucleic acids and microorganisms in body fluids using single-molecule tethering. Nat. Commun. 2020, 11, 4774. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef] [Green Version]
- Fan, X.; Li, L.; Zhao, Y.; Liu, Y.; Liu, C.; Wang, Q.; Dong, Y.; Wang, S.; Chi, T.; Song, F.; et al. Clinical validation of two recombinase-based isothermal amplification assays (RPA/RAA) for the rapid detection of African swine fever virus. Front. Microbiol. 2020, 11, 1696. [Google Scholar] [CrossRef]
- Hassan, M.; Grist, L.; Poirier, A.; La Ragione, R. JMM profile: Loop-mediated isothermal amplification (LAMP): For the rapid detection of nucleic acid targets in resource-limited settings. J. Med. Microbiol. 2022, 71. [Google Scholar] [CrossRef]
- Ali, Z.; Aman, R.; Mahas, A.; Rao, G.S.; Tehseen, M.; Marsic, T.; Salunke, R.; Subudhi, A.K.; Hala, S.M.; Hamdan, S.M.; et al. iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 2020, 288, 198129. [Google Scholar] [CrossRef]
- Fang, X.; Liu, Y.; Kong, J.; Jiang, X. Loop-mediated isothermal amplification integrated on microfluidic chips for point-of-care quantitative detection of pathogens. Anal. Chem. 2010, 7, 3002–3006. [Google Scholar] [CrossRef]
- Li, R.; Chen, J.; Zhang, X.; Cui, J.; Tao, S.-C.; Yang, L. Mini-disk capillary array coupling with LAMP for visual detection of multiple nucleic acids using genetically modified organism analysis as an example. J. Agric. Food Chem. 2019, 68, 899–906. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Chen, Y.; Yang, Q.; Peng, C.; Wang, X.; Zhang, M.; Qian, S.; Xu, J.; Wu, J. A reversible valve-assisted chip coupling with integrated sample treatment and CRISPR/Cas12a for visual detection of Vibrio parahaemolyticus. Biosens. Bioelectron. 2021, 188, 113352. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yu, H.; Ji, Y.; Wei, W.; Peng, C.; Wang, X.; Xu, X.; Sun, M.; Xu, J. Development and Application of a Visual Duck Meat Detection Strategy for Molecular Diagnosis of Duck-Derived Components. Foods 2022, 11, 1895. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Nolder, D.; Reboud, J.; Oguike, M.C.; van Schalkwyk, D.A.; Sutherland, C.J.; Cooper, J.M. Paper-origami-based multiplexed malaria diagnostics from whole blood. Angew. Chem. 2016, 128, 15476–15479. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Wang, W.; Yang, Z.; Wang, X.; Xu, W.; Huang, K.; Luo, Y.; He, X.; Cheng, N. A portable 3D-printed biosensing device for rapid detection of genetically modified maize MON810. Sens. Actuators Chem. 2021, 349, 130748. [Google Scholar] [CrossRef]
- Wang, X.; Chen, Y.; Chen, X.; Peng, C.; Wang, L.; Xu, X.; Wu, J.; Wei, W.; Xu, J. A highly integrated system with rapid DNA extraction, recombinase polymerase amplification, and lateral flow biosensor for on-site detection of genetically modified crops. Anal. Chim. Acta 2020, 1109, 158–168. [Google Scholar] [CrossRef]
- Rogan, G.; Dudin, Y.; Lee, T.; Magin, K.; Astwood, J.; Bhakta, N.; Leach, J.; Sanders, P.; Fuchs, R. Immunodiagnostic methods for detection of 5-enolpyruvylshikimate-3-phosphate synthase in Roundup Ready® soybeans. Food Control. 1999, 10, 407–414. [Google Scholar] [CrossRef]
- Tang, R.; Li, M.; Yan, X.; Xie, M.; Na Liu, L.; Li, Z.; Xu, F. Comparison of paper-based nucleic acid extraction materials for point-of-care testing applications. Cellulose 2022, 29, 2479–2495. [Google Scholar] [CrossRef]
- Bulcke, M.V.D.; De Schrijver, A.; De Bernardi, D.; Devos, Y.; MbongoMbella, G.; Casi, A.L.; Moens, W.; Sneyers, M. Sneyers, Detection of genetically modified plant products by protein strip testing: An evaluation of real-life samples. Eur. Food Res. Technol. 2007, 225, 49–57. [Google Scholar] [CrossRef] [Green Version]
- Rott, M.E.; Lawrence, T.S.; Wall, A.E.M.; Green, M.J. Detection and quantification of roundup ready soy in foods by conventional and real-time polymerase chain reaction. J. Agric. Food Chem. 2004, 52, 5223–5232. [Google Scholar] [CrossRef]
- Berdal, K.G.; Holst-Jensen, A. Roundup Ready® soybean event-specific real-time quantitative PCR assay and estimation of the practical detection and quantification limits in GMO analyses. Eur. Food Res. Technol. 2001, 213, 432–438. [Google Scholar] [CrossRef]
- Mason, M.G.; Botella, J.R. Rapid (30-second), equipment-free purification of nucleic acids using easy-to-make dipsticks. Nat. Protoc. 2020, 15, 3663–3677. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, Y.; Lin, L.; Wu, T.; Zhao, Z.; Ying, B.; Chang, L. A finger-driven disposable micro-platform based on isothermal amplification for the application of multiplexed and point-of-care diagnosis of tuberculosis. Biosens. Bioelectron. 2022, 195, 113663. [Google Scholar] [CrossRef] [PubMed]

| Analytical Method | Target | a Instrument | b Time for Detection | Sensitivity | Suitability for Controlling the Temperature | Suitability for On-Site-Testing | Ref. |
|---|---|---|---|---|---|---|---|
| ELISA | Protein | Need | >5 h | 0.25% | Unsuitable | Unsuitable | [17] |
| Protein strip test | Protein | No need | 3–5 min | 0.1% | Unsuitable | Suitable | [19] |
| Conventional PCR | DNA | Need | >3 h | 8 copies | Suitable | Unsuitable | [20] |
| RT-PCR | DNA | Need | >2.5 h | 1 copy | Suitable | Unsuitable | [21] |
| DRPA-LFB | DNA | No need | About 30 min | 10 copies | Unsuitable | Suitable | [16] |
| Event-specific RPA | DNA | No need | 25 min | 0.1% | Unsuitable | Suitable | [15] |
| Our method | DNA | No need | ≤25 min | 50 copies | Suitable | Suitable | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, J.-Y.; Wang, X.-F.; Peng, C.; Chen, X.-Y.; Xu, X.-L.; Wei, W.; Yang, L.; Cai, J.; Xu, J.-F. SMART: On-Site Rapid Detection of Nucleic Acid from Plants, Animals, and Microorganisms in under 25 Minutes. Biosensors 2023, 13, 82. https://doi.org/10.3390/bios13010082
Ma J-Y, Wang X-F, Peng C, Chen X-Y, Xu X-L, Wei W, Yang L, Cai J, Xu J-F. SMART: On-Site Rapid Detection of Nucleic Acid from Plants, Animals, and Microorganisms in under 25 Minutes. Biosensors. 2023; 13(1):82. https://doi.org/10.3390/bios13010082
Chicago/Turabian StyleMa, Jun-Yuan, Xiao-Fu Wang, Cheng Peng, Xiao-Yun Chen, Xiao-Li Xu, Wei Wei, Lei Yang, Jian Cai, and Jun-Feng Xu. 2023. "SMART: On-Site Rapid Detection of Nucleic Acid from Plants, Animals, and Microorganisms in under 25 Minutes" Biosensors 13, no. 1: 82. https://doi.org/10.3390/bios13010082
APA StyleMa, J.-Y., Wang, X.-F., Peng, C., Chen, X.-Y., Xu, X.-L., Wei, W., Yang, L., Cai, J., & Xu, J.-F. (2023). SMART: On-Site Rapid Detection of Nucleic Acid from Plants, Animals, and Microorganisms in under 25 Minutes. Biosensors, 13(1), 82. https://doi.org/10.3390/bios13010082







