Biosensors for Cancer Biomarkers Based on Mesoporous Silica Nanoparticles
Abstract
1. Introduction
2. Cancer Biomarkers
2.1. General Discussion on Cancer Biomarkers and Relevant Biosensors
2.2. Specific Cancer Biomarkers Targeted by MSN-Based Biosensors
3. Detection Methods
3.1. Electrochemical Detection Methods
3.2. Optical Detection Methods
4. MSN-Based Biosensors by Cancer Type
4.1. Lung Cancer
4.2. Breast Cancer
4.3. Prostate Cancer
4.4. Cervical Cancer
4.5. Pancreatic Cancer
4.6. Ovarian Cancer
4.7. Other Cancers
5. Perspectives and Outlook
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, S.-H.; Mou, C.-Y.; Lin, H.-P. Synthesis of Mesoporous Silica Nanoparticles. Chem. Soc. Rev. 2013, 42, 3862–3875. [Google Scholar] [CrossRef]
- Narayan, R.; Nayak, U.Y.; Raichur, A.M.; Garg, S. Mesoporous Silica Nanoparticles: A Comprehensive Review on Synthesis and Recent Advances. Pharmaceutics 2018, 10, 118. [Google Scholar] [CrossRef] [PubMed]
- Nooney, R.I.; Thirunavukkarasu, D.; Chen, Y.; Josephs, R.; Ostafin, A.E. Synthesis of Nanoscale Mesoporous Silica Spheres with Controlled Particle Size. Chem. Mater. 2002, 14, 4721–4728. [Google Scholar] [CrossRef]
- Kankala, R.K.; Han, Y.; Na, J.; Lee, C.; Sun, Z.; Wang, S.; Kimura, T.; Ok, Y.S.; Yamauchi, Y.; Chen, A.; et al. Nanoarchitectured Structure and Surface Biofunctionality of Mesoporous Silica Nanoparticles. Adv. Mater. 2020, 32, 1907035. [Google Scholar] [CrossRef] [PubMed]
- Sancenón, F.; Pascual, L.; Oroval, M.; Aznar, E.; Martínez-Máñez, R. Gated Silica Mesoporous Materials in Sensing Applications. ChemistryOpen 2015, 4, 418–437. [Google Scholar] [CrossRef] [PubMed]
- Manzano, M.; Vallet-Regí, M. Mesoporous Silica Nanoparticles in Nanomedicine Applications. J. Mater. Sci. Mater. Med. 2018, 29, 65. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, A.; Tripathi, D.K.; Yadav, S.; Chauhan, D.K.; Živčák, M.; Ghorbanpour, M.; El-Sheery, N.I.; Brestic, M. Application of Silicon Nanoparticles in Agriculture. 3 Biotech 2019, 9, 90. [Google Scholar] [CrossRef]
- Zamboulis, A.; Moitra, N.; Moreau, J.J.E.; Cattoën, X.; Wong Chi Man, M. Hybrid Materials: Versatile Matrices for Supporting Homogeneous Catalysts. J. Mater. Chem. 2010, 20, 9322. [Google Scholar] [CrossRef]
- Aguilar, Z.P. Nanobiosensors. In Nanomaterials for Medical Applications; Elsevier: Amsterdam, The Netherlands, 2013; pp. 127–179. ISBN 978-0-12-385089-8. [Google Scholar]
- Bhalla, N.; Jolly, P.; Formisano, N.; Estrela, P. Introduction to Biosensors. Essays Biochem. 2016, 60, 1–8. [Google Scholar] [CrossRef]
- Naresh, V.; Lee, N. A Review on Biosensors and Recent Development of Nanostructured Materials-Enabled Biosensors. Sensors 2021, 21, 1109. [Google Scholar] [CrossRef]
- Tothill, I.E. Biosensors for Cancer Markers Diagnosis. Semin. Cell Dev. Biol. 2009, 20, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Singhal, J.; Verma, S.; Kumar, S.; Mehrotra, D. Recent Advances in Nano-Bio-Sensing Fabrication Technology for the Detection of Oral Cancer. Mol. Biotechnol. 2021, 63, 339–362. [Google Scholar] [CrossRef] [PubMed]
- Jafari, S.; Derakhshankhah, H.; Alaei, L.; Fattahi, A.; Varnamkhasti, B.S.; Saboury, A.A. Mesoporous Silica Nanoparticles for Therapeutic/Diagnostic Applications. Biomed. Pharmacother. 2019, 109, 1100–1111. [Google Scholar] [CrossRef] [PubMed]
- Kholafazad Kordasht, H.; Pazhuhi, M.; Pashazadeh-Panahi, P.; Hasanzadeh, M.; Shadjou, N. Multifunctional Aptasensors Based on Mesoporous Silica Nanoparticles as an Efficient Platform for Bioanalytical Applications: Recent Advances. TrAC Trends Anal. Chem. 2020, 124, 115778. [Google Scholar] [CrossRef]
- Qasim Almajidi, Y.; Althomali, R.H.; Gandla, K.; Uinarni, H.; Sharma, N.; Hussien, B.M.; Alhassan, M.S.; Mireya Romero-Parra, R.; Singh Bisht, Y. Multifunctional Immunosensors Based on Mesoporous Silica Nanomaterials as Efficient Sensing Platforms in Biomedical and Food Safety Analysis: A Review of Current Status and Emerging Applications. Microchem. J. 2023, 191, 108901. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Tabassum, D.P.; Polyak, K. Tumorigenesis: It Takes a Village. Nat. Rev. Cancer 2015, 15, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Wodarz, A.; Näthke, I. Cell Polarity in Development and Cancer. Nat. Cell Biol. 2007, 9, 1016–1024. [Google Scholar] [CrossRef]
- Soper, S.A.; Brown, K.; Ellington, A.; Frazier, B.; Garcia-Manero, G.; Gau, V.; Gutman, S.I.; Hayes, D.F.; Korte, B.; Landers, J.L.; et al. Point-of-Care Biosensor Systems for Cancer Diagnostics/Prognostics. Biosens. Bioelectron. 2006, 21, 1932–1942. [Google Scholar] [CrossRef]
- Henry, N.L.; Hayes, D.F. Cancer Biomarkers. Mol. Oncol. 2012, 6, 140–146. [Google Scholar] [CrossRef]
- Barhoum, A.; Altintas, Z.; Devi, K.S.S.; Forster, R.J. Electrochemiluminescence Biosensors for Detection of Cancer Biomarkers in Biofluids: Principles, Opportunities, and Challenges. Nano Today 2023, 50, 101874. [Google Scholar] [CrossRef]
- Wagner, P.D.; Verma, M.; Srivastava, S. Challenges for Biomarkers in Cancer Detection. Ann. N. Y. Acad. Sci. 2004, 1022, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Jayanthi, V.S.P.K.S.A.; Das, A.B.; Saxena, U. Recent Advances in Biosensor Development for the Detection of Cancer Biomarkers. Biosens. Bioelectron. 2017, 91, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Qu, X. Cancer Biomarker Detection: Recent Achievements and Challenges. Chem. Soc. Rev. 2015, 44, 2963–2997. [Google Scholar] [CrossRef] [PubMed]
- Gan, S.D.; Patel, K.R. Enzyme Immunoassay and Enzyme-Linked Immunosorbent Assay. J. Investig. Dermatol. 2013, 133, e12. [Google Scholar] [CrossRef] [PubMed]
- Kubista, M.; Andrade, J.M.; Bengtsson, M.; Forootan, A.; Jonák, J.; Lind, K.; Sindelka, R.; Sjöback, R.; Sjögreen, B.; Strömbom, L.; et al. The Real-Time Polymerase Chain Reaction. Mol. Asp. Med. 2006, 27, 95–125. [Google Scholar] [CrossRef] [PubMed]
- Land, K.J.; Boeras, D.I.; Chen, X.-S.; Ramsay, A.R.; Peeling, R.W. REASSURED Diagnostics to Inform Disease Control Strategies, Strengthen Health Systems and Improve Patient Outcomes. Nat. Microbiol. 2018, 4, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.; Shah, M.R.; Barek, J.; Malik, M.I. Cancer Biomarkers and Their Biosensors: A Comprehensive Review. TrAC Trends Anal. Chem. 2023, 158, 116813. [Google Scholar] [CrossRef]
- Füzéry, A.K.; Levin, J.; Chan, M.M.; Chan, D.W. Translation of Proteomic Biomarkers into FDA Approved Cancer Diagnostics: Issues and Challenges. Clin. Proteom. 2013, 10, 13. [Google Scholar] [CrossRef]
- Wu, J.; Fu, Z.; Yan, F.; Ju, H. Biomedical and Clinical Applications of Immunoassays and Immunosensors for Tumor Markers. TrAC Trends Anal. Chem. 2007, 26, 679–688. [Google Scholar] [CrossRef]
- Sanchez-Carbayo, M. Recent Advances in Bladder Cancer Diagnostics. Clin. Biochem. 2004, 37, 562–571. [Google Scholar] [CrossRef] [PubMed]
- Gann, P.H. A Prospective Evaluation of Plasma Prostate-Specific Antigen for Detection of Prostatic Cancer. JAMA 1995, 273, 289. [Google Scholar] [CrossRef] [PubMed]
- Gold, P.; Freedman, S.O. Demonstration of Tumor-Specific Antigens in Human Colonic Carcinomata by Immunological Tolerance and Absorption Techniques. J. Exp. Med. 1965, 121, 439–462. [Google Scholar] [CrossRef] [PubMed]
- Mordente, A.; Meucci, E.; Martorana, G.E.; Silvestrini, A. Cancer Biomarkers Discovery and Validation: State of the Art, Problems and Future Perspectives. In Advances in Cancer Biomarkers; Scatena, R., Ed.; Advances in Experimental Medicine and Biology; Springer: Dordrecht, The Netherlands, 2015; Volume 867, pp. 9–26. ISBN 978-94-017-7214-3. [Google Scholar]
- Ross, J.S.; Fletcher, J.A.; Bloom, K.J.; Linette, G.P.; Stec, J.; Symmans, W.F.; Pusztai, L.; Hortobagyi, G.N. Targeted Therapy in Breast Cancer. Mol. Cell. Proteom. 2004, 3, 379–398. [Google Scholar] [CrossRef]
- Nasrollahpour, H.; Mahdipour, M.; Isildak, I.; Rashidi, M.-R.; Naseri, A.; Khalilzadeh, B. A Highly Sensitive Electrochemiluminescence Cytosensor for Detection of SKBR-3 Cells as Metastatic Breast Cancer Cell Line: A Constructive Phase in Early and Precise Diagnosis. Biosens. Bioelectron. 2021, 178, 113023. [Google Scholar] [CrossRef]
- Zheng, T.; Pierre-Pierre, N.; Yan, X.; Huo, Q.; Almodovar, A.J.O.; Valerio, F.; Rivera-Ramirez, I.; Griffith, E.; Decker, D.D.; Chen, S.; et al. Gold Nanoparticle-Enabled Blood Test for Early Stage Cancer Detection and Risk Assessment. ACS Appl. Mater. Interfaces 2015, 7, 6819–6827. [Google Scholar] [CrossRef]
- Epstein, J.I. PSA and PAP as Immunohistochemical Markers IN Prostate Cancer. Urol. Clin. N. Am. 1993, 20, 757–770. [Google Scholar] [CrossRef]
- Li, X.; Tan, J.; Yu, J.; Feng, J.; Pan, A.; Zheng, S.; Wu, J. Use of a Porous Silicon–Gold Plasmonic Nanostructure to Enhance Serum Peptide Signals in MALDI-TOF Analysis. Anal. Chim. Acta 2014, 849, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Schuerle, S.; Dudani, J.S.; Christiansen, M.G.; Anikeeva, P.; Bhatia, S.N. Magnetically Actuated Protease Sensors for in Vivo Tumor Profiling. Nano Lett. 2016, 16, 6303–6310. [Google Scholar] [CrossRef]
- Rao, H.; Wu, H.; Huang, Q.; Yu, Z.; Zhang, Q.; Zhong, Z. Clinical Diagnostic Value for Colorectal Cancer Based on Serum CEA, CA24-2 and CA19-9. Clin. Lab. 2021, 67, 4. [Google Scholar] [CrossRef]
- Tokunaga, R.; Sakamoto, Y.; Nakagawa, S.; Yoshida, N.; Baba, H. The Utility of Tumor Marker Combination, Including Serum P53 Antibody, in Colorectal Cancer Treatment. Surg. Today 2017, 47, 636–642. [Google Scholar] [CrossRef] [PubMed]
- Lu, N.; Gao, A.; Dai, P.; Mao, H.; Zuo, X.; Fan, C.; Wang, Y.; Li, T. Ultrasensitive Detection of Dual Cancer Biomarkers with Integrated CMOS-Compatible Nanowire Arrays. Anal. Chem. 2015, 87, 11203–11208. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chang, S.; Wang, N.; Song, P.; Wei, H.; Liu, J. Clinical Utility of Six Serum Tumor Markers for the Diagnosis of Lung Cancer. iLABMED 2023, 1, 132–141. [Google Scholar] [CrossRef]
- Qiao, Z.; Perestrelo, R.; Reyes-Gallardo, E.M.; Lucena, R.; Cárdenas, S.; Rodrigues, J.; Câmara, J.S. Octadecyl Functionalized Core–Shell Magnetic Silica Nanoparticle as a Powerful Nanocomposite Sorbent to Extract Urinary Volatile Organic Metabolites. J. Chromatogr. A 2015, 1393, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.W.; Doerksen, J.D.; Kang, S.; Walsh, D.; Yang, Q.; Hong, D.; Liu, J.T.C. Multiplexed Molecular Imaging of Fresh Tissue Surfaces Enabled by Convection-Enhanced Topical Staining with SERS-Coded Nanoparticles. Small 2016, 12, 5612–5621. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, B.; He, M.; Zhang, Y.; Peng, L.; Hu, B. Boronic Acid Recognition Based-Gold Nanoparticle-Labeling Strategy for the Assay of Sialic Acid Expression on Cancer Cell Surface by Inductively Coupled Plasma Mass Spectrometry. Analyst 2016, 141, 1286–1293. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.-J.; Ju, Q.; Li, G.-C. Tumor Markers for Hepatocellular Carcinoma. Mol. Clin. Oncol. 2013, 1, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Smith, D.A.; Guler, E.; Kikano, E.G.; Rajdev, M.A.; Yoest, J.M.; Ramaiya, N.H.; Tirumani, S.H. Past, Present, and Future of Serum Tumor Markers in Management of Ovarian Cancer: A Guide for the Radiologist. RadioGraphics 2021, 41, 1839–1856. [Google Scholar] [CrossRef] [PubMed]
- Khong, H.T.; Rosenberg, S.A. Pre-Existing Immunity to Tyrosinase-Related Protein (TRP)-2, a New TRP-2 Isoform, and the NY-ESO-1 Melanoma Antigen in a Patient with a Dramatic Response to Immunotherapy. J. Immunol. 2002, 168, 951–956. [Google Scholar] [CrossRef]
- Manne, U.; Srivastava, R.-G.; Srivastava, S. Keynote Review: Recent Advances in Biomarkers for Cancer Diagnosis and Treatment. Drug Discov. Today 2005, 10, 965–976. [Google Scholar] [CrossRef]
- Balendiran, G.K.; Dabur, R.; Fraser, D. The Role of Glutathione in Cancer. Cell Biochem. Funct. 2004, 22, 343–352. [Google Scholar] [CrossRef]
- Morris, P.E.; Bernard, G.R. Significance of Glutathione in Lung Disease and Implications for Therapy. Am. J. Med. Sci. 1994, 307, 119–127. [Google Scholar] [CrossRef]
- Kennedy, L.; Sandhu, J.K.; Harper, M.-E.; Cuperlovic-Culf, M. Role of Glutathione in Cancer: From Mechanisms to Therapies. Biomolecules 2020, 10, 1429. [Google Scholar] [CrossRef] [PubMed]
- Gamcsik, M.P.; Kasibhatla, M.S.; Teeter, S.D.; Colvin, O.M. Glutathione Levels in Human Tumors. Biomarkers 2012, 17, 671–691. [Google Scholar] [CrossRef] [PubMed]
- Moll, R. Epithelial Tumor Markers: Cytokeratins and Tissue Polypeptide Antigen (TPA). In Morphological Tumor Markers; Seifer, G., Ed.; Current Topics in Pathology; Springer: Berlin/Heidelberg, Germany, 1987; Volume 77, pp. 71–101. ISBN 978-3-642-71358-3. [Google Scholar]
- Sundström, B.E.; Stigbrand, T.I. Cytokeratins and Tissue Polypeptide Antigen. Int. J. Biol. Markers 1994, 9, 102–108. [Google Scholar] [CrossRef]
- Wieskopf, B.; Demangeat, C.; Purohit, A.; Stenger, R.; Gries, P.; Kreisman, H.; Quoix, E. Cyfra 21-1 as a Biologic Marker of Non-Small Cell Lung Cancer. Chest 1995, 108, 163–169. [Google Scholar] [CrossRef]
- Duffy, M.J.; Shering, S.; Sherry, F.; McDermott, E.; O’Higgins, N. CA 15–3: A Prognostic Marker in Breast Cancer. Int. J. Biol. Markers 2000, 15, 330–333. [Google Scholar] [CrossRef]
- Mitri, Z.; Constantine, T.; O’Regan, R. The HER2 Receptor in Breast Cancer: Pathophysiology, Clinical Use, and New Advances in Therapy. Chemother. Res. Pract. 2012, 2012, 743193. [Google Scholar] [CrossRef] [PubMed]
- Maetzel, D.; Denzel, S.; Mack, B.; Canis, M.; Went, P.; Benk, M.; Kieu, C.; Papior, P.; Baeuerle, P.A.; Munz, M.; et al. Nuclear Signalling by Tumour-Associated Antigen EpCAM. Nat. Cell Biol. 2009, 11, 162–171. [Google Scholar] [CrossRef]
- Went, P.T.H.; Lugli, A.; Meier, S.; Bundi, M.; Mirlacher, M.; Sauter, G.; Dirnhofer, S. Frequent EpCam Protein Expression in Human Carcinomas. Hum. Pathol. 2004, 35, 122–128. [Google Scholar] [CrossRef]
- Gutman, A.B.; Gutman, E.B. An “Acid” Phosphatase Occurring in The Serum Of Patients with Metastasizing Carcinoma of the Prostate Gland. J. Clin. Investig. 1938, 17, 473–478. [Google Scholar] [CrossRef]
- Henneberry, M.O.; Engel, G.; Grayhack, J.T. Acid Phosphatase. Urol. Clin. N. Am. 1979, 6, 629–641. [Google Scholar] [CrossRef]
- Singh, J.; Pasi, D.K.; Bala, M.; Kumar, A.; Singh, A.; Jakhar, R.; Sharma, A.; Saini, A. Evaluation of prostate-specific antigen and total serum acid phosphatase in prostatic carcinoma. Natl. J. Physiol. Pharm. Pharmacol. 2023, 13, 1065–1071. [Google Scholar]
- Lilja, H. A Kallikrein-like Serine Protease in Prostatic Fluid Cleaves the Predominant Seminal Vesicle Protein. J. Clin. Investig. 1985, 76, 1899–1903. [Google Scholar] [CrossRef]
- Gjertson, C.K.; Albertsen, P.C. Use and Assessment of PSA in Prostate Cancer. Med. Clin. N. Am. 2011, 95, 191–200. [Google Scholar] [CrossRef]
- Burd, E.M. Human Papillomavirus and Cervical Cancer. Clin. Microbiol. Rev. 2003, 16, 1–17. [Google Scholar] [CrossRef]
- Chang, Y.-F.; Yan, G.-J.; Liu, G.-C.; Hong, Y.; Chen, H.-L.; Jiang, S.; Zhong, Y.; Xiyang, Y.-B.; Hu, T. HPV16 E6 Promotes the Progression of HPV Infection-Associated Cervical Cancer by Upregulating Glucose-6-Phosphate Dehydrogenase Expression. Front. Oncol. 2021, 11, 718781. [Google Scholar] [CrossRef] [PubMed]
- Ghittoni, R.; Accardi, R.; Hasan, U.; Gheit, T.; Sylla, B.; Tommasino, M. The Biological Properties of E6 and E7 Oncoproteins from Human Papillomaviruses. Virus Genes 2010, 40, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Torigoe, T. Radioimmunoassay for Tumor Antigen of Human Cervical Squamous Cell Carcinoma. Cancer 1977, 40, 1621–1628. [Google Scholar] [CrossRef]
- Ohara, K. Assessment of Cervical Cancer Radioresponse by Serum Squamous Cell Carcinoma Antigen and Magnetic Resonance Imaging. Obstet. Gynecol. 2002, 100, 781–787. [Google Scholar] [CrossRef]
- Hasan, S.; Jacob, R.; Manne, U.; Paluri, R. Advances in Pancreatic Cancer Biomarkers. Oncol. Rev. 2019, 13, 69–76. [Google Scholar] [CrossRef]
- Duan, L.; Hu, X.; Feng, D.; Lei, S.; Hu, G. GPC-1 May Serve as a Predictor of Perineural Invasion and a Prognosticator of Survival in Pancreatic Cancer. Asian J. Surg. 2013, 36, 7–12. [Google Scholar] [CrossRef]
- Atallah, G.A.; Abd Aziz, N.H.; Teik, C.K.; Shafiee, M.N.; Kampan, N.C. New Predictive Biomarkers for Ovarian Cancer. Diagnostics 2021, 11, 465. [Google Scholar] [CrossRef] [PubMed]
- Cesewski, E.; Johnson, B.N. Electrochemical Biosensors for Pathogen Detection. Biosens. Bioelectron. 2020, 159, 112214. [Google Scholar] [CrossRef]
- Tanwar, A.; Gandhi, H.A.; Kushwaha, D.; Bhattacharya, J. A Review on Microelectrode Array Fabrication Techniques and Their Applications. Mater. Today Chem. 2022, 26, 101153. [Google Scholar] [CrossRef]
- Grieshaber, D.; MacKenzie, R.; Vörös, J.; Reimhult, E. Electrochemical Biosensors—Sensor Principles and Architectures. Sensors 2008, 8, 1400–1458. [Google Scholar] [CrossRef]
- Chaubey, A.; Malhotra, B.D. Mediated Biosensors. Biosens. Bioelectron. 2002, 17, 441–456. [Google Scholar] [CrossRef]
- Cho, I.H.; Kim, D.H.; Park, S. Electrochemical biosensors: Perspective on functional nanomaterials for on-site analysis. Biomater. Res. 2020, 24, 6. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Matos Pires, N.M.; Yang, Z.; Jiang, Z. Advances in Electrochemical Biosensors Based on Nanomaterials for Protein Biomarker Detection in Saliva. Adv. Sci. 2023, 10, 2205429. [Google Scholar] [CrossRef]
- Huang, X.; Zhu, Y.; Kianfar, E. Nano Biosensors: Properties, Applications and Electrochemical Techniques. J. Mater. Res. Technol. 2021, 12, 1649–1672. [Google Scholar] [CrossRef]
- Deng, K.; Zhang, Y.; Tong, X. Sensitive Electrochemical Detection of microRNA-21 Based on Propylamine-Functionalized Mesoporous Silica with Glucometer Readout. Anal. Bioanal. Chem. 2018, 410, 1863–1871. [Google Scholar] [CrossRef] [PubMed]
- Lv, F.; Wang, M.; Ma, H.; Hu, L.; Wei, Q.; Wu, D. Sensitive Detection of CYFRA21-1 by a Controlled Release Sensor Based on Personal Glucose Meter. Sens. Actuators B Chem. 2022, 371, 132543. [Google Scholar] [CrossRef]
- Gai, P.; Gu, C.; Hou, T.; Li, F. Integration of Biofuel Cell-Based Self-Powered Biosensing and Homogeneous Electrochemical Strategy for Ultrasensitive and Easy-To-Use Bioassays of MicroRNA. ACS Appl. Mater. Interfaces 2018, 10, 9325–9331. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Puig, S.; Lazo-Fraga, A.R.; Korgel, B.A.; Oza, G.; Dutt, A.; Vallejo-Becerra, V.; Valdés-González, A.C.; Chávez-Ramírez, A.U. Molecularly Imprinted Polymer-Silica Nanocomposite Based Potentiometric Sensor for Early Prostate Cancer Detection. Mater. Lett. 2022, 309, 131324. [Google Scholar] [CrossRef]
- Tvorynska, S.; Barek, J.; Josypcuk, B. High-Performance Amperometric Biosensor for Flow Injection Analysis Consisting of a Replaceable Lactate Oxidase-Based Mini-Reactor and a Silver Amalgam Screen-Printed Electrode. Electrochim. Acta 2023, 445, 142033. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, Y.; Yu, H.; Wu, D.; Ma, H.; Li, H.; Du, B.; Wei, Q. Label-Free Electrochemical Immunosensor for Prostate-Specific Antigen Based on Silver Hybridized Mesoporous Silica Nanoparticles. Anal. Biochem. 2013, 434, 123–127. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Li, Y.; Li, C.; Yang, J.; Liu, D.; Wang, H.; Xu, R.; Zhang, Y.; Wei, Q. An Ultrasensitive Split-Type Electrochemical Immunosensor Based on Controlled-Release Strategy for Detection of CA19-9. Biosens. Bioelectron. 2023, 227, 115180. [Google Scholar] [CrossRef]
- Krishnan, S.; He, X.; Zhao, F.; Zhang, Y.; Liu, S.; Xing, R. Dual Labeled Mesoporous Silica Nanospheres Based Electrochemical Immunosensor for Ultrasensitive Detection of Carcinoembryonic Antigen. Anal. Chim. Acta 2020, 1133, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Wen, T.; Xia, C.; Yu, Q.; Yu, Y.; Li, S.; Zhou, C.; Sun, K.; Yue, S. A Dual-Signal Electrochemical Immunosensor for the Detection of HPV16 E6 Oncoprotein Based on PdBP Dendritic Ternary Nanospheres and MBSi-Chi Nanocomposites. Analyst 2022, 147, 2272–2279. [Google Scholar] [CrossRef]
- Jamei, H.R.; Rezaei, B.; Ensafi, A.A. An Ultrasensitive Electrochemical Anti-Lysozyme Aptasensor with Biorecognition Surface Based on Aptamer/Amino-rGO/Ionic Liquid/Amino-Mesosilica Nanoparticles. Colloids Surf. B Biointerfaces 2019, 181, 16–24. [Google Scholar] [CrossRef]
- Chen, H.; Wang, Z.; Liu, Z.; Niu, Q.; Wang, X.; Miao, Z.; Zhang, H.; Wei, J.; Wan, M.; Mao, C. Construction of 3D Electrochemical Cytosensor by Layer-by-Layer Assembly for Ultra-Sensitive Detection of Cancer Cells. Sens. Actuators B Chem. 2021, 329, 128995. [Google Scholar] [CrossRef]
- Yola, M.L.; Atar, N.; Özcan, N. A Novel Electrochemical Lung Cancer Biomarker Cytokeratin 19 Fragment Antigen 21-1 Immunosensor Based on Si3N4/MoS2 Incorporated MWCNTs and Core–Shell Type Magnetic Nanoparticles. Nanoscale 2021, 13, 4660–4669. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-X.; Lv, C.-L.; Tang, C.; Jiang, L.-Y.; Wang, A.-J.; Feng, J.-J. Ultrasensitive Sandwich-Typed Electrochemical Immunoassay for Detection of Squamous Cell Carcinoma Antigen Based on Highly Branched PtCo Nanocrystals and Dendritic Mesoporous SiO2@AuPt Nanoparticles. Microchim. Acta 2022, 189, 416. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Li, W.; Ma, Z. Improved Sandwich-Format Electrochemical Immunosensor Based on “Smart” SiO2@polydopamine Nanocarrier. Biosens. Bioelectron. 2018, 109, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Li, W.; Duan, S.; Peng, J.; Liu, J.; Ma, W.; Wang, H.; He, X.; Wang, K. Mesoporous Silica Containers and Programmed Catalytic Hairpin Assembly/Hybridization Chain Reaction Based Electrochemical Sensing Platform for MicroRNA Ultrasensitive Detection with Low Background. Anal. Chem. 2019, 91, 10672–10678. [Google Scholar] [CrossRef] [PubMed]
- Sadi, S.; Khalilzadeh, B.; Mahdipour, M.; Sokouti Nasimi, F.; Isildak, I.; Davaran, S.; Rashidi, M.-R.; Bani, F. Early Stage Evaluation of Cancer Stem Cells Using Platinum Nanoparticles/CD133+ Enhanced Nanobiocomposite. Cancer Nano 2023, 14, 55. [Google Scholar] [CrossRef]
- Kovarova, A.; Kastrati, G.; Pekarkova, J.; Metelka, R.; Drbohlavova, J.; Bilkova, Z.; Selesovska, R.; Korecka, L. Biosensor with Electrochemically Active Nanocomposites for Signal Amplification and Simultaneous Detection of Three Ovarian Cancer Biomarkers. Electrochim. Acta 2023, 469, 143213. [Google Scholar] [CrossRef]
- Xiao, G.; Ge, H.; Yang, Q.; Zhang, Z.; Cheng, L.; Cao, S.; Ji, J.; Zhang, J.; Yue, Z. Light-Addressable Photoelectrochemical Sensors for Multichannel Detections of GPC1, CEA and GSH and Its Applications in Early Diagnosis of Pancreatic Cancer. Sens. Actuators B Chem. 2022, 372, 132663. [Google Scholar] [CrossRef]
- Rasheed, S.; Kanwal, T.; Ahmad, N.; Fatima, B.; Najam-ul-Haq, M.; Hussain, D. Advances and Challenges in Portable Optical Biosensors for Onsite Detection and Point-of-Care Diagnostics. TrAC Trends Anal. Chem. 2024, 173, 117640. [Google Scholar] [CrossRef]
- Zhao, X.; Dai, X.; Zhao, S.; Cui, X.; Gong, T.; Song, Z.; Meng, H.; Zhang, X.; Yu, B. Aptamer-Based Fluorescent Sensors for the Detection of Cancer Biomarkers. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 247, 119038. [Google Scholar] [CrossRef]
- Wang, H.; Wu, T.; Li, M.; Tao, Y. Recent Advances in Nanomaterials for Colorimetric Cancer Detection. J. Mater. Chem. B 2021, 9, 921–938. [Google Scholar] [CrossRef] [PubMed]
- Khansili, N.; Rattu, G.; Krishna, P.M. Label-Free Optical Biosensors for Food and Biological Sensor Applications. Sens. Actuators B Chem. 2018, 265, 35–49. [Google Scholar] [CrossRef]
- Xia, N.; Deng, D.; Mu, X.; Liu, A.; Xie, J.; Zhou, D.; Yang, P.; Xing, Y.; Liu, L. Colorimetric Immunoassays Based on Pyrroloquinoline Quinone-Catalyzed Generation of Fe(II)-Ferrozine with Tris(2-Carboxyethyl)Phosphine as the Reducing Reagent. Sens. Actuators B Chem. 2020, 306, 127571. [Google Scholar] [CrossRef]
- Son, M.H.; Park, S.W.; Sagong, H.Y.; Jung, Y.K. Recent Advances in Electrochemical and Optical Biosensors for Cancer Biomarker Detection. BioChip J. 2023, 17, 44–67. [Google Scholar] [CrossRef]
- Chen, S.; Yu, Y.-L.; Wang, J.-H. Inner Filter Effect-Based Fluorescent Sensing Systems: A Review. Anal. Chim. Acta 2018, 999, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Zhang, C. Electrogenerated Chemiluminescence Biosensing. Anal. Chem. 2020, 92, 524–534. [Google Scholar] [CrossRef] [PubMed]
- Kaur, B.; Kumar, S.; Kaushik, B.K. Recent Advancements in Optical Biosensors for Cancer Detection. Biosens. Bioelectron. 2022, 197, 113805. [Google Scholar] [CrossRef]
- Piliarik, M.; Vaisocherová, H.; Homola, J. Surface Plasmon Resonance Biosensing. In Biosensors and Biodetection; Rasooly, A., Herold, K.E., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2009; Volume 503, pp. 65–88. ISBN 978-1-60327-566-8. [Google Scholar]
- Lin, C.; Li, Y.; Peng, Y.; Zhao, S.; Xu, M.; Zhang, L.; Huang, Z.; Shi, J.; Yang, Y. Recent Development of Surface-Enhanced Raman Scattering for Biosensing. J. Nanobiotechnol. 2023, 21, 149. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Lao, Y.-H.; Mintz, R.L.; Chen, Z.; Shao, D.; Hu, H.; Wang, H.-X.; Tao, Y.; Leong, K.W. A Multifunctional Mesoporous Silica–Gold Nanocluster Hybrid Platform for Selective Breast Cancer Cell Detection Using a Catalytic Amplification-Based Colorimetric Assay. Nanoscale 2019, 11, 2631–2636. [Google Scholar] [CrossRef]
- Zhang, Y.; Meng, W.; Li, X.; Wang, D.; Shuang, S.; Dong, C. Dendritic Mesoporous Silica Nanoparticle-Tuned High-Affinity MnO2 Nanozyme for Multisignal GSH Sensing and Target Cancer Cell Detection. ACS Sustain. Chem. Eng. 2022, 10, 5911–5921. [Google Scholar] [CrossRef]
- Chen, S.; Li, Z.; Xue, R.; Huang, Z.; Jia, Q. Confining Copper Nanoclusters in Three Dimensional Mesoporous Silica Particles: Fabrication of an Enhanced Emission Platform for “Turn off-on” Detection of Acid Phosphatase Activity. Anal. Chim. Acta 2022, 1192, 339387. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.; Pan, Y.; Luan, X.; Gao, Y.; Yang, J.; Wang, Y.; Song, Y. Copper Metal-Organic Framework Incorporated Mesoporous Silica as a Bioorthogonal Biosensor for Detection of Glutathione. Sens. Actuators B Chem. 2021, 345, 130382. [Google Scholar] [CrossRef]
- Bagheri Hashkavayi, A.; Cha, B.S.; Hwang, S.H.; Kim, J.; Park, K.S. Highly Sensitive Electrochemical Detection of Circulating EpCAM-Positive Tumor Cells Using a Dual Signal Amplification Strategy. Sens. Actuators B Chem. 2021, 343, 130087. [Google Scholar] [CrossRef]
- Chang, Z.; Feng, J.; Zheng, X. A Highly Sensitive Fluorescence Sensor Based on Lucigenin/Chitosan/SiO 2 Composite Nanoparticles for microRNA Detection Using Magnetic Separation. Luminescence 2020, 35, 835–844. [Google Scholar] [CrossRef]
- Esmaeili, Y.; Khavani, M.; Bigham, A.; Sanati, A.; Bidram, E.; Shariati, L.; Zarrabi, A.; Jolfaie, N.A.; Rafienia, M. Mesoporous Silica@chitosan@gold Nanoparticles as “on/off” Optical Biosensor and pH-Sensitive Theranostic Platform against Cancer. Int. J. Biol. Macromol. 2022, 202, 241–255. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Fan, J.; Cui, L.; Ke, W.; Zheng, F.; Zhao, Y. Fluorometric Nanoprobes for Simultaneous Aptamer-Based Detection of Carcinoembryonic Antigen and Prostate Specific Antigen. Microchim. Acta 2019, 186, 152. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Cao, J.; Ding, S.-N. Simultaneous Detection of Two Ovarian Cancer Biomarkers in Human Serums with Biotin-Enriched Dendritic Mesoporous Silica Nanoparticles-Labeled Multiplex Lateral Flow Immunoassay. Sens. Actuators B Chem. 2022, 371, 132597. [Google Scholar] [CrossRef]
- Liu, S.; Li, J.; Zou, Y.; Jiang, Y.; Wu, L.; Deng, Y. Construction of Magnetic Core–Large Mesoporous Satellite Immunosensor for Long-Lasting Chemiluminescence and Highly Sensitive Tumor Marker Determination. Small 2023, 19, 2304631. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Z.; Fang, L.; Weng, A.; Luo, F.; Guo, L.; Qiu, B.; Lin, Z. Electrochemiluminescence Sensor for Cancer Cell Detection Based on H2O2-Triggered Stimulus Response System. J. Anal. Test. 2020, 4, 128–135. [Google Scholar] [CrossRef]
- Wang, W.; Feng, D.; Wang, Y.; Kan, X. Ruthenium Poly(Ethylenimine)/Gold Nanoparticles Immobilized on Dendritic Mesoporous Silica Nanoparticles for a CA15-3 Electrochemiluminescence Immunosensor via Cu 2 O@PDA Dual Quenching. ACS Appl. Nano Mater. 2023, 6, 19271–19278. [Google Scholar] [CrossRef]
- Huang, Z.; Zhao, L.; Li, Y.; Wang, H.; Ma, H.; Wei, Q.; Wu, D. Glucose Oxidation Induced pH Stimuli Response Controlled Release Electrochemiluminescence Biosensor for Ultrasensitive Detection of CYFRA 21-1. Talanta 2024, 266, 124955. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Du, Y.; Ru, Z.; Fan, D.; Yang, L.; Ren, X.; Wei, Q. Aggregation-Induced Electrochemiluminescence Frame of Silica-Confined Tetraphenylethylene Derivative Matrixes for CD44 Detection via Peptide Recognition. Anal. Chem. 2023, 95, 6725–6731. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Yoon, H.J.; Lee, S.E.; Lee, L.P. Multifunctional Cellular Targeting, Molecular Delivery, and Imaging by Integrated Mesoporous-Silica with Optical Nanocrescent Antenna: MONA. ACS Nano 2022, 16, 2013–2023. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Cui, M.; Zhou, H.; Zhang, S. DNA-Hybrid-Gated Functional Mesoporous Silica for Sensitive DNA Methyltransferase SERS Detection. Chem. Commun. 2015, 51, 13983–13985. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Vaccarella, S.; Morgan, E.; Li, M.; Etxeberria, J.; Chokunonga, E.; Manraj, S.S.; Kamate, B.; Omonisi, A.; Bray, F. Global Variations in Lung Cancer Incidence by Histological Subtype in 2020: A Population-Based Study. Lancet Oncol. 2023, 24, 1206–1218. [Google Scholar] [CrossRef]
- Mundžić, M.; Lazović, J.; Mladenović, M.; Pavlović, A.; Ultimo, A.; Gobbo, O.L.; Ruiz-Hernandez, E.; Santos-Martinez, M.J.; Knežević, N.Ž. MRI-Based Sensing of pH-Responsive Content Release from Mesoporous Silica Nanoparticles. J. Sol-Gel Sci. Technol. 2024. [Google Scholar] [CrossRef]
- Mladenović, M.; Morgan, I.; Ilić, N.; Saoud, M.; Pergal, M.V.; Kaluđerović, G.N.; Knežević, N.Ž. pH-Responsive Release of Ruthenium Metallotherapeutics from Mesoporous Silica-Based Nanocarriers. Pharmaceutics 2021, 13, 460. [Google Scholar] [CrossRef] [PubMed]
- Miyajima, T.; Abry, S.; Zhou, W.; Albela, B.; Bonneviot, L.; Oumi, Y.; Sano, T.; Yoshitake, H. Estimation of Spacing between 3-Bromopropyl Functions Grafted on Mesoporous Silica Surfaces by a Substitution Reaction Using Diamine Probe Molecules. J. Mater. Chem. 2007, 17, 3901. [Google Scholar] [CrossRef]
- Yi, S.; Zheng, J.; Lv, P.; Zhang, D.; Zheng, X.; Zhang, Y.; Liao, R. Controlled Drug Release from Cyclodextrin-Gated Mesoporous Silica Nanoparticles Based on Switchable Host–Guest Interactions. Bioconjugate Chem. 2018, 29, 2884–2891. [Google Scholar] [CrossRef]
- Cheng, C.-A.; Deng, T.; Lin, F.-C.; Cai, Y.; Zink, J.I. Supramolecular Nanomachines as Stimuli-Responsive Gatekeepers on Mesoporous Silica Nanoparticles for Antibiotic and Cancer Drug Delivery. Theranostics 2019, 9, 3341–3364. [Google Scholar] [CrossRef]
- Kim, C.; Yoon, S.; Lee, J.H. Facile large-scale synthesis of mesoporous silica nanoparticles at room temperature in a monophasic system with fine size control. Microporous Mesoporous Mater. 2019, 288, 109595. [Google Scholar] [CrossRef]
- Zhang, K.; Xu, L.-L.; Jiang, J.-G.; Calin, N.; Lam, K.-F.; Zhang, S.-J.; Wu, H.-H.; Wu, G.-D.; Albela, B.; Bonneviot, L.; et al. Facile Large-Scale Synthesis of Monodisperse Mesoporous Silica Nanospheres with Tunable Pore Structure. J. Am. Chem. Soc. 2013, 135, 2427–2430. [Google Scholar] [CrossRef] [PubMed]
- Jundale, R.B.; Sonawane, J.R.; Palghadmal, A.V.; Jaiswal, H.K.; Deore, H.S.; Kulkarni, A.A. Scaling-up continuous production of mesoporous silica particles at kg scale: Design & operational strategies. React. Chem. Eng. 2024, 9, 1914–1923. [Google Scholar] [CrossRef]
| Cancer Biomarker | Cancer Type | Reference |
|---|---|---|
| PSA | Prostate | [37] |
| IgG | Prostate | [38] |
| PAP, PSA | Prostate | [39] |
| Peptide fragments | Colorectal | [40] |
| MMP | Colorectal | [41] |
| CEA, CA 19-9, CA A24-2 | Colorectal and pancreatic | [42] |
| P53 gene | Colorectal | [43] |
| CYFRA 21-1 | Lung | [44] |
| CEA, CA 19-9, SCC antigen, NSE | Lung | [45] |
| EVOM | Breast | [46] |
| EGFR, HER2, transmembrane glycoproteins CD44 and CD24 | Breast | [47] |
| Sialic acid | Breast and liver | [48] |
| AFP | Liver | [49] |
| CA 125, HE4 | Ovarian | [50] |
| TRP-2, NY-ESO-1 melanoma Antigen | Melanoma | [51] |
| Technique | Method | MSN Role | Target Biomarker | Key Performances | Reference |
|---|---|---|---|---|---|
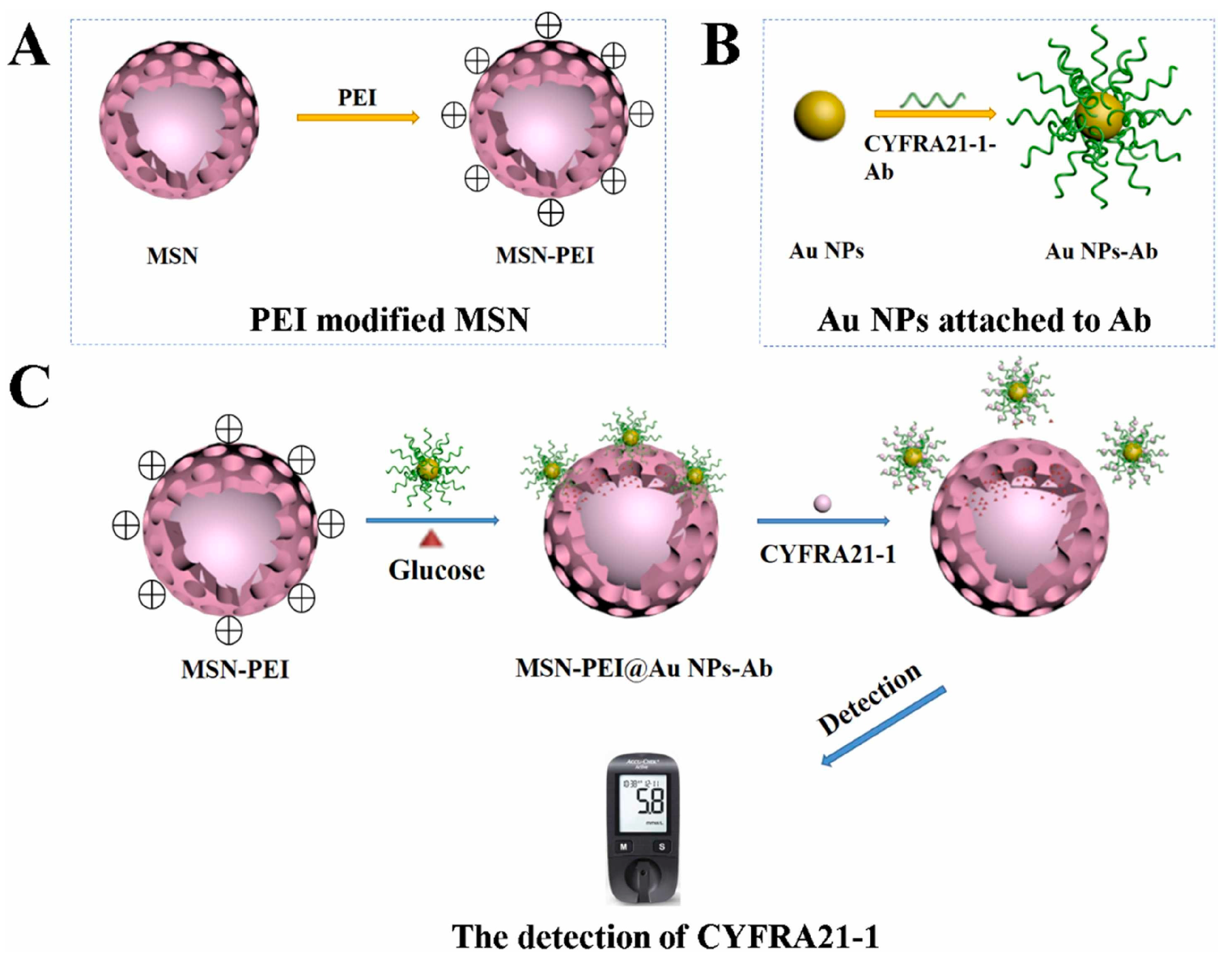
| Potentiometry | Commercial glucometer | Release of glucose upon target cDNA hybridization | miRNA-21 | 50 pM–5 nM 1 19 pM 2 | [84] |
| Release of glucose upon target binding to antibody | CYFRA 21-1 | 1.3–160 ng/mL 1 | [85] | ||
| Open circuit voltage | Release of [Fe(CN)6]3− upon target cDNA hybridization | miRNA-21 | 10 aM–1 pM 1 | [86] | |
| Chrono-potentiometry | MIP performance improvement | Sarcosine | 10 nM–10 μM 1 7.8 Nm 2 | [87] | |
| Amperometry | Chrono-amperometry | Lactate oxidase immobilization | Lactic acid | 40–500 μM 1 | [88] |
| Voltammetry | Cyclic voltammetry | Antibody immobilization with AgNP for electron transfer improvement | PSA | 50 pg/mL–50 ng/mL 1 15 pg/mL 2 | [89] |
| Differential pulse voltammetry | Release of glucose from target-bound MSNs | CA 19-9 | 0.01–100 U/mL 1 0.0005 U/mL 2 | [90] | |
| Dual-labeled MSNs with AuNRs and HRP for signal enhancement | CEA | 0.1–5 pg/mL 1 5.25 fg/mL 2 | [91] | ||
| Sandwich-type immunoassay with MB@MSNs for signal enhancement | HPV16 E6 oncoprotein | 50 fg/mL–4 ng/mL 1 | [92] | ||
| Amino-MSNs in composite with Amino-rGO and IL for signal enhancement | Lysozyme | 20 fM–50 nM 1 | [93] | ||
| SNA-loaded MSNs for improved capture of target | MCF-7 cancer cells | 1−1.0 × 107 cells/mL 1 4 cells/mL 2 | [94] | ||
| Sandwich-type immunoassay with MMSN@AuNP-Ab2 for signal enhancement | CYFRA 21-1 | 0.01–1.0 pg/mL 1 2 fg/mL 2 | [95] | ||
| Sandwich-type immunoassay with thionine-loaded MSNs for signal enhancement | SCCA | 0.01–120 ng/mL 1 0.33 pg/mL 2 | [96] | ||
| Square wave voltammetry | Sandwich-type immunoassay with MB-loaded MSNs for signal production by controlled MB release | PSA | 10 fg/mL–100 ng/mL 1 1.25 fg/mL 2 | [97] | |
| Release of MB from programmed target-enabled CHA for HCR signal amplification | miRNA-21 | 0.1 fM–5 pM 1 | [98] | ||
| Sensitivity improvement by MSNs/PtNPs | CD133 | 5–20 cells/5 μL 1 | [99] | ||
| Square wave anodic/cathodic stripping voltammetry | Nanocomposites for signal development and enhancement: PbS-QD@MSNs, CdTe-QD@MSNs, and AuNPs@MSNs | HE4, CA-125, and AFP | HE4: 0.02–20 pM 1; LOD 5.07 pM CA-125: 0.45–450 IU/L 1; LOD 3.1 IU/L AFP: 0.1–500 ng/L 1; LOD 2.44 pg/L | [100] | |
| Impedimetry | Electrochemical impedance spectroscopy | Amino-MSNs in composite with Amino-rGO and IL for signal enhancement | Lysozyme | 10 fM–200 nM 1 | [93] |
| Photoelectrochemical method | Chrono-amperometry | CD@MSB for improved sensitivity | Glutathione | 34.9 nM 2 | [101] |
| Type | Method | MSN Role | Target Biomarker | Key Performances | Reference |
|---|---|---|---|---|---|
| Colorimetric | Enzyme based | AuNC-loaded MSNs for improved signal | HER2 | 10–1000 cells 1 10 cells 2 | [113] |
| Non-enzyme based | DMSN-enabled signal development using CPT/DM-FA nanozyme | GSH | 5–80 μM 1 0.654 μM 2 | [114] | |
| PQQ-decorated MSNs for sandwich-type signal enhancer | PSA | 5–500 pg/mL 1 1 pg/mL 2 | [106] | ||
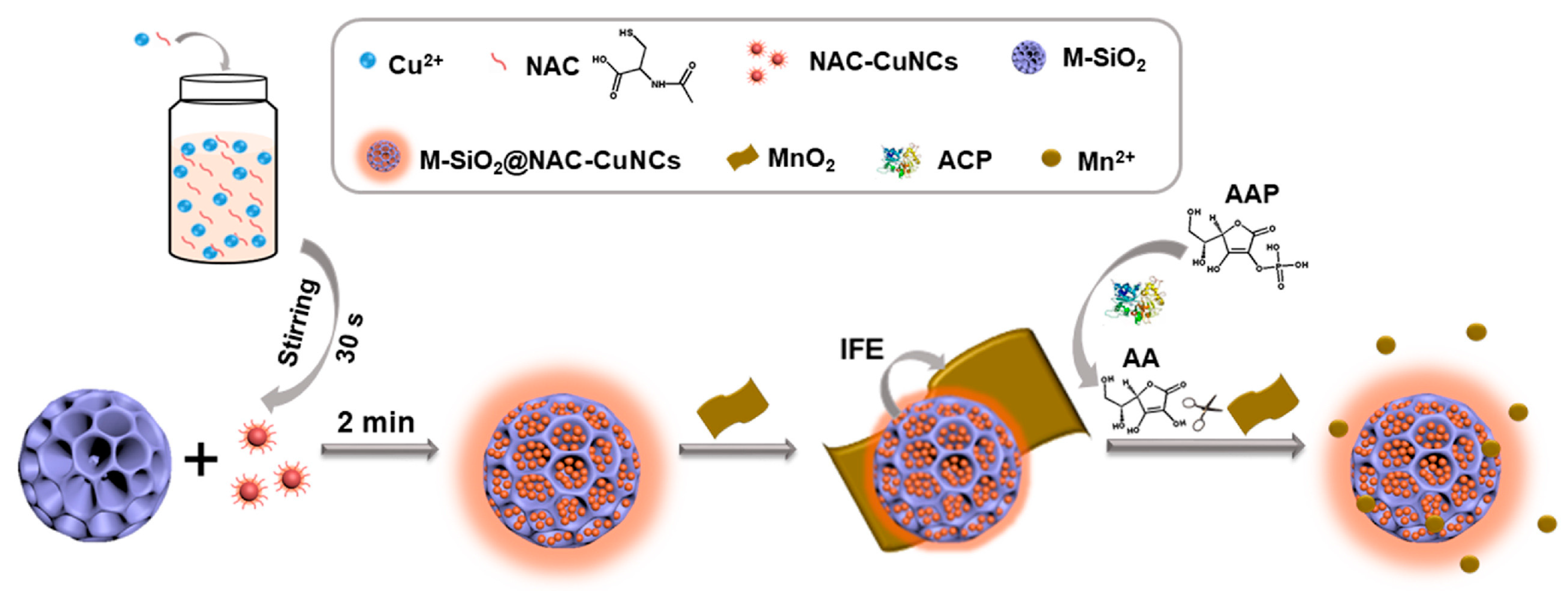
| Fluorescence | Inner filter effect | CuNC-loaded MSNs for improved fluorescence signal | ACP | 0.5–28 U/L 1 0.47 U/L 2 | [115] |
| Nanoreactor based on Cu-MOF-MSNs for signal enhancement | GSH | 0–0.1 mM 1 25 μM 2 | [116] | ||
| Release of Rh6G from MSNs upon ssDNA-AuNP cleaving by target | Flap endonuclease 1 | 0.05–1.75 U 1 0.03 U 2 | [117] | ||
| Hybridization-manipulated signal on Luc/CS/MSNs | let-7a (miRNA) | 30 fM–9 pM 1 10 fM 2 | [118] | ||
| Forster resonance energy transfer (FRET) | Aptamer-enabled signal on/off in MSN nanosystem with CS(cur)NPs and AuNPs | MUC-1 (CA 15-3) | - | [119] | |
| Aptamer-enabled signal development using QD@MSNs | PSA and CEA | PSA: 1 fg/mL–0.1 ng/mL 1; 0.9 fg/mL 2 CEA: 1 fg/mL–10 pg/mL 1; 0.7 fg/mL 2 | [120] | ||
| Lateral-flow immunoassay | Sandwich-type signal development using BDMSNs | CA 125 and HE4 | CA125: 0.1–1000 U/mL 1; 5 U/mL 2 HE4: 1–1000 pM 1; 5 pM 2 | [121] | |
| Chemiluminescence | Signal amplification by HRP-Ab1@MSNs | CEA | 10 pg/mL–20 ng/mL 1 3 pg/mL 2 | [122] | |
| Electrochemiluminescence | Signal enhancement by CS-Lu-modified SBMMs | SKBR-3 | 20–2000 cells/mL 1 20 cells/mL 2 | [37] | |
| DMSN-enabled signal development using CPT/DM-FA nanozyme | GSH | 10–250 μM 1 0.654 μM 2 | [114] | ||
| Controlled release of Ru(dcbpy)32+ from PBA-MSNs | MCF-7 | 3 × 102–105 cells 208 cells | [123] | ||
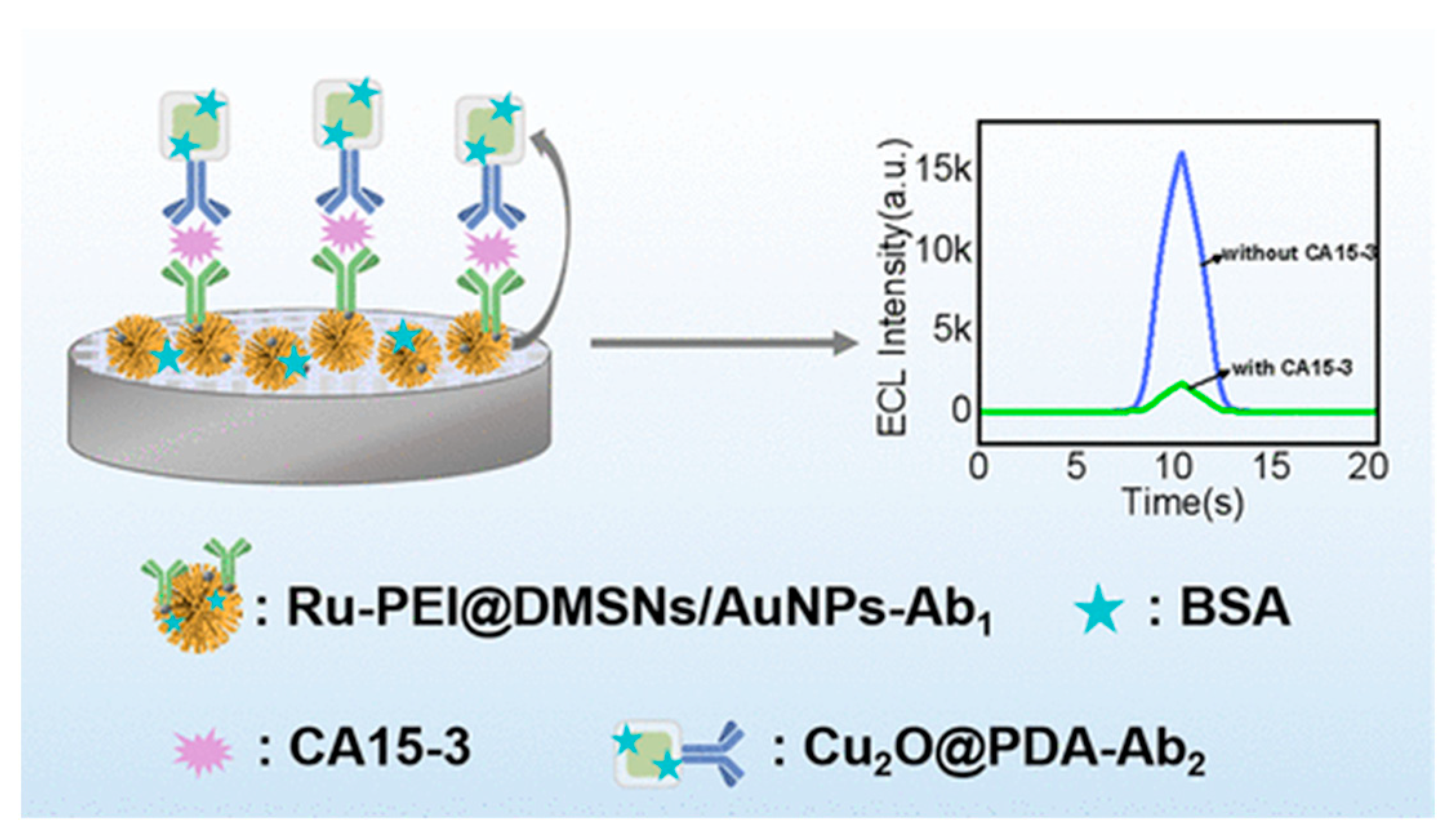
| Ru(dcbpy)32+-loaded MSNs with dual-quenching signal development | CA 15-3 | 5.0 × 10–5–6.0 × 102 U/mL 1 2.4 × 10–6 U/mL 2 | [124] | ||
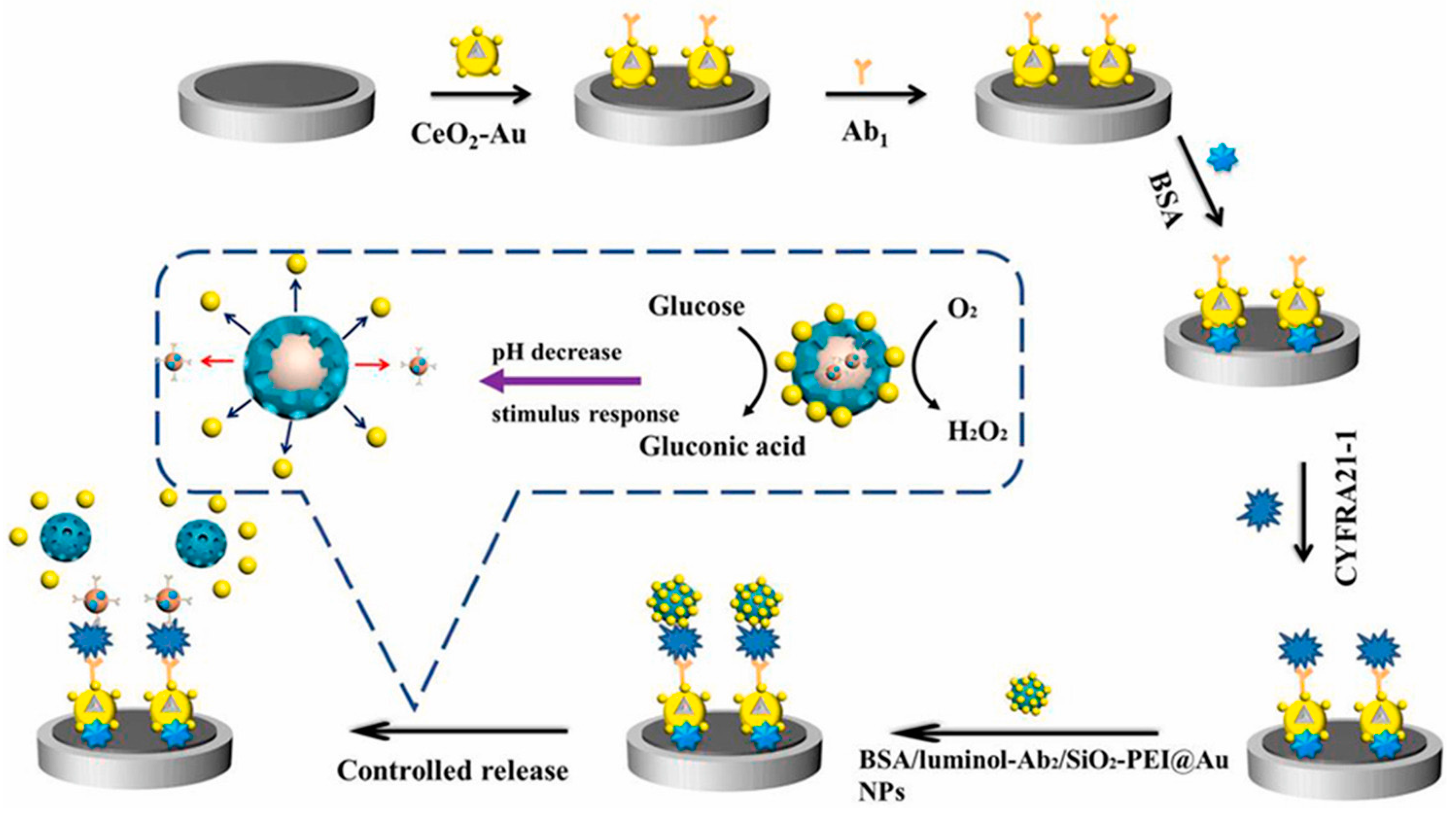
| Controlled release of luminol-Ab2 from MSN-PEI upon target binding and pH-stimuli response | CYFRA 21-1 | 1 fg/mL–100 ng/mL 1 0.4 fg/mL 2 | [125] | ||
| TPE-TEA-encapsulated MSNs for signal enhancement using DNA strand displacement strategy | MCF-7 cells | 10 pg/mL–100 ng/mL 1 | [126] | ||
| Surface plasmon resonance | Plasmonic energy resonance transfer | MSN-enabled Au nanocrescent antenna (MONA) | MCF-7 cancer cells | - | [127] |
| Other | UV-Vis spectrometry | DMSN-enabled signal development using CPT/DM-FA nanozyme | GSH | 2–60 μΜ 1 0.654 μM 2 | [114] |
| Surface-enhanced Raman spectroscopy | Target-enabled signal development by specific DNA release from MSNs | Methyltransferase | 0.1–10 U/mL 1 0.02 U/mL 2 | [128] |
| Properties | Benefits | Challenges | Applications Related to Sensing Cancer Biomarkers |
|---|---|---|---|
| High surface area | Surface functionalization with different molecules. | Controlling the amount and distribution of surface functional groups. | High amount of receptors for interaction with analytes or for attachment to sensing surfaces for optical or electrochemical detection with low LOD. |
| Porosity | Uniform distribution of pores with small diameter (2–3 nm), which can be used to load and entrap cargo molecules. | Optimization of porous structure to enhance the capacity for storing and entrapping molecules. | Loading signaling molecules (analytes) and their controlled release for optical or electrochemical sensing. |
| High stability | Facile formation of stable covalent linkages in reaction with organosilanes. Stability in testing media. | Achieving enhanced degradation for in vivo applications. Long-term stability in weakly alkaline media can present a challenge to achieving sensors for prolonged operation. | Formation of stable sensing surfaces for possible reusable detection. |
| Biocompatibility | Due to its biocompatibility, the use of silica is approved for cosmetics use. | Achieving approvement for in vivo diagnostics. | Possible construction of wearable biosensors. |
| Low costs | Highly scalable synthesis with cheap reactants and does not require high purity of chemicals. | The need for the use of expensive recognition elements in post-synthesis modification for specific and selective sensing. | Possible application for affordable POCT detection. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mladenović, M.; Jarić, S.; Mundžić, M.; Pavlović, A.; Bobrinetskiy, I.; Knežević, N.Ž. Biosensors for Cancer Biomarkers Based on Mesoporous Silica Nanoparticles. Biosensors 2024, 14, 326. https://doi.org/10.3390/bios14070326
Mladenović M, Jarić S, Mundžić M, Pavlović A, Bobrinetskiy I, Knežević NŽ. Biosensors for Cancer Biomarkers Based on Mesoporous Silica Nanoparticles. Biosensors. 2024; 14(7):326. https://doi.org/10.3390/bios14070326
Chicago/Turabian StyleMladenović, Minja, Stefan Jarić, Mirjana Mundžić, Aleksandra Pavlović, Ivan Bobrinetskiy, and Nikola Ž. Knežević. 2024. "Biosensors for Cancer Biomarkers Based on Mesoporous Silica Nanoparticles" Biosensors 14, no. 7: 326. https://doi.org/10.3390/bios14070326
APA StyleMladenović, M., Jarić, S., Mundžić, M., Pavlović, A., Bobrinetskiy, I., & Knežević, N. Ž. (2024). Biosensors for Cancer Biomarkers Based on Mesoporous Silica Nanoparticles. Biosensors, 14(7), 326. https://doi.org/10.3390/bios14070326






