Directed Evolution of Protein-Based Sensors for Anaerobic Biological Activation of Methane
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Strains and Plasmids
2.3. Library Construction
2.4. Fluorescence-Activated Cell Sorting (FACS)
2.5. Rescreening and Characterization of Isolated Variants
2.6. Structural Modeling and Molecular Docking
3. Results and Discussion
3.1. Directed Evolution of MS-Responsive ItcR Variants
3.2. Characterization of MS Biosensors
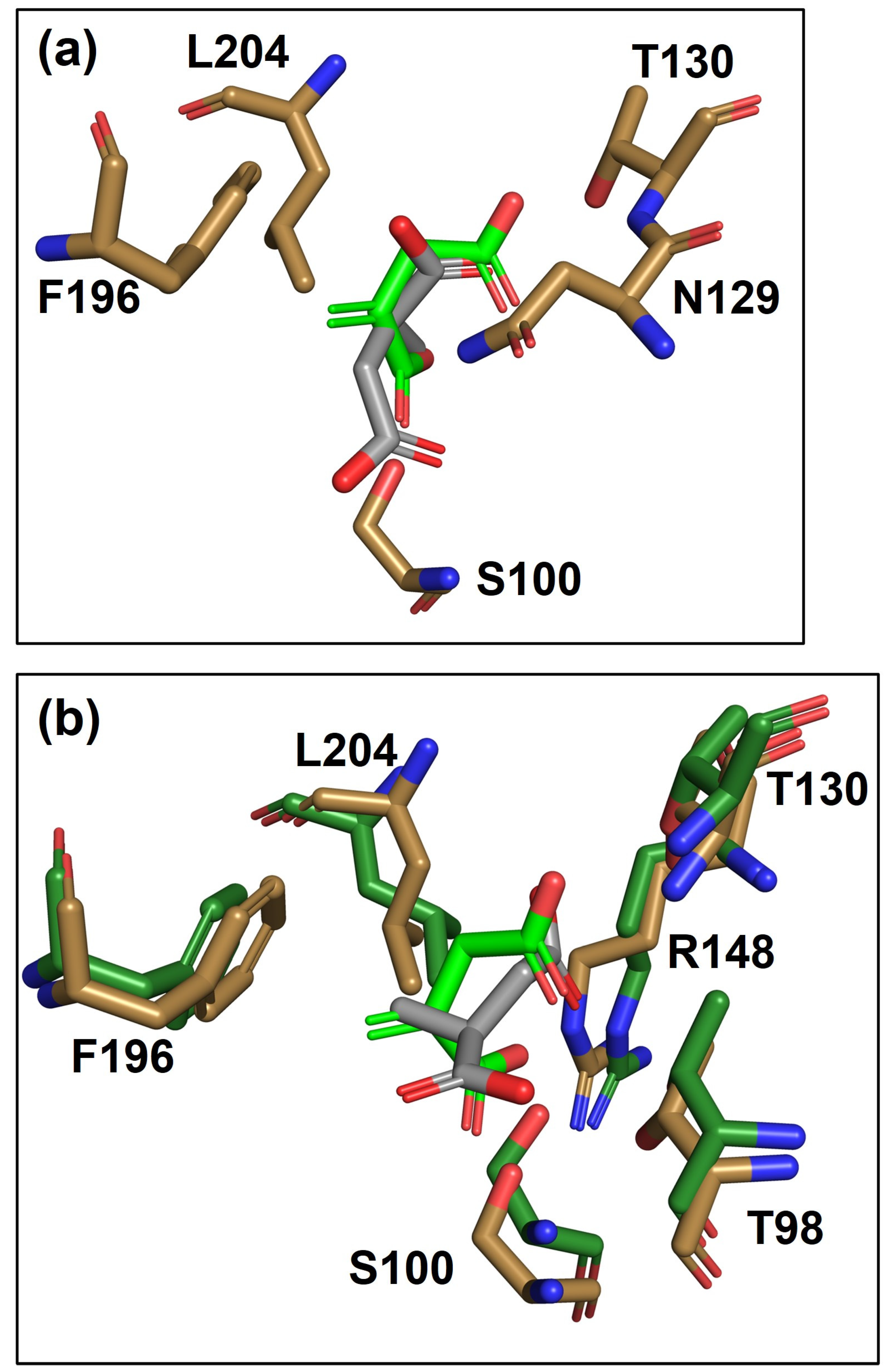
3.3. Binding-Site Modeling and Analysis: Variant 7 vs. WT-ItcR
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Haynes, C.A.; Gonzalez, R. Rethinking biological activation of methane and conversion to liquid fuels. Nat. Chem. Biol. 2014, 10, 331–339. [Google Scholar] [CrossRef]
- Hwang, I.Y.; Nguyen, A.D.; Nguyen, T.T.; Nguyen, L.T.; Lee, O.K.; Lee, E.Y. Biological conversion of methane to chemicals and fuels: Technical challenges and issues. Appl. Microbiol. Biotechnol. 2018, 102, 3071–3080. [Google Scholar] [CrossRef]
- Musat, F. The anaerobic degradation of gaseous, nonmethane alkanes—From in situ processes to microorganisms. Comput. Struct. Biotechnol. J. 2015, 13, 222–228. [Google Scholar] [CrossRef]
- Wang, Y.; Nguyen, N.; Lee, S.H.; Wang, Q.; May, J.A.; Gonzalez, R.; Cirino, P.C. Engineering Escherichia coli for anaerobic alkane activation: Biosynthesis of (1-methylalkyl)succinates. Biotechnol. Bioeng. 2022, 119, 315–320. [Google Scholar] [CrossRef]
- Wu, M.; Li, J.; Lai, C.-Y.; Leu, A.O.; Sun, S.; Gu, R.; Erler, D.V.; Liu, L.; Li, L.; Tyson, G.W.; et al. Nitrate-driven anaerobic oxidation of ethane and butane by bacteria. ISME J. 2024, 18, wrad011. [Google Scholar] [CrossRef]
- Jarling, R.; Kühner, S.; Basílio Janke, E.; Gruner, A.; Drozdowska, M.; Golding, B.T.; Rabus, R.; Wilkes, H. Versatile transformations of hydrocarbons in anaerobic bacteria: Substrate ranges and regio- and stereo-chemistry of activation reactions†. Front. Microbiol. 2015, 6, 00880. [Google Scholar] [CrossRef]
- Küppers, J.; Mitschke, N.; Heyen, S.; Rabus, R.; Wilkes, H.; Christoffers, J. Metabolites of the Anaerobic Degradation of n-Hexane by Denitrifying Betaproteobacterium Strain HxN1. ChemBioChem 2020, 21, 373–380. [Google Scholar] [CrossRef]
- Singh, R.; Guzman, M.S.; Bose, A. Anaerobic Oxidation of Ethane, Propane, and Butane by Marine Microbes: A Mini Review. Front. Microbiol. 2017, 8, 02056. [Google Scholar] [CrossRef]
- Kniemeyer, O.; Musat, F.; Sievert, S.M.; Knittel, K.; Wilkes, H.; Blumenberg, M.; Michaelis, W.; Classen, A.; Bolm, C.; Joye, S.B.; et al. Anaerobic oxidation of short-chain hydrocarbons by marine sulphate-reducing bacteria. Nature 2007, 449, 898–901. [Google Scholar] [CrossRef]
- Qian, S.; Cirino, P.C. Using metabolite-responsive gene regulators to improve microbial biosynthesis. Curr. Opin. Chem. Eng. 2016, 14, 93–102. [Google Scholar] [CrossRef]
- Zhou, G.J.; Zhang, F. Applications and Tuning Strategies for Transcription Factor-Based Metabolite Biosensors. Biosensors 2023, 13, 428. [Google Scholar] [CrossRef] [PubMed]
- Tellechea-Luzardo, J.; Stiebritz, M.T.; Carbonell, P. Transcription factor-based biosensors for screening and dynamic regulation. Front. Bioeng. Biotechnol. 2023, 11, 1118702. [Google Scholar] [CrossRef] [PubMed]
- Hanko, E.K.R.; Minton, N.P.; Malys, N. A Transcription Factor-Based Biosensor for Detection of Itaconic Acid. ACS Synth. Biol. 2018, 7, 1436–1446. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Shaner, N.C.; Campbell, R.E.; Steinbach, P.A.; Giepmans, B.N.G.; Palmer, A.E.; Tsien, R.Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004, 22, 1567–1572. [Google Scholar] [CrossRef] [PubMed]
- Siegele, D.A.; Hu, J.C. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc. Natl. Acad. Sci. USA 1997, 94, 8168–8172. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Song, J.; Zhang, H.; Lin, Y.; Han, S.; Huang, Y.; Zheng, S. Development of a Transcription Factor-Based Diamine Biosensor in Corynebacterium glutamicum. ACS Synth. Biol. 2021, 10, 3074–3083. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Wang, J.; Jia, A.; Lin, Y.; Zheng, S. Development of a Transcriptional Factor PuuR-Based Putrescine-Specific Biosensor in Corynebacterium glutamicum. Bioengineering 2023, 10, 157. [Google Scholar] [CrossRef]
- Zhang, J.; Barajas, J.F.; Burdu, M.; Ruegg, T.L.; Dias, B.; Keasling, J.D. Development of a Transcription Factor-Based Lactam Biosensor. ACS Synth. Biol. 2017, 6, 439–445. [Google Scholar] [CrossRef]
- Wang, Z.; Doshi, A.; Chowdhury, R.; Wang, Y.; Maranas, C.D.; Cirino, P.C. Engineering sensitivity and specificity of AraC-based biosensors responsive to triacetic acid lactone and orsellinic acid. Protein. Eng. Des. Sel. 2020, 33, gzaa027. [Google Scholar] [CrossRef]
- FlowJo™; Version 10.8.0; Becton, Dickinson and Company: Ashland, OR, USA, 2021.
- Sun, P.; Zhang, Z.; Wang, B.; Liu, C.; Chen, C.; Liu, P.; Li, X. A genetically encoded fluorescent biosensor for detecting itaconate with subcellular resolution in living macrophages. Nat. Commun. 2022, 13, 6562. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Schrödinger Release 2022-4: Maestro; Schrödinger, LLC: New York, NY, USA, 2022.
- Schrödinger Release 2022-4: Glide; Schrödinger, LLC: New York, NY, USA, 2022.
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef]
- The PyMOL Molecular Graphics System; Version 2.0; Schrödinger, LLC: New York, NY, USA, 2015.
- Gadagkar, S.R.; Call, G.B. Computational tools for fitting the Hill equation to dose-response curves. J. Pharmacol. Toxicol. Methods 2015, 71, 68–76. [Google Scholar] [CrossRef]
- Samanta, D.; Sani, R.K. Methane Oxidation via Chemical and Biological Methods: Challenges and Solutions. Methane 2023, 2, 279–303. [Google Scholar] [CrossRef]



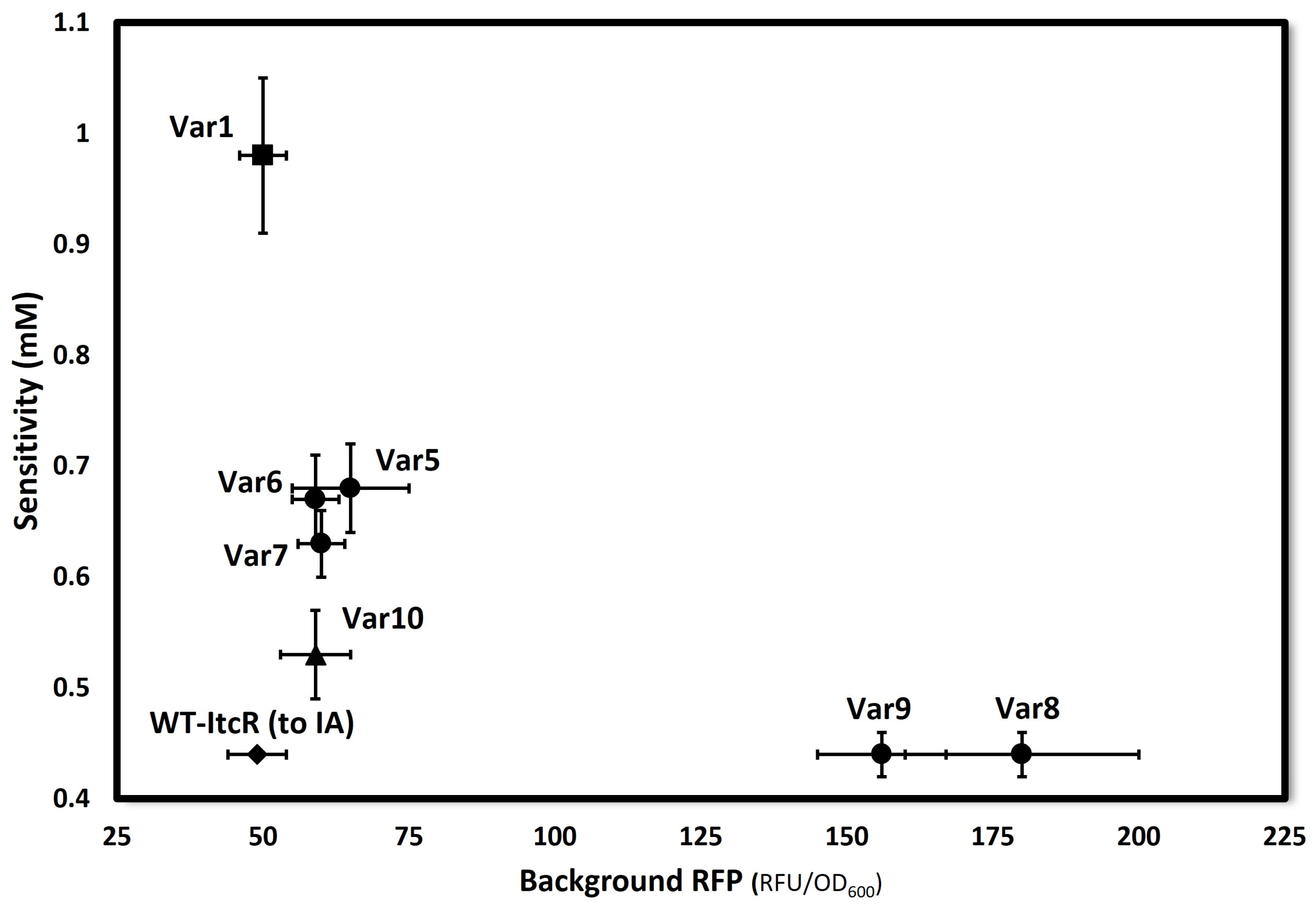
| Variant | a | b | k (mM) | n |
|---|---|---|---|---|
| Var1 | 50 ± 4 | 870 ± 50 | 0.98 ± 0.10 | 1.9 ± 0.1 |
| Var5 | 65 ± 10 | 1500 ± 100 | 0.68 ± 0.04 | 1.5 ± 0.1 |
| Var6 | 59 ± 4 | 1350 ± 20 | 0.67 ± 0.04 | 1.7 ± 0.1 |
| Var7 | 60 ± 4 | 1500 ± 50 | 0.63 ± 0.03 | 1.7 ± 0.1 |
| Var8 | 180 ± 20 | 2300 ± 90 | 0.44 ± 0.02 | 1.3 ± 0.1 |
| Var9 | 160 ± 10 | 2300 ± 70 | 0.44 ± 0.02 | 1.4 ± 0.1 |
| Var10 | 59 ± 6 | 1200 ± 40 | 0.53 ± 0.04 | 1.7 ± 0.3 |
| WT-ItcR (to IA) | 49 ± 5 | 2700 ± 20 | 0.44 ± 0.01 | 1.7 ± 0.1 |
| MS Concentration (mM) | WT | Var1 | Var5 | Var6 | Var7 | Var8 | Var9 | Var10 |
|---|---|---|---|---|---|---|---|---|
| 0.1 | 1.0 ± 0.1 | 1.2 ± 0.2 | 2.2 ± 0.4 | 1.8 ± 0.2 | 2.2 ± 0.6 | 2.7 ± 0.1 | 2.4 ± 0.2 | 1.9 ± 0.2 |
| 1 | 1.0 ± 0.1 | 9.3 ± 0.9 | 15 ± 2 | 15 ± 1 | 17 ± 2 | 9.6 ± 0.9 | 11 ± 1 | 15 ± 2 |
| 5 | 2.5 ± 0.2 | 17 ± 1 | 23 ± 3 | 23 ± 1 | 25 ± 3 | 13 ± 1 | 14 ± 1 | 20 ± 2 |
| WT | Var1 | Var5 | Var6 | Var7 | Var8 | Var9 | Var10 | |
|---|---|---|---|---|---|---|---|---|
| MS/IA | 0.030 ± 0.002 | 1.4 ± 0.1 | 1.0 ± 0.1 | 2.3 ± 0.1 | 1.4 ± 0.1 | 1.8 ± 0.1 | 0.8 ± 0.1 | 1.1 ± 0.1 |
| MS/fumarate | 1.8 ± 0.1 | 15 ± 1 | 23 ± 4 | 21 ± 4 | 23 ± 3 | 12 ± 2 | 12 ± 1 | 15 ± 2 |
| MS/succinate | 1.9 ± 0.3 | 17 ± 2 | 22 ± 3 | 25 ± 2 | 27 ± 6 | 13 ± 3 | 13 ± 1 | 16 ± 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bahrami Moghadam, E.; Nguyen, N.; Wang, Y.; Cirino, P.C. Directed Evolution of Protein-Based Sensors for Anaerobic Biological Activation of Methane. Biosensors 2024, 14, 325. https://doi.org/10.3390/bios14070325
Bahrami Moghadam E, Nguyen N, Wang Y, Cirino PC. Directed Evolution of Protein-Based Sensors for Anaerobic Biological Activation of Methane. Biosensors. 2024; 14(7):325. https://doi.org/10.3390/bios14070325
Chicago/Turabian StyleBahrami Moghadam, Ehsan, Nam Nguyen, Yixi Wang, and Patrick C. Cirino. 2024. "Directed Evolution of Protein-Based Sensors for Anaerobic Biological Activation of Methane" Biosensors 14, no. 7: 325. https://doi.org/10.3390/bios14070325





