Surface Functionalization and Escherichia coli Detection Using Surface-Enhanced Raman Spectroscopy Driven by Functional Organic Polymer/Gold Nanofilm-Based Microfluidic Chip
Abstract
:1. Introduction
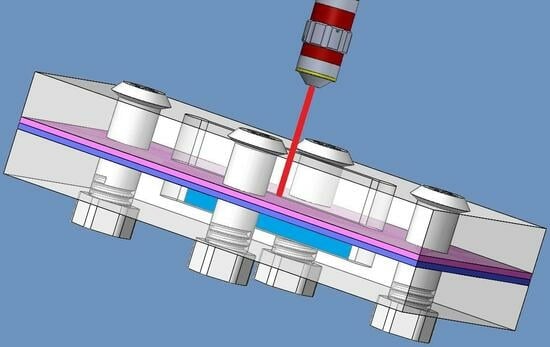
2. Design and Simulation in COMSOL
2.1. Design Considerations
2.2. Simulation Setup
3. Materials and Methods
3.1. FOP Synthesis
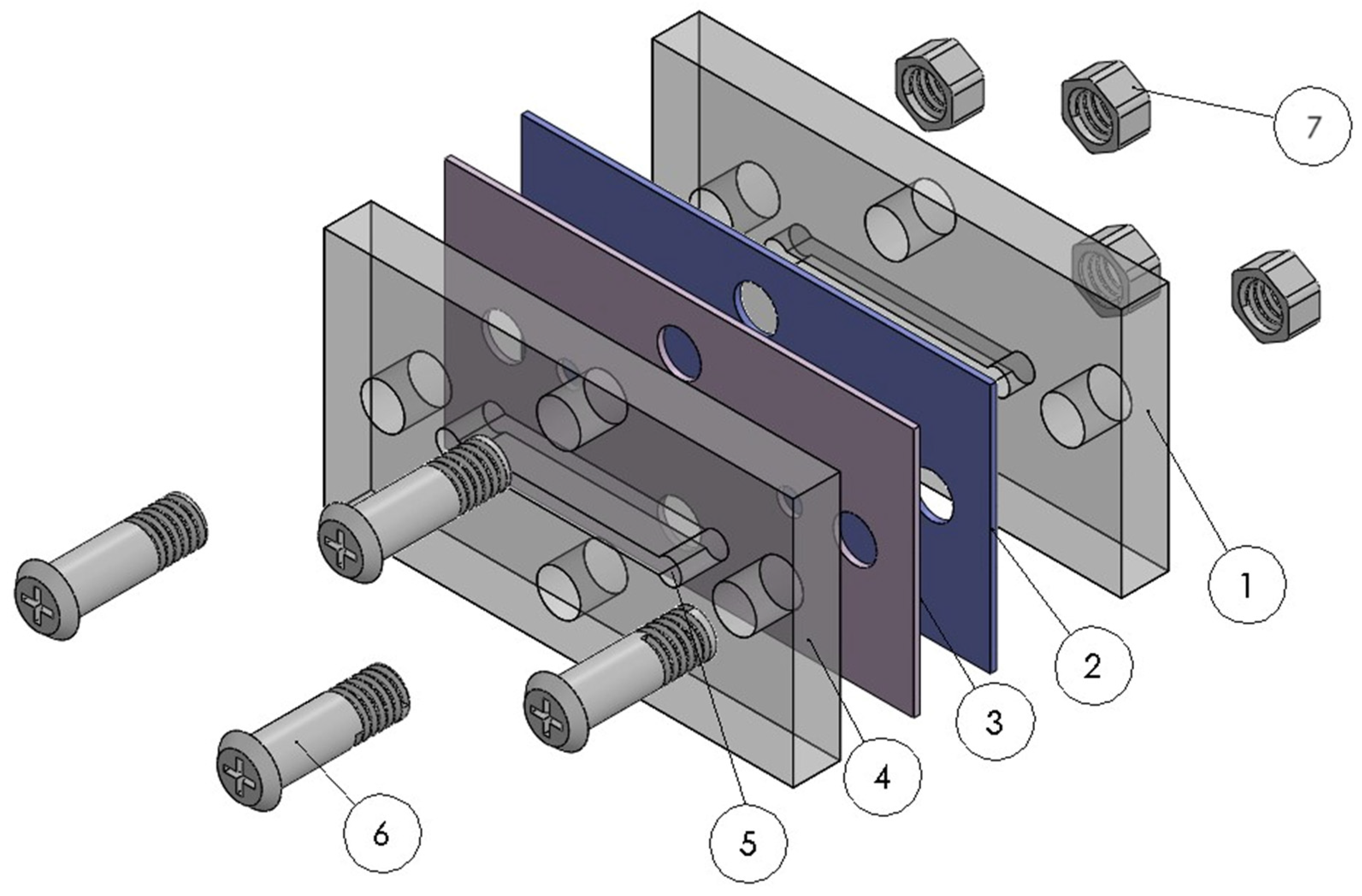
3.2. Prototype Manufacturing
3.3. Cysteamine Functionalization
3.4. Bacterium Detection Using As-Fabricated Prototype
3.5. Measurements
4. Results
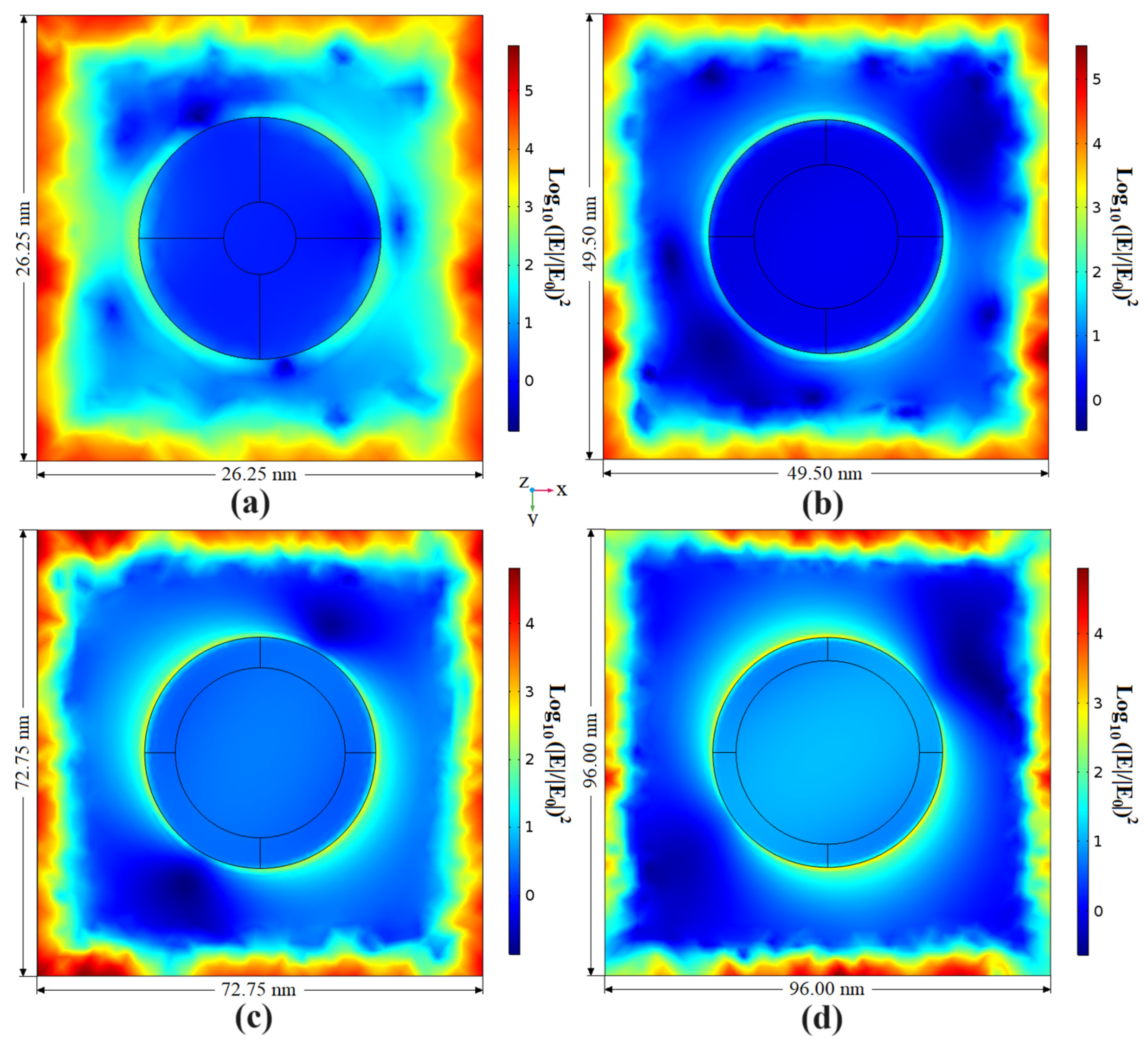
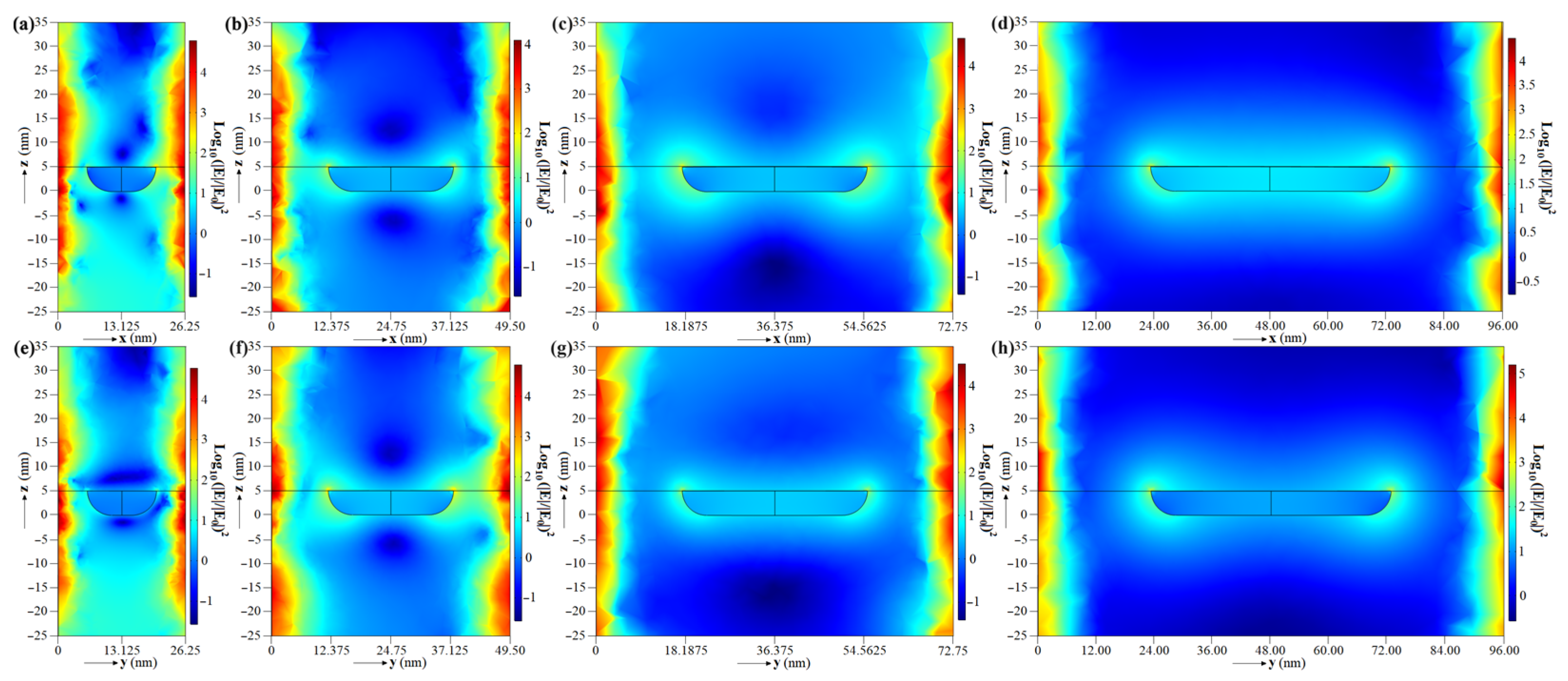
4.1. Simulation Results
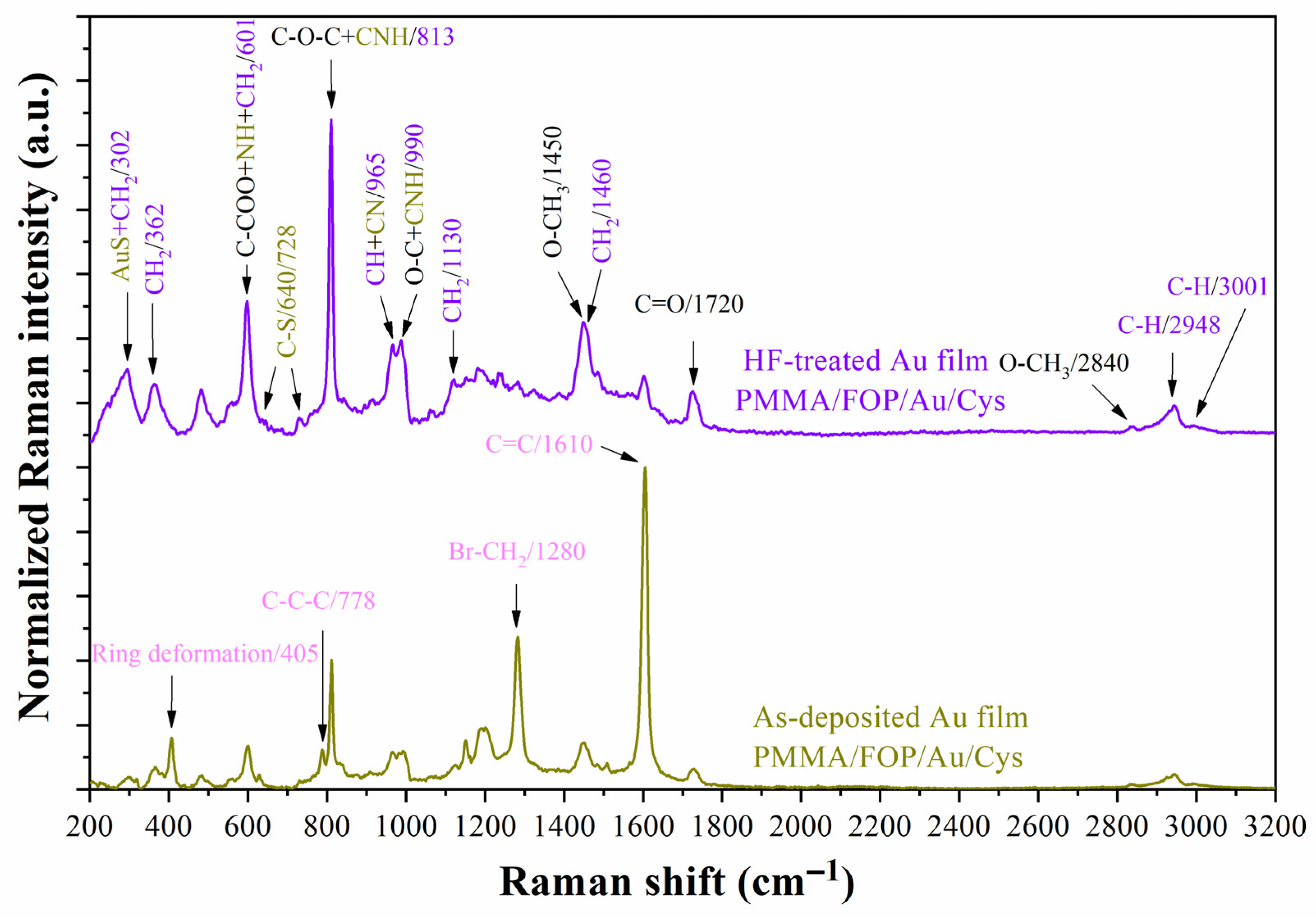
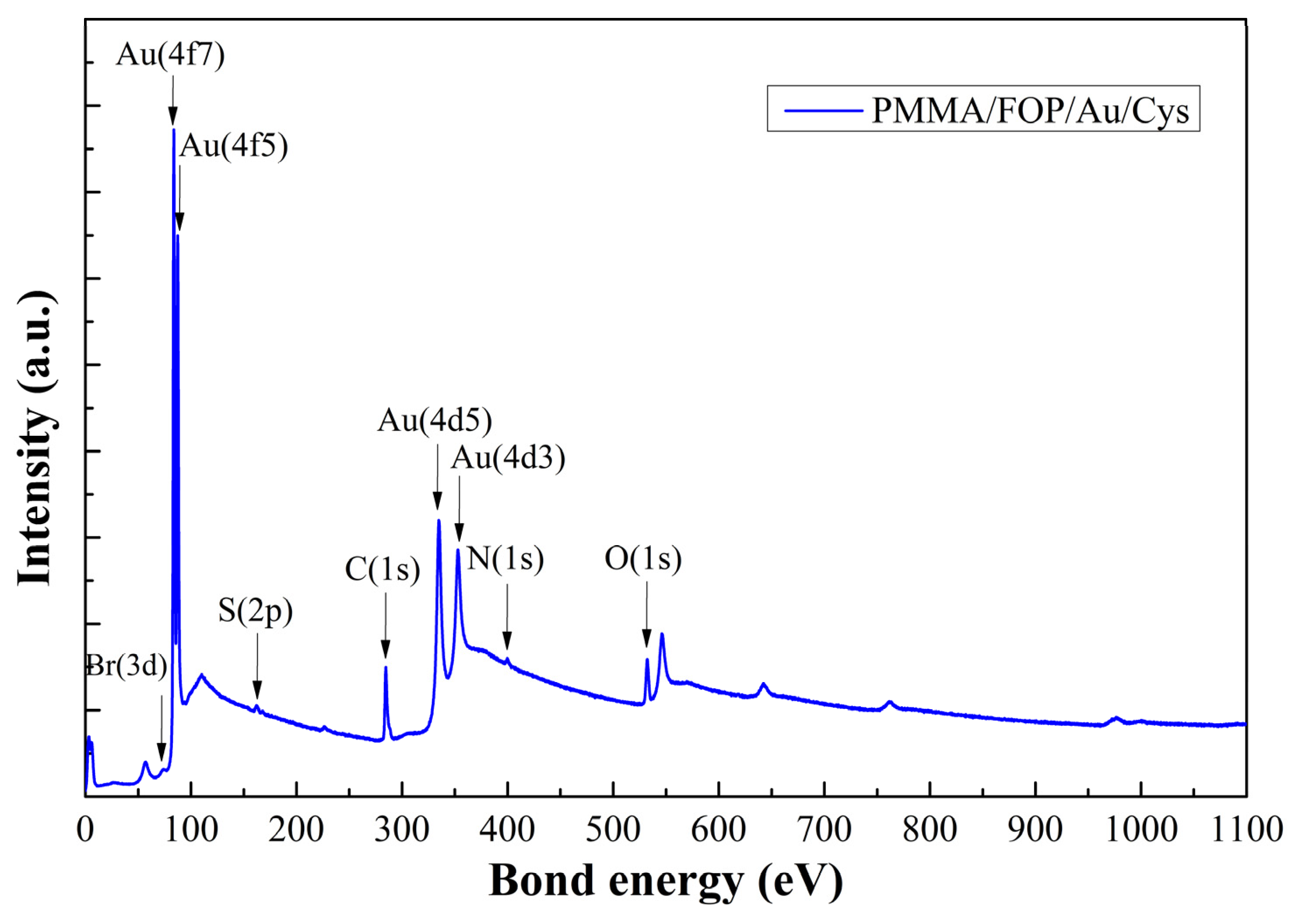
4.2. Manufacturing Step Tracking
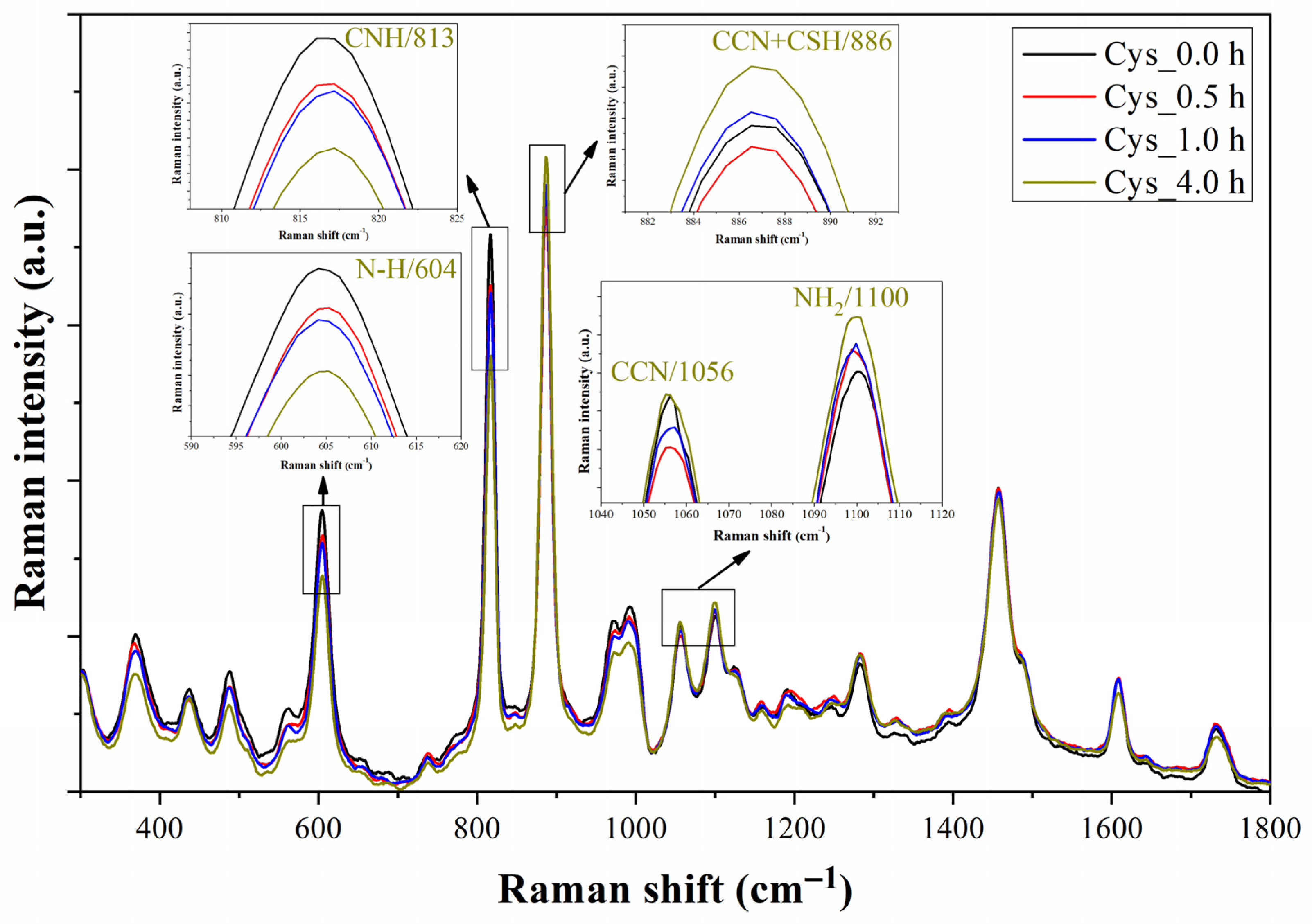
4.3. Monitoring of the Functionalization Process
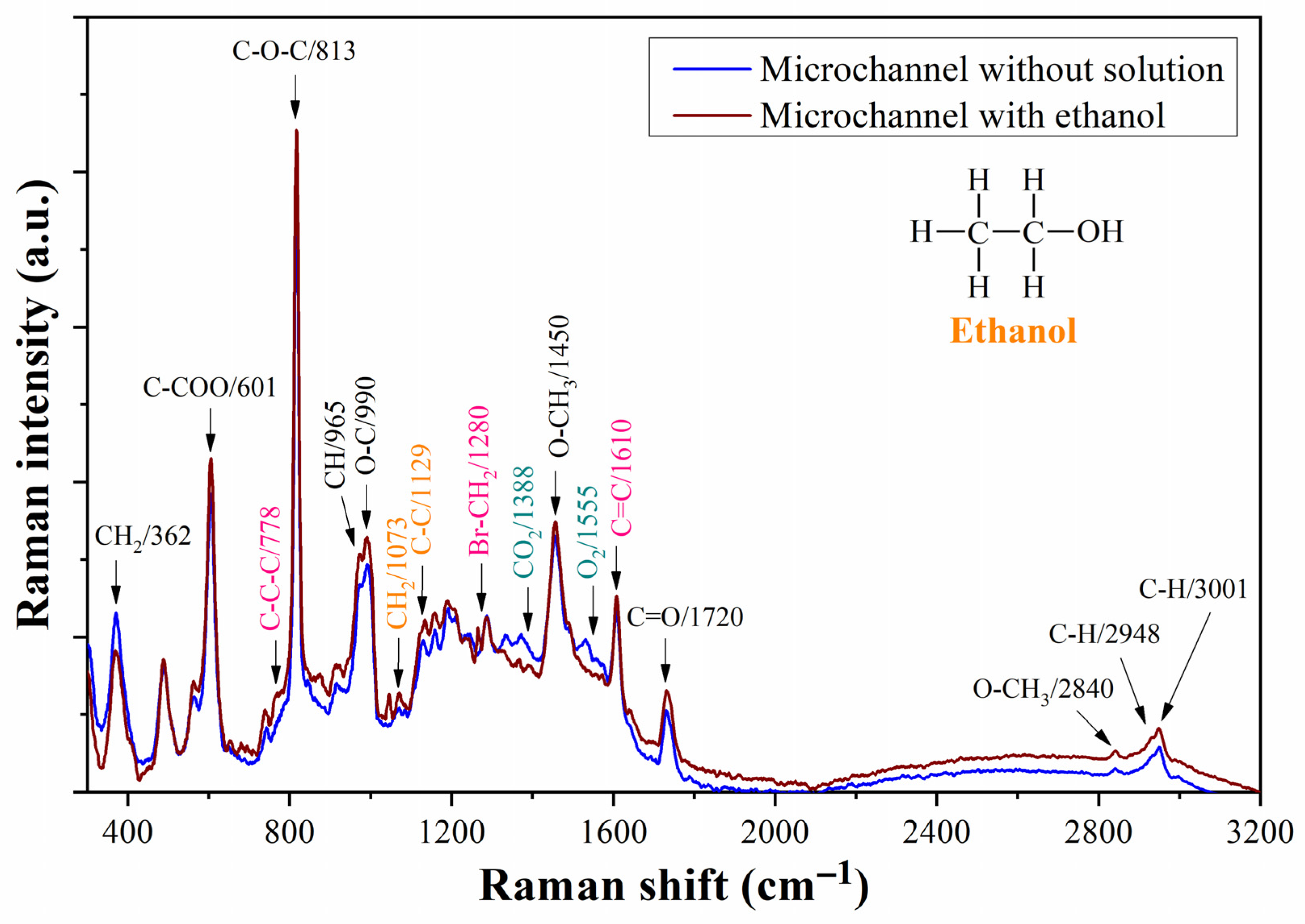
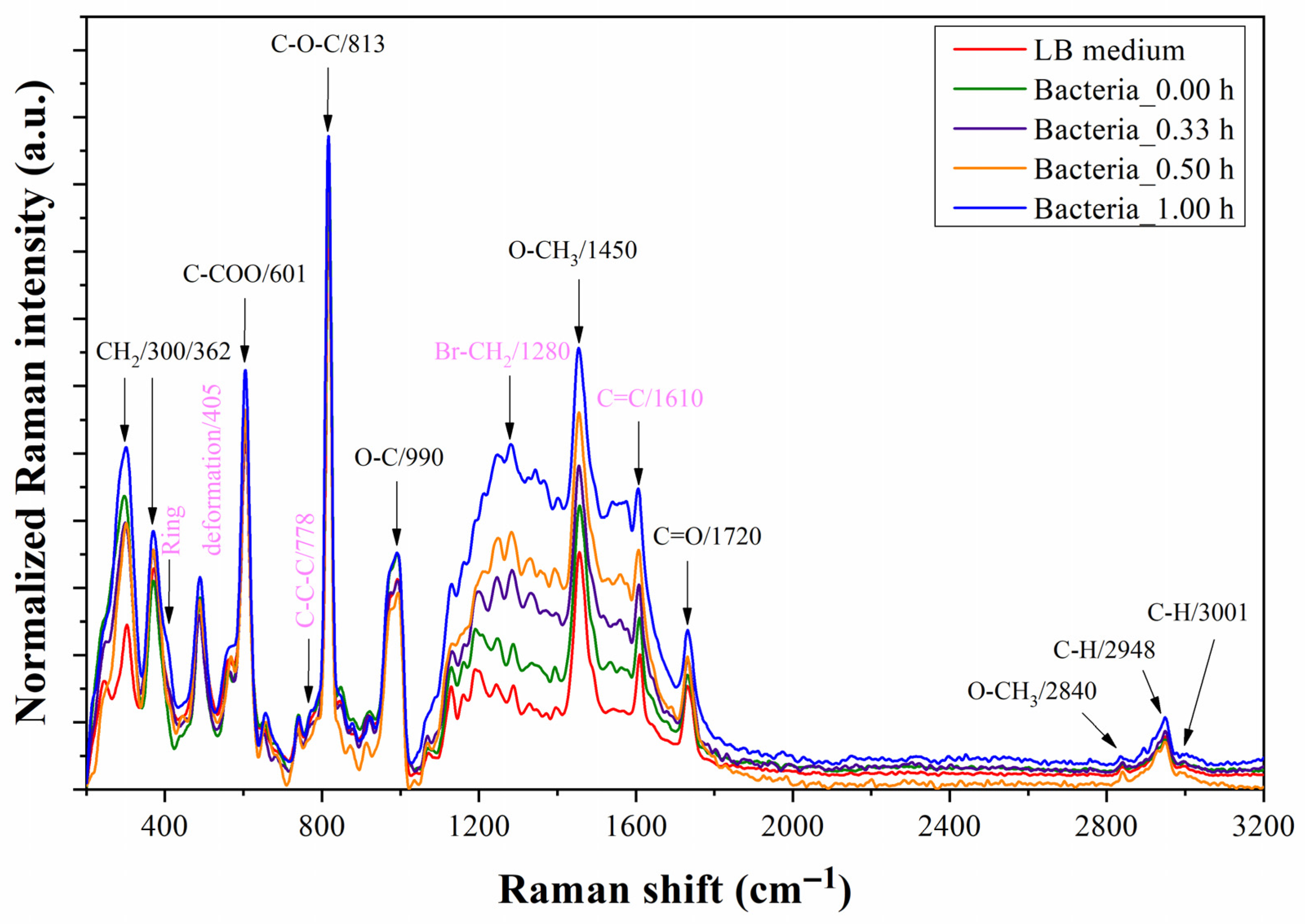
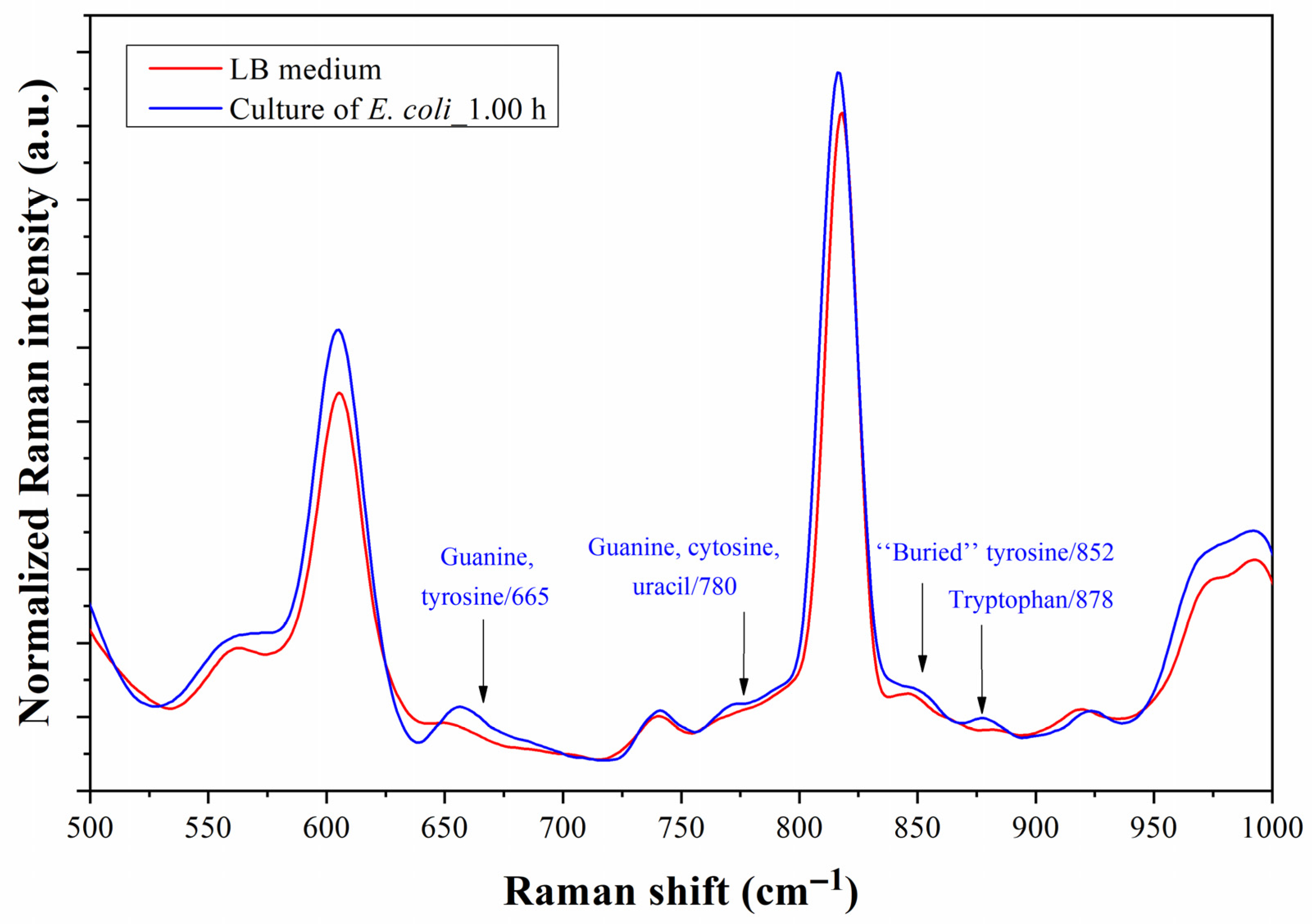
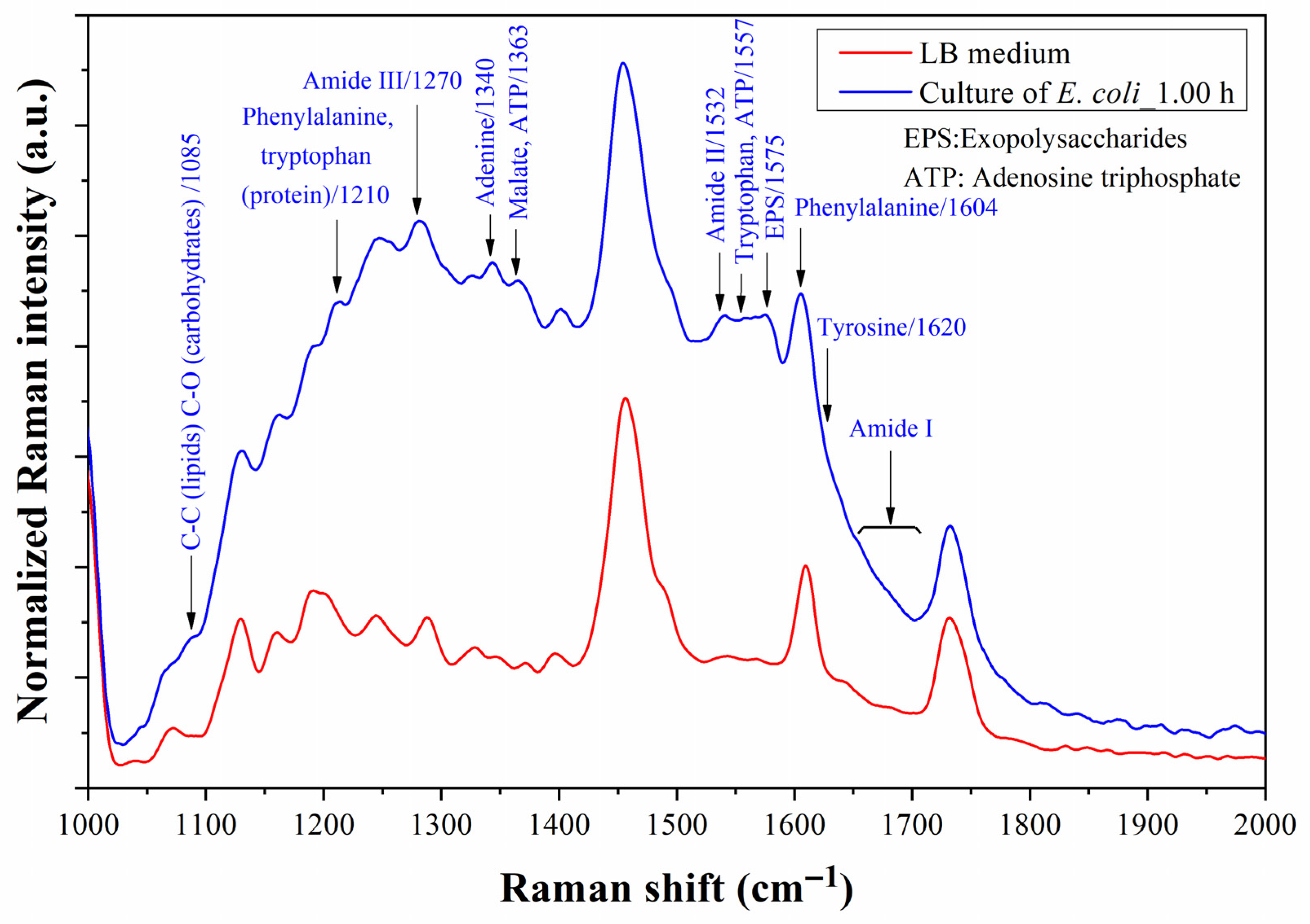
4.4. Bacterium Detection with As-Fabricated Prototype
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Tack, D.M.; Kisselburgh, H.M.; Richardson, L.C.; Geissler, A.; Griffin, P.M.; Payne, D.C.; Gleason, B.L. Shiga Toxin-Producing Escherichia coli Outbreaks in the United States, 2010–2017. Microorganisms 2021, 9, 1529. [Google Scholar] [CrossRef]
- Hannah, S.; Dobrea, A.; Lasserre, P.; Blair, E.O.; Alcorn, D.; Hoskisson, P.A.; Corrigan, D.K. Development of a rapid, antimicrobial susceptibility test for E. coli based on low-cost, screen-printed electrodes. Biosensors 2020, 10, 153. [Google Scholar] [CrossRef]
- World Health Organization. World Health Statistics 2022: Monitoring Health for the SDGs, Sustainable Development Goals; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- World Health Organization. Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report 2022; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- Nurliyana, M.R.; Sahdan, M.; Wibowo, K.; Muslihati, A.; Saim, H.; Ahmad, S.; Sari, Y.; Mansor, Z. The Detection Method of Escherichia coli in Water Resources: A Review. J. Phys. Conf. Ser. 2018, 995, 012065. [Google Scholar] [CrossRef]
- Srivastava, S.K.; Hamo, H.B.; Kushmaro, A.; Marks, R.S.; Grüner, C.; Rauschenbach, B.; Abdulhalim, I. Highly sensitive and specific detection of E. coli by a SERS nanobiosensor chip utilizing metallic nanosculptured thin films. Analyst 2015, 140, 3201–3209. [Google Scholar] [CrossRef]
- Wang, C.; Madiyar, F.; Yu, C.; Li, J. Detection of extremely low concentration waterborne pathogen using a multiplexing self-referencing SERS microfluidic biosensor. J. Biol. Eng. 2017, 11, 9. [Google Scholar] [CrossRef]
- Whang, K.; Lee, J.-H.; Shin, Y.; Lee, W.; Kim, Y.W.; Kim, D.; Lee, L.P.; Kang, T. Plasmonic bacteria on a nanoporous mirror via hydrodynamic trapping for rapid identification of waterborne pathogens. Light. Sci. Appl. 2018, 7, 68. [Google Scholar] [CrossRef]
- Witkowska, E.; Korsak, D.; Kowalska, A.; Księżopolska-Gocalska, M.; Niedziółka-Jönsson, J.; Roźniecka, E.; Michałowicz, W.; Albrycht, P.; Podrażka, M.; Hołyst, R.; et al. Surface-enhanced Raman spectroscopy introduced into the International Standard Organization (ISO) regulations as an alternative method for detection and identification of pathogens in the food industry. Anal. Bioanal. Chem. 2017, 409, 1555–1567. [Google Scholar] [CrossRef]
- Ionescu, R. Biosensor platforms for rapid detection of E. coli bacteria. In Recent Advances on Physiology, Pathogenesis and Biotechnological Applications; InTechOpen: London, UK, 2017; pp. 275–289. [Google Scholar]
- Cao, Z.; Li, C.; Yang, X.; Wang, S.; Zhang, X.; Zhao, C.; Xue, B.; Gao, C.; Zhou, H.; Yang, Y.; et al. Rapid quantitative detection of live Escherichia coli based on chronoamperometry. Biosensors 2022, 12, 845. [Google Scholar] [CrossRef]
- Zadeh, S.H.; Kashanian, S.; Nazari, M. A label-free carbohydrate-based electrochemical sensor to detect Escherichia coli pathogenic bacteria using d-mannose on a glassy carbon electrode. Biosensors 2023, 13, 619. [Google Scholar] [CrossRef]
- Gunasekaran, D.; Gerchman, Y.; Vernick, S. Electrochemical detection of waterborne bacteria using bi-functional magnetic nanoparticle conjugates. Biosensors 2022, 12, 36. [Google Scholar] [CrossRef]
- Mosier-Boss, P.A. Review on SERS of Bacteria. Biosensors 2017, 7, 51. [Google Scholar] [CrossRef]
- Su, L.; Zhang, P.; Zheng, D.-W.; Wang, Y.-J.; Zhong, R.-G. Rapid detection of Escherichia coli and Salmonella typhimurium by surface-enhanced Raman scattering. Optoelectron. Lett. 2015, 11, 157–160. [Google Scholar] [CrossRef]
- Wei, C.; Li, M.; Zhao, X. Surface-enhanced Raman scattering (SERS) with silver nano substrates synthesized by microwave for rapid detection of foodborne pathogens. Front. Microbiol. 2018, 9, 2857. [Google Scholar] [CrossRef]
- Yang, D.; Zhou, H.; Haisch, C.; Niessner, R.; Ying, Y. Reproducible E. coli detection based on label-free SERS and mapping. Talanta 2016, 146, 457–463. [Google Scholar] [CrossRef]
- Chisanga, M.; Muhamadali, H.; Kimber, R.; Goodacre, R. Quantitative detection of isotopically enriched E. coli cells by SERS. Faraday Discuss. 2017, 205, 331–343. [Google Scholar] [CrossRef]
- Boca, S.; Leordean, C.; Astilean, S.; Farcau, C. Chemiresistive/SERS dual sensor based on densely packed gold nanoparticles. Nanotechnol 2015, 6, 2498–2503. [Google Scholar] [CrossRef]
- Gao, W.; Li, B.; Yao, R.; Li, Z.; Wang, X.; Dong, X.; Qu, H.; Li, Q.; Li, N.; Chi, H.; et al. Intuitive label-free SERS detection of bacteria using aptamer-based in situ silver nanoparticles synthesis. Anal. Chem. 2017, 89, 9836–9842. [Google Scholar] [CrossRef]
- Comsol, A.B. Comsol Multiphysics-Wave Optics Module, User’s Guide; COMSOL: Stockholm, Sweden, 2018. [Google Scholar]
- Hernández-Cruz, O.; Avila-Gutierrez, L.; Zolotukhin, M.G.; Gonzalez, G.; Monroy, B.M.; Montiel, R.; Vera-Graziano, R.; Romero-Ibarra, J.E.; Novelo-Peralta, O.; Rojas, F.A.M. Spontaneous, Solvent-Free, Polymer-Templated, Solid–Solid Transformation of Thin Metal Films into Nanoparticles. Nano Lett. 2016, 16, 5420–5425. [Google Scholar] [CrossRef]
- Ding, S.-Y.; You, E.-M.; Tian, Z.-Q.; Moskovits, M. Electromagnetic theories of surface-enhanced Raman spectroscopy. Chem. Soc. Rev. 2017, 46, 4042–4076. [Google Scholar] [CrossRef]
- Klenke, C.M.; Brawley, Z.T.; Bauman, S.J.; Darweesh, A.A.; Debu, D.T.; Herzog, J.B. Dataset for SERS plasmonic array: Width, spacing, and thin film oxide thickness optimization. Data 2018, 3, 37. [Google Scholar] [CrossRef]
- Zhang, X.; Qiu, J.; Li, X.; Zhao, J.; Liu, L.H. Complex refractive indices measurements of polymers in visible and near-infrared bands. Appl. Opt. 2020, 59, 2337–2344. [Google Scholar] [CrossRef]
- Vogel, A.I.; Cresswell, W.T.; Jeffery, G.J.; Leicester, J. Bond refractions and bond parachors. Chem. Ind. 1950, 18, 358. [Google Scholar]
- Vogel, A.I.; Cresswell, W.T.; Jeffery, G.H.; Leicester, J. Calculation of the refractive indices of liquid organic compounds: Bond molecular refraction coefficients. Chem. Ind. 1951, 19, 376. [Google Scholar]
- Kedenburg, S.; Vieweg, M.; Gissibl, T.; Giessen, H. Linear refractive index and absorption measurements of nonlinear optical liquids in the visible and near-infrared spectral region. Opt. Mater. Express 2012, 2, 1588–1611. [Google Scholar] [CrossRef]
- Milazzo, R.G.; Mio, A.M.; D’arrigo, G.; Smecca, E.; Alberti, A.; Fisichella, G.; Giannazzo, F.; Spinella, C.; Rimini, E. Influence of hydrofluoric acid treatment on electroless deposition of Au clusters. Beilstein J. Nanotechnol. 2017, 8, 183–189. [Google Scholar] [CrossRef]
- Zangmeister, R.A.; Tarlov, M.J. UV graft polymerization of polyacrylamide hydrogel plugs in microfluidic channels. Langmuir 2003, 19, 6901–6904. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, Z.; Liu, H.; Perea-López, N.; Ranasinghe, J.C.; Bepete, G.; Minns, A.M.; Rossi, R.M.; Lindner, S.E.; Huang, S.X.; et al. Understanding the excitation wavelength dependence and thermal stability of the SARS-CoV-2 receptor-binding domain using surface-enhanced raman scattering and machine learning. ACS Photonics 2022, 9, 2963–2972. [Google Scholar] [CrossRef]
- Larkin, P. Infrared and Raman Spectroscopy: Principles and Spectral Interpretation; Elsevier Science and Technology Books: Stamford, Fairfield, 2017. [Google Scholar]
- Thomas, K.J.; Sheeba, M.; Nampoori, V.P.N.; Vallabhan, C.P.G.; Radhakrishnan, P. Raman spectra of polymethyl methacrylate optical fibres excited by a 532 nm diode pumped solid state laser. J. Opt. A Pure Appl. Opt. 2008, 10, 055303. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, R.-S.; Chen, X.-J. Vibrational Properties of p-Terphenyl. J. Phys. Chem. A 2018, 122, 6903–6908. [Google Scholar] [CrossRef]
- Varnholt, B.; Oulevey, P.; Luber, S.; Kumara, C.; Dass, A.; Bürgi, T. Structural information on the Au–S interface of thiolate-protected gold clusters: A Raman spectroscopy study. J. Phys. Chem. C 2014, 118, 9604–9611. [Google Scholar] [CrossRef]
- Dikmen, G. Investigation of Tautomeric Structures of 6-Aza-2-Thiouracil-5-Carboxylic Acid Using Vibrational Spectroscopy Along with Dft Theoretical Method. Eskişehir Tech. Univ. J. Sci. Technol. A—Appl. Sci. Eng. 2021, 22, 23–35. [Google Scholar] [CrossRef]
- Bloxham, S.; Eicher-Lorka, O.; Jakubėnas, R.; Niaura, G. Adsorption of Cysteamine at Copper Electrodes as Studied by Surface-Enhanced Raman Spectroscopy. Spectrosc. Lett. 2003, 36, 211–226. [Google Scholar] [CrossRef]
- Riauba, L.; Niaura, G.; Eicher-Lorka, O.; Butkus, E. A Study of cysteamine ionization in solution by Raman spectroscopy and theoretical modeling. J. Phys. Chem. A 2006, 110, 13394–13404. [Google Scholar] [CrossRef]
- Schwartzkopf, M.; Buffet, A.; Körstgens, V.; Metwalli, E.; Schlage, K.; Benecke, G.; Perlich, J.; Rawolle, M.; Rothkirch, A.; Heidmann, B.; et al. From atoms to layers: In situ gold cluster growth kinetics during sputter deposition. Nanoscale 2013, 5, 5053–5062. [Google Scholar] [CrossRef]
- Darmandeh, H.; Löffler, J.; Tzouras, N.V.; Dereli, B.; Scherpf, T.; Feichtner, K.; Broeck, S.V.; Van Hecke, K.; Saab, M.; Cazin, C.S.J.; et al. Au⋅⋅⋅H−C Hydrogen Bonds as Design Principle in Gold(I) Catalysis. Angew. Chem. 2021, 133, 21182–21192. [Google Scholar] [CrossRef]
- Rance, G.A.; Khlobystov, A.N. Interactions of carbon nanotubes and gold nanoparticles: The effects of solvent dielectric constant and temperature on controlled assembly of superstructures. Dalton Trans. 2014, 43, 7400–7406. [Google Scholar] [CrossRef]
- Casaletto, M.P.; Longo, A.; Martorana, A.; Prestianni, A.; Venezia, A.M. XPS study of supported gold catalysts: The role of Au0 and Au+δ species as active sites. Surf. Interface Anal. 2006, 38, 215–218. [Google Scholar] [CrossRef]
- Rignanese, G.-M.; Pasquarello, A.; Charlier, J.-C.; Gonze, X.; Car, R. Nitrogen Incorporation at Si(001)−SiO2 Interfaces: Relation between N1sCore-Level Shifts and Microscopic Structure. Phys. Rev. Lett. 1997, 79, 5174. [Google Scholar] [CrossRef]
- Chastain, J.; King, R.C., Jr. Handbook of X-ray Photoelectron Spectroscopy; Perkin-Elmer Corporation: Eden Prairie, MN, USA, 1992; Volume 40. [Google Scholar]
- Matrosov, I.I.; Petrov, D.V.; Tikhomirov, A.A. Improvement of the sensitivity of a stationary Raman gas analyzer. In Proceedings of the 20th International Symposium on Atmospheric and Ocean Optics: Atmospheric Physics, Novosibirsk, Russian, 25 November 2014; pp. 542–546. [Google Scholar]
- Paret, M.L.; Sharma, S.K.; Green, L.M.; Alvarez, A.M. Biochemical Characterization of Gram-Positive and Gram-Negative Plant-Associated Bacteria with Micro-Raman Spectroscopy. Appl. Spectrosc. 2010, 64, 433–441. [Google Scholar] [CrossRef]
- Berger, A.J.; Zhu, Q. Identification of oral bacteria by raman microspectroscopy. J. Mod. Opt. 2003, 50, 2375–2380. [Google Scholar] [CrossRef]
- Félix-Rivera, H.; Hernández-Rivera, S.P. Raman spectroscopy techniques for the detection of biological samples in suspensions and as aerosol particles: A review. Sens. Imaging 2012, 13, 1–25. [Google Scholar] [CrossRef]
- Nguyen, E.P.; Chrimes, A.F.; Brkljača, R.; Ou, J.Z.; Berean, K.; Zhuiykov, S.; Kalantarzadeh, K. Assessment of a Raman micro-spectroscopy/microfluidics unit using a model E. coli/glucose bio-system. In Proceedings of the 7th IEEE International Conference on Nano/Molecular Medicine and Engineering, Phuket, Thailand, 10–13 November 2013; pp. 157–162. [Google Scholar]
- Azeem, M.; Qaisar, R.; Karim, A.; Ranade, A.; Elmoselhi, A. Density Functional Model of Raman Spectra for the Seminiferous Tubules of Hind Limb Unloaded Mouse; Social Science Research Network: London, UK, 2021. [Google Scholar] [CrossRef]
- Yan, B.-X.; Zhu, Y.-Y.; Wei, Y.; Pei, H. Study on surface enhanced Raman scattering of Au and Au@Al2O3 spherical dimers based on 3D finite element method. Sci. Rep. 2021, 11, 8391. [Google Scholar] [CrossRef]
- Chien, F.-C.; Huang, W.Y.; Shiu, J.-Y.; Kuo, C.W.; Chen, P. Revealing the spatial distribution of the site enhancement for the surface enhanced Raman scattering on the regular nanoparticle arrays. Opt. Express 2009, 17, 13974–13981. [Google Scholar] [CrossRef]
- Zhu, Z.; Li, Q.; Bai, B.; Fan, S. Reusable three-dimensional nanostructured substrates for surface-enhanced Raman scattering. Nanoscale Res. Lett. 2014, 9, 25. [Google Scholar] [CrossRef]
- Wang, H.; Wang, Y.; Wang, Y.; Xu, W.; Xu, S. Modulation of hot regions in waveguide-based evanescent-field-coupled localized surface plasmons for plasmon-enhanced spectroscopy. Photonics Res. 2017, 5, 527–535. [Google Scholar] [CrossRef]
- Solís, D.M.; Taboada, J.M.; Obelleiro, F.; Liz-Marzán, L.M.; de Abajo, F.J.G. Optimization of Nanoparticle-Based SERS Substrates through Large-Scale Realistic Simulations. ACS Photonics 2017, 4, 329–337. [Google Scholar] [CrossRef]
- Knorr, I.; Christou, K.; Meinertz, J.; Selle, A.; Ihlemann, J.; Marowsky, G. Prediction and Optimization of Surface Enhanced Raman Scattering Geometries using COMSOL Multiphysics. In Proceedings of the COMSOL Conference, Hannover, Germany, 4–6 January 2008; pp. 1–7. [Google Scholar]
- Chirumamilla, A.; Moise, I.-M.; Cai, Z.; Ding, F.; Jensen, K.B.; Wang, D.; Kristensen, P.K.; Jensen, L.R.; Fojan, P.; Popok, V.; et al. Lithography-free fabrication of scalable 3D nanopillars as ultrasensitive SERS substrates. Appl. Mater. Today 2023, 31, 101763. [Google Scholar] [CrossRef]
- Malyshev, D.; Öberg, R.; Dahlberg, T.; Wiklund, K.; Landström, L.; Andersson, P.O.; Andersson, M. Laser induced degradation of bacterial spores during micro-Raman spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 265, 120381. [Google Scholar] [CrossRef]
- Pavel, I.; McCarney, E.; Elkhaled, A.; Morrill, A.; Plaxco, K.; Moskovits, M. Label-free SERS detection of small proteins modified to act as bifunctional linkers. J. Phys. Chem. C 2008, 112, 4880–4883. [Google Scholar] [CrossRef]





| Material | Refractive Index | Extinction Coefficient |
|---|---|---|
| Air | 1 | 0 |
| PMMA a | 1.4860 | 1.2650 × 10−7 |
| FOP b | 1.4522 | 0 |
| Au c | 0.4708 | 4.9876 |
| H2O d | 1.3284 | 1.5430 × 10−7 |
| Day | EF in xy Plane z = 5 nm | EF in xz Plane x = [−0.1, 0.1] nm z = [–25, 35] nm | EF in yz Plane y = [−0.1, 0.1] nm z = [−25, 35] nm | |||
|---|---|---|---|---|---|---|
| Average | Maximum | Average | Maximum | Average | Maximum | |
| 6 | 5.6195 × 109 | 7.7637 × 1011 | 4.7876 × 107 | 2.3894 × 109 | 5.3242 × 107 | 2.2011 × 109 |
| 12 | 5.5212 × 108 | 1.0411 × 1011 | 6.5086 × 106 | 1.2200 × 108 | 8.1982 × 106 | 5.8675 × 108 |
| 18 | 8.4126 × 107 | 3.5902 × 1010 | 6.9237 × 105 | 4.1541 × 107 | 5.7364 × 106 | 6.0196 × 108 |
| 24 | 3.9312 × 107 | 1.3120 × 1010 | 2.0354 × 104 | 5.9255 × 105 | 7.8220 × 106 | 2.3317 × 108 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cortes-Cano, H.; Olvera, L.I.; Méndez-Aguilar, E.M.; España-Sánchez, B.L.; Arriaga, L.G.; Oza, G.; Herrera-Celis, J. Surface Functionalization and Escherichia coli Detection Using Surface-Enhanced Raman Spectroscopy Driven by Functional Organic Polymer/Gold Nanofilm-Based Microfluidic Chip. Biosensors 2023, 13, 994. https://doi.org/10.3390/bios13120994
Cortes-Cano H, Olvera LI, Méndez-Aguilar EM, España-Sánchez BL, Arriaga LG, Oza G, Herrera-Celis J. Surface Functionalization and Escherichia coli Detection Using Surface-Enhanced Raman Spectroscopy Driven by Functional Organic Polymer/Gold Nanofilm-Based Microfluidic Chip. Biosensors. 2023; 13(12):994. https://doi.org/10.3390/bios13120994
Chicago/Turabian StyleCortes-Cano, Hugo, Lilian Iraís Olvera, Emilia M. Méndez-Aguilar, Beatriz Liliana España-Sánchez, Luis Gerardo Arriaga, Goldie Oza, and José Herrera-Celis. 2023. "Surface Functionalization and Escherichia coli Detection Using Surface-Enhanced Raman Spectroscopy Driven by Functional Organic Polymer/Gold Nanofilm-Based Microfluidic Chip" Biosensors 13, no. 12: 994. https://doi.org/10.3390/bios13120994
APA StyleCortes-Cano, H., Olvera, L. I., Méndez-Aguilar, E. M., España-Sánchez, B. L., Arriaga, L. G., Oza, G., & Herrera-Celis, J. (2023). Surface Functionalization and Escherichia coli Detection Using Surface-Enhanced Raman Spectroscopy Driven by Functional Organic Polymer/Gold Nanofilm-Based Microfluidic Chip. Biosensors, 13(12), 994. https://doi.org/10.3390/bios13120994