Advances of 3D Cell Co-Culture Technology Based on Microfluidic Chips
Abstract
:1. Introduction
2. Design of Cell Co-Culture System On-Chip
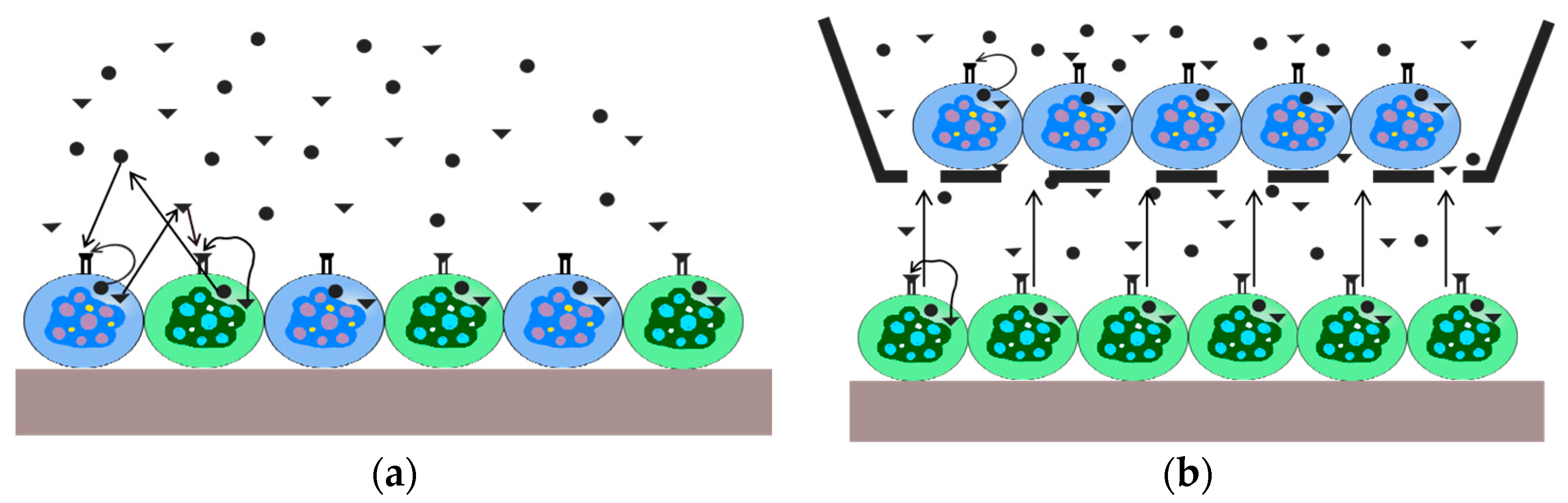
2.1. Direct Co-Culture of Cells
2.2. Indirect Co-Culture of Cells
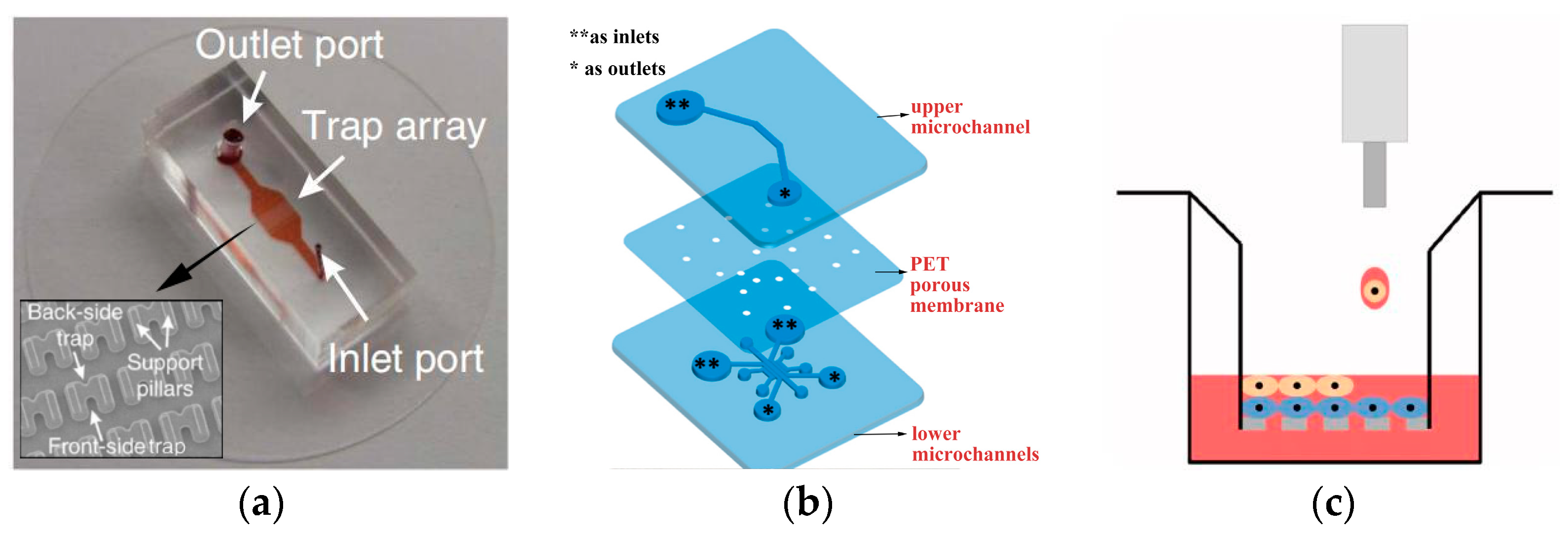
3. Detection of Cell Co-Culture System by Microfluidics
4. Application
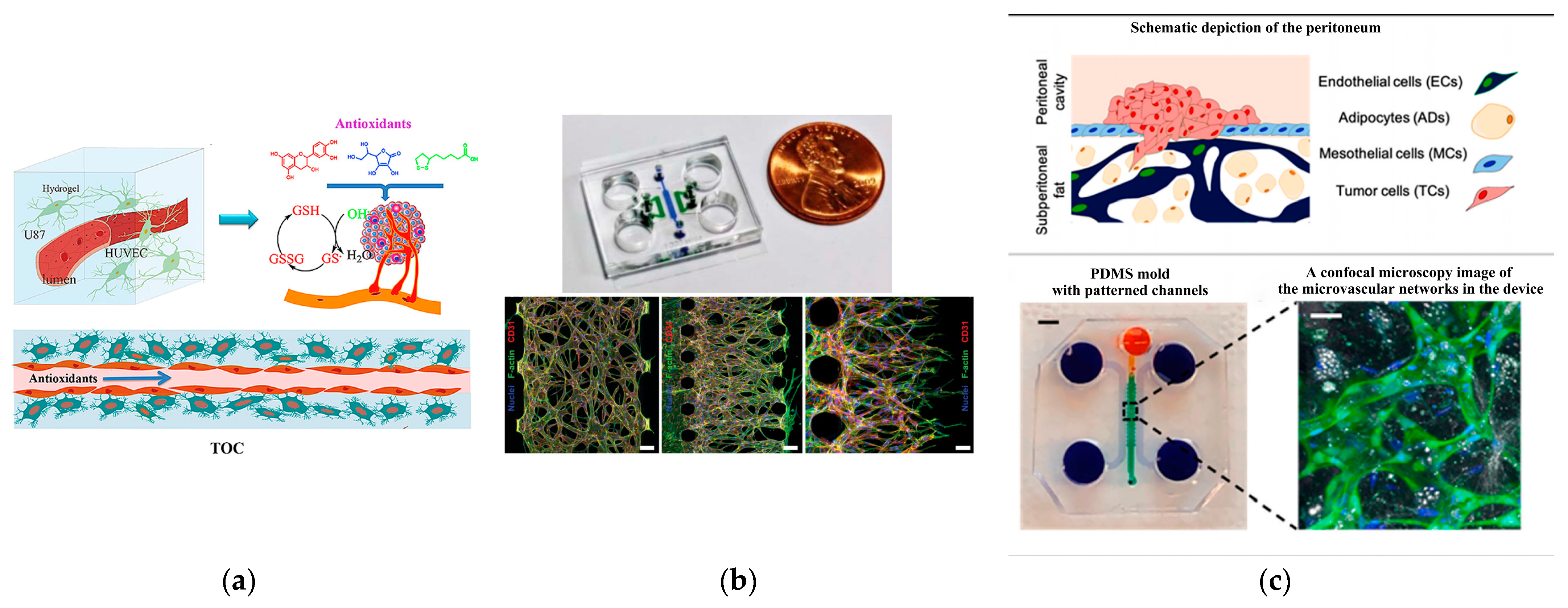
4.1. Angiogenesis
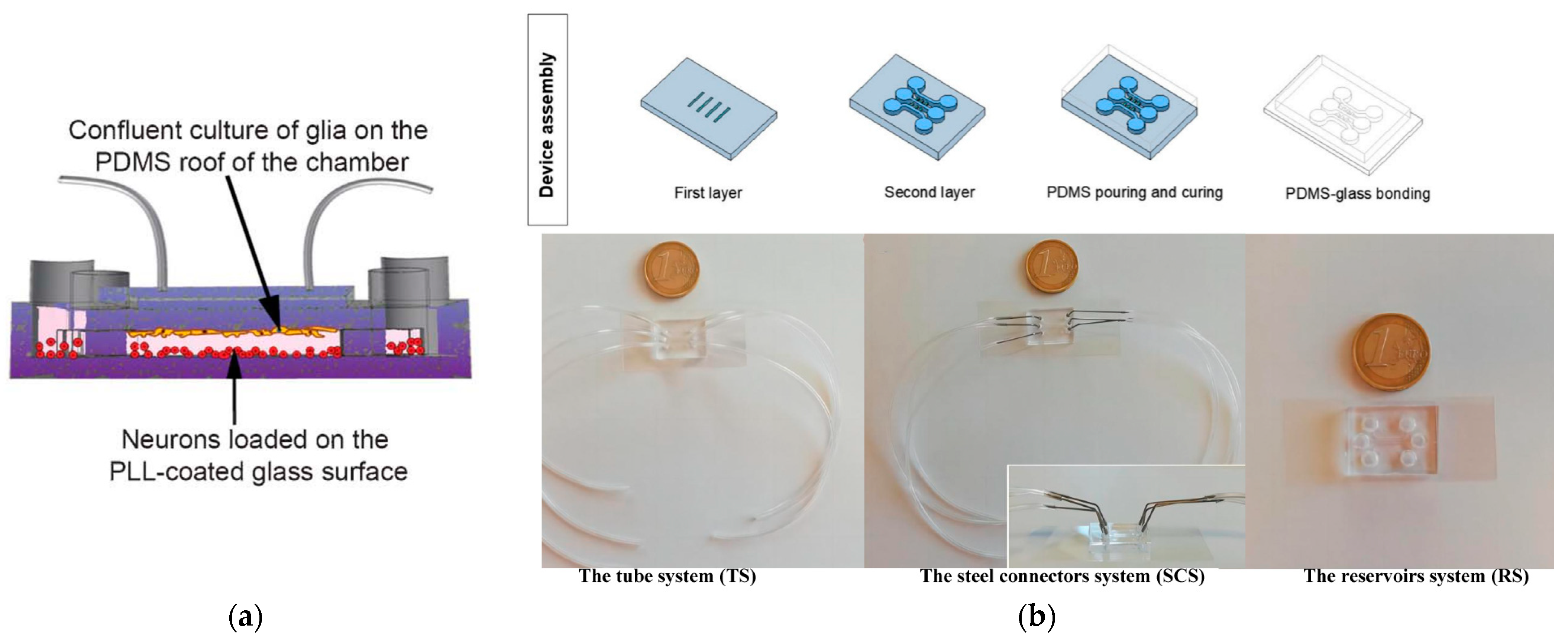
4.2. Blood–Brain Barrier
4.3. Blood–Gas Barrier
4.4. Other Organoid Chips
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Du, H.; Bartleson, J.M.; Butenko, S.; Alonso, V.; Liu, W.F.; Winer, D.A.; Butte, M.J. Tuning immunity through tissue mechanotransduction. Nat. Rev. Immunol. 2023, 23, 174–188. [Google Scholar] [CrossRef] [PubMed]
- Vu, T.Q.; de Castro, R.M.; Qin, L. Bridging the gap: Microfluidic devices for short and long distance cell-cell communication. Lab Chip 2017, 17, 1009–1023. [Google Scholar] [CrossRef] [PubMed]
- Vera, D.; García-Díaz, M.; Torras, N.; Álvarez, M.; Villa, R.; Martinez, E. Engineering Tissue Barrier Models on Hydrogel Microfluidic Platforms. ACS Appl. Mater. Interfaces 2021, 13, 13920–13933. [Google Scholar] [CrossRef]
- Yuan, J.; Li, X.; Yu, S. Cancer organoid co-culture model system: Novel approach to guide precision medicine. Front. Immunol. 2023, 13, 1061388. [Google Scholar] [CrossRef]
- Goshi, N.; Morgan, R.K.; Lein, P.J.; Seker, E. A primary neural cell culture model to study neuron, astrocyte, and microglia interactions in neuroinflammation. J. Neuroinflamm. 2020, 17, 155. [Google Scholar] [CrossRef] [PubMed]
- Arora, D.; Gupta, P.; Jaglan, S.; Roullier, C.; Grovel, O.; Bertrand, S. Expanding the chemical diversity through microorganisms co-culture: Current status and outlook. Biotechnol. Adv. 2020, 40, 107521. [Google Scholar] [CrossRef]
- Ribeiro-Filho, A.C.; Levy, D.; Ruiz, J.L.M.; Mantovani, M.D.C.; Bydlowski, S.P. Traditional and Advanced Cell Cultures in Hematopoietic Stem Cell Studies. Cells 2019, 8, 1628. [Google Scholar] [CrossRef]
- Yin, S.; Lu, R.; Liu, C.; Zhu, S.; Wan, H.; Lin, Y.; Wang, Q.; Qu, X.; Li, J. Composite Microfluidic Petri Dish-Chip (MPD-Chip) without Protein Coating for 2D Cell Culture. Langmuir 2023, 39, 15643–15652. [Google Scholar] [CrossRef]
- Ravi, M.; Paramesh, V.; Kaviya, S.R.; Anuradha, E.; Solomon, F.D. 3D cell culture systems: Advantages and applications. J. Cell. Physiol. 2015, 230, 16–26. [Google Scholar] [CrossRef]
- Edmondson, R.; Broglie, J.J.; Adcock, A.F.; Yang, L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev. Technol. 2014, 12, 207–218. [Google Scholar] [CrossRef]
- Guo, F.; French, J.B.; Li, P.; Zhao, H.; Chan, C.Y.; Fick, J.R.; Benkovic, S.J.; Huang, T.J. Probing cell-cell communication with microfluidic devices. Lab Chip 2013, 13, 3152–3162. [Google Scholar] [CrossRef] [PubMed]
- Nahavandi, S.; Tang, S.Y.; Baratchi, S.; Soffe, R.; Nahavandi, S.; Kalantar-zadeh, K.; Mitchell, A.; Khoshmanesh, K. Microfluidic platforms for the investigation of intercellular signaling mechanisms. Small 2014, 10, 4810–4826. [Google Scholar] [CrossRef] [PubMed]
- Kirsch, M.; Rach, J.; Handke, W.; Seltsam, A.; Pepelanova, I.; Strauß, S.; Vogt, P.; Scheper, T.; Lavrentieva, A. Comparative Analysis of Mesenchymal Stem Cell Cultivation in Fetal Calf Serum, Human Serum, and Platelet Lysate in 2D and 3D Systems. Front. Bioeng. Biotechnol. 2021, 8, 598389. [Google Scholar] [CrossRef]
- Sackmann, E.K.; Fulton, A.L.; Beebe, D.J. The present and future role of microfluidics in biomedical research. Nature 2014, 507, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Filippi, M.; Buchner, T.; Yasa, O.; Weirich, S.; Katzschmann, R.K. Microfluidic Tissue Engineering and Bio-Actuation. Adv. Mater. 2022, 34, 2108427. [Google Scholar] [CrossRef] [PubMed]
- Sart, S.; Ronteix, G.; Jain, S.; Amselem, G.; Baroud, C.N. Cell Culture in Microfluidic Droplets. Chem. Rev. 2022, 122, 7061–7096. [Google Scholar] [CrossRef] [PubMed]
- de Jongh, R.; Spijkers, X.M.; Pasteuning-Vuhman, S.; Vulto, P.; Pasterkamp, R.J. Neuromuscular junction-on-a-chip: ALS disease modeling and read-out development in microfluidic devices. J. Neurochem. 2021, 157, 393–412. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, A.; Sethumadhavan, A.; Krishnan, U.M. Toward Building the Neuromuscular Junction: In Vitro Models To Study Synaptogenesis and Neurodegeneration. ACS Omega 2019, 4, 12969–12977. [Google Scholar] [CrossRef] [PubMed]
- Habibey, R.; Rojo Arias, J.E.; Striebel, J.; Busskamp, V. Microfluidics for Neuronal Cell and Circuit Engineering. Chem. Rev. 2022, 122, 14842–14880. [Google Scholar] [CrossRef]
- Mehta, P.; Rahman, Z.; Ten Dijke, P.; Boukany, P.E. Microfluidics meets 3D cancer cell migration. Trends Cancer 2022, 8, 683–697. [Google Scholar] [CrossRef]
- Li, C.; He, W.; Wang, N.; Xi, Z.; Deng, R.; Liu, X.; Kang, R.; Xie, L.; Liu, X. Application of microfluidics in detection of circulating tumor cells. Front. Bioeng. Biotechnol. 2022, 10, 907232. [Google Scholar] [CrossRef] [PubMed]
- Limongi, T.; Guzzi, F.; Parrotta, E.; Candeloro, P.; Scalise, S.; Lucchino, V.; Gentile, F.; Tirinato, L.; Coluccio, M.L.; Torre, B.; et al. Microfluidics for 3D Cell and Tissue Cultures: Microfabricative and Ethical Aspects Updates. Cells 2022, 11, 1699. [Google Scholar] [CrossRef] [PubMed]
- Saorin, G.; Caligiuri, I.; Rizzolio, F. Microfluidic organoids-on-a-chip: The future of human models. Semin. Cell Dev. Biol. 2023, 144, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Knoška, J.; Adriano, L.; Awel, S.; Beyerlein, K.R.; Yefanov, O.; Oberthuer, D.; Murillo, G.E.P.; Roth, N.; Sarrou, I.; Villanueva-Perez, P.; et al. Ultracompact 3D microfluidics for time-resolved structural biology. Nat. Commun. 2020, 11, 657. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Yan, S.; Miao, C.; Li, L.; Shi, W.; Liu, X.; Luo, Y.; Liu, T.; Lin, B.; Wu, W.; et al. Paper Microfluidics for Cell Analysis. Adv. Healthc. Mater. 2019, 8, 1801084. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Meng, X.; Yu, X.; Wang, G.; Dong, Z.; Zhou, Z.; Qi, M.; Yu, X.; Ji, T.; Wang, F. From 2D to 3D Co-Culture Systems: A Review of Co-Culture Models to Study the Neural Cells Interaction. Int. J. Mol. Sci. 2022, 23, 13116. [Google Scholar] [CrossRef] [PubMed]
- Palikuqi, B.; Nguyen, D.T.; Li, G.; Schreiner, R.; Pellegata, A.F.; Liu, Y.; Redmond, D.; Geng, F.; Lin, Y.; Gómez-Salinero, J.M.; et al. Adaptable haemodynamic endothelial cells for organogenesis and tumorigenesis. Nature 2020, 585, 426–432. [Google Scholar] [CrossRef] [PubMed]
- Khoo, B.L.; Grenci, G.; Lim, Y.B.; Lee, S.C.; Han, J.; Lim, C.T. Expansion of patient-derived circulating tumor cells from liquid biopsies using a CTC microfluidic culture device. Nat. Protoc. 2018, 13, 34–58. [Google Scholar] [CrossRef] [PubMed]
- Park, S.E.; Georgescu, A.; Huh, D. Organoids-on-a-chip. Science 2019, 364, 960–965. [Google Scholar] [CrossRef]
- Hofer, M.; Lutolf, M.P. Engineering organoids. Nat. Rev. Mater. 2021, 6, 402–420. [Google Scholar] [CrossRef]
- Nashimoto, Y.; Okada, R.; Hanada, S.; Arima, Y.; Nishiyama, K.; Miura, T.; Yokokawa, R. Vascularized cancer on a chip: The effect of perfusion on growth and drug delivery of tumor spheroid. Biomaterials 2020, 229, 119547. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Mun, H.; Sung, C.O.; Cho, E.J.; Jeon, H.J.; Chun, S.M.; Jung, D.J.; Shin, T.H.; Jeong, G.S.; Kim, D.K.; et al. Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat. Commun. 2019, 10, 3991. [Google Scholar] [CrossRef] [PubMed]
- Schuster, B.; Junkin, M.; Kashaf, S.S.; Romero-Calvo, I.; Kirby, K.; Matthews, J.; Weber, C.R.; Rzhetsky, A.; White, K.P.; Tay, S. Automated microfluidic platform for dynamic and combinatorial drug screening of tumor organoids. Nat. Commun. 2020, 11, 5271–5283. [Google Scholar] [CrossRef] [PubMed]
- Youhanna, S.; Lauschke, V.M. The Past, Present and Future of Intestinal In Vitro Cell Systems for Drug Absorption Studies. J. Pharm. Sci. 2021, 110, 50–65. [Google Scholar] [CrossRef] [PubMed]
- Moarefian, M.; Davalos, R.V.; Tafti, D.K.; Achenie, L.E.; Jones, C.N. Modeling iontophoretic drug delivery in a microfluidic device. Lab Chip 2020, 20, 3310–3321. [Google Scholar] [CrossRef] [PubMed]
- Mark, D.; Haeberle, S.; Roth, G.; von Stetten, F.; Zengerle, R. Microfluidic lab-on-a-chip platforms: Requirements, characteristics and applications. Chem. Soc. Rev. 2010, 39, 1153–1182. [Google Scholar] [CrossRef] [PubMed]
- Amirifar, L.; Shamloo, A.; Nasiri, R.; de Barros, N.R.; Wang, Z.Z.; Unluturk, B.D.; Libanori, A.; Ievglevskyi, O.; Diltemiz, S.E.; Sances, S.; et al. Brain-on-a-chip: Recent advances in design and techniques for microfluidic models of the brain in health and disease. Biomaterials 2022, 285, 121531. [Google Scholar] [CrossRef] [PubMed]
- Adriani, G.; Ma, D.; Pavesi, A.; Kamm, R.D.; Goh, E.L. A 3D neurovascular microfluidic model consisting of neurons, astrocytes and cerebral endothelial cells as a blood-brain barrier. Lab Chip 2017, 17, 448–459. [Google Scholar] [CrossRef] [PubMed]
- Bang, S.; Lee, S.; Choi, N.; Kim, H.N. Emerging Brain-Pathophysiology-Mimetic Platforms for Studying Neurodegenerative Diseases: Brain Organoids and Brains-on-a-Chip. Adv. Healthc. Mater. 2021, 10, 2002119–2002145. [Google Scholar] [CrossRef]
- Noel, G.; Baetz, N.W.; Staab, J.F.; Donowitz, M.; Kovbasnjuk, O.; Pasetti, M.F.; Zachos, N.C. A primary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Sci. Rep. 2017, 7, 45270. [Google Scholar] [CrossRef]
- Rahman, S.; Ghiboub, M.; Donkers, J.M.; van de Steeg, E.; van Tol, E.A.F.; Hakvoort, T.B.M.; de Jonge, W.J. The Progress of Intestinal Epithelial Models from Cell Lines to Gut-On-Chip. Int. J. Mol. Sci. 2021, 22, 13472. [Google Scholar] [CrossRef] [PubMed]
- Borciani, G.; Montalbano, G.; Baldini, N.; Cerqueni, G.; Vitale-Brovarone, C.; Ciapetti, G. Co-culture systems of osteoblasts and osteoclasts: Simulating in vitro bone remodeling in regenerative approaches. Acta Biomater. 2020, 108, 22–45. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, F.H.; Sharma, P.; Gibbons, A.; Goggans, T.; Erzurum, S.C.; Haque, S.J. Human airway epithelial cell determinants of survival and functional phenotype for primary human mast cells. Proc. Natl. Acad. Sci. USA 2005, 102, 14380–14385. [Google Scholar] [CrossRef] [PubMed]
- Hou, A.; Hou, K.; Huang, Q.; Lei, Y.; Chen, W. Targeting Myeloid-Derived Suppressor Cell, a Promising Strategy to Overcome Resistance to Immune Checkpoint Inhibitors. Front. Immunol. 2020, 11, 783. [Google Scholar] [CrossRef]
- Shi, M.; Majumdar, D.; Gao, Y.; Brewer, B.M.; Goodwin, C.R.; McLean, J.A.; Li, D.; Webb, D.J. Glia co-culture with neurons in microfluidic platforms promotes the formation and stabilization of synaptic contacts. Lab Chip 2013, 13, 3008–3021. [Google Scholar] [CrossRef] [PubMed]
- Bayik, D.; Lathia, J.D. Cancer stem cell-immune cell crosstalk in tumour progression. Nat. Rev. Cancer 2021, 21, 526–536. [Google Scholar] [CrossRef] [PubMed]
- Dura, B.; Dougan, S.K.; Barisa, M.; Hoehl, M.M.; Lo, C.T.; Ploegh, H.L.; Voldman, J. Profiling lymphocyte interactions at the single-cell level by microfluidic cell pairing. Nat. Commun. 2015, 6, 5940. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, B.; Wang, L.; Peng, J.; Wu, K.; Liu, T. The development of droplet-based microfluidic virus detection technology for human infectious diseases. Anal. Methods 2024, 16, 971–978. [Google Scholar] [CrossRef]
- Deng, Y.; Guo, Y.; Xu, B. Recent development of microfluidic technology for cell trapping in single cell analysis: A review. Processes 2020, 8, 1253. [Google Scholar] [CrossRef]
- Zhang, P.; Liu, C.; Modavi, C.; Abate, A.; Chen, H. Printhead on a chip: Empowering droplet-based bioprinting with microfluidics. Trends Biotechnol. 2023, 42, 353–388. [Google Scholar] [CrossRef]
- Chen, L.J.; Raut, B.; Nagai, N.; Abe, T.; Kaji, H. Prototyping a Versatile Two-Layer Multi-Channel Microfluidic Device for Direct-Contact Cell-Vessel Co-Culture. Micromachines 2020, 11, 79. [Google Scholar] [CrossRef]
- Dudman, J.; Ferreira, A.M.; Gentile, P.; Wang, X.; Dalgarno, K. Microvalve Bioprinting of MSC-Chondrocyte Co-Cultures. Cells 2021, 10, 3329. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, S.I.; Bogetofte, H.; Ritter, L.; Agergaard, J.B.; Hammerich, D.; Kabiljagic, A.A.; Wlodarczyk, A.; Lopez, S.G.; Sørensen, M.D.; Jørgensen, M.L.; et al. Microglia-Secreted Factors Enhance Dopaminergic Differentiation of Tissue- and iPSC-Derived Human Neural Stem Cells. Stem Cell Rep. 2021, 16, 281–294. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, L.; Long, Z.; Zeng, L.; Wu, Y. Protoplasmic astrocytes enhance the ability of neural stem cells to differentiate into neurons in vitro. PLoS ONE 2012, 7, e38243. [Google Scholar] [CrossRef] [PubMed]
- Kempuraj, D.; Khan, M.M.; Thangavel, R.; Xiong, Z.; Yang, E.; Zaheer, A. Glia maturation factor induces interleukin-33 release from astrocytes: Implications for neurodegenerative diseases. J. Neuroimmune Pharmacol. 2013, 8, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Saadeldin, I.M.; Abdelfattah-Hassan, A.; Swelum, A.A. Feeder Cell Type Affects the Growth of In Vitro Cultured Bovine Trophoblast Cells. Biomed. Res. Int. 2017, 1061589. [Google Scholar] [CrossRef]
- Le-Bel, G.; Cortez Ghio, S.; Guérin, L.P.; Bisson, F.; Germain, L.; Guérin, S.L. Irradiated Human Fibroblasts as a Substitute Feeder Layer to Irradiated Mouse 3T3 for the Culture of Human Corneal Epithelial Cells: Impact on the Stability of the Transcription Factors Sp1 and NFI. Int. J. Mol. Sci. 2019, 20, 6296. [Google Scholar] [CrossRef] [PubMed]
- Trettner, S.; Findeisen, A.; Taube, S.; Horn, P.A.; Sasaki, E.; zur Nieden, N.I. Osteogenic induction from marmoset embryonic stem cells cultured in feeder-dependent and feeder-independent conditions. Osteoporos. Int. 2014, 25, 1255–1266. [Google Scholar] [CrossRef] [PubMed]
- López-Fagundo, C.; Livi, L.L.; Ramchal, T.; Darling, E.M.; Hoffman-Kim, D. A biomimetic synthetic feeder layer supports the proliferation and self-renewal of mouse embryonic stem cells. Acta Biomater. 2016, 39, 55–64. [Google Scholar] [CrossRef]
- Li, R.; Zhang, X.; Lv, X.; Geng, L.; Li, Y.; Qin, K.; Deng, Y. Microvalve controlled multi-functional microfluidic chip for divisional cell co-culture. Anal. Biochem. 2017, 539, 48–53. [Google Scholar] [CrossRef]
- Yang, K.; Park, H.J.; Han, S.; Lee, J.; Ko, E.; Kim, J.; Lee, J.S.; Yu, J.H.; Song, K.Y.; Cheong, E.; et al. Recapitulation of in vivo-like paracrine signals of human mesenchymal stem cells for functional neuronal differentiation of human neural stem cells in a 3D microfluidic system. Biomaterials 2015, 63, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Cui, F.; Chen, H.; Yang, Y.; Li, G.; Mao, H.; Lyu, X. A Microfluidic Cell Co-Culture Chip for the Monitoring of Interactions between Macrophages and Fibroblasts. Biosensors 2022, 13, 70. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Cai, B.; Wei, X.; Wang, Z.; Rao, L.; Meng, Q.F.; Liao, Q.; Liu, W.; Guo, S.; Zhao, X. A valve-based microfluidic device for on-chip single cell treatments. Electrophoresis 2019, 40, 961–968. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Wetzel, I.; Marriott, I.; Dréau, D.; D’Avanzo, C.; Kim, D.Y.; Tanzi, R.E.; Cho, H. A 3D human triculture system modeling neurodegeneration and neuroinflammation in Alzheimer’s disease. Nat. Neurosci. 2018, 21, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Sakthivel, K.; O’Brien, A.; Kim, K.; Hoorfar, M. Microfluidic analysis of heterotypic cellular interactions: A review of techniques and applications. Trends Anal. Chem. 2019, 117, 166–185. [Google Scholar] [CrossRef]
- Zahavi, E.E.; Ionescu, A.; Gluska, S.; Gradus, T.; Ben-Yaakov, K.; Perlson, E. A compartmentalized microfluidic neuromuscular co-culture system reveals spatial aspects of GDNF functions. J. Cell Sci. 2015, 128, 1241–1252. [Google Scholar] [PubMed]
- Chung, H.H.; Mireles, M.; Kwarta, B.J.; Gaborski, T.R. Use of porous membranes in tissue barrier and co-culture models. Lab Chip 2018, 18, 1671–1689. [Google Scholar] [CrossRef]
- De Vitis, E.; La Pesa, V.; Gervaso, F.; Romano, A.; Quattrini, A.; Gigli, G.; Moroni, L.; Polini, A. A microfabricated multi-compartment device for neuron and Schwann cell differentiation. Sci. Rep. 2021, 11, 7019–7030. [Google Scholar] [CrossRef] [PubMed]
- Son, M.; Wang, A.G.; Kenna, E.; Tay, S. High-throughput co-culture system for analysis of spatiotemporal cell-cell signaling. Biosens. Bioelectron. 2023, 225, 115089–115098. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Chen, W.; Deng, S.; Xie, L.; Feng, J.; Geng, J.; Jiang, D.; Dai, H.; Wang, C. Modeling alveolar injury using microfluidic co-cultures for monitoring bleomycin-induced epithelial/fibroblastic cross-talk disorder. RSC Adv. 2017, 7, 42738–42749. [Google Scholar] [CrossRef]
- Saygili, E.; Yildiz-Ozturk, E.; Green, M.J.; Ghaemmaghami, A.M.; Yesil-Celiktas, O. Human lung-on-chips: Advanced systems for respiratory virus models and assessment of immune response. Biomicrofluidics 2021, 15, 021501. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, Y.; Chen, P.; Su, H.; Du, W.; Liu, B.F. Highly efficient microfluidic device for cell trapping and pairing towards cell-cell communication analysis. Sens. Actuators B Chem. 2019, 283, 685–692. [Google Scholar] [CrossRef]
- Liu, Y.; Fan, Z.; Qiao, L.; Liu, B. Advances in microfluidic strategies for single-cell research. Trends Analyt. Chem. 2022, 157, 116822. [Google Scholar] [CrossRef]
- Anggraini, D.; Ota, N.; Shen, Y.; Tang, T.; Tanaka, Y.; Hosokawa, Y.; Li, M.; Yalikun, Y. Recent advances in microfluidic devices for single-cell cultivation: Methods and applications. Lab Chip 2022, 22, 1438–1468. [Google Scholar] [CrossRef]
- Jie, M.; Li, H.F.; Lin, L.; Zhang, J.; Lin, J.M. Integrated microfluidic system for cell co-culture and simulation of drug metabolism. RSC Adv. 2016, 6, 54564–54572. [Google Scholar] [CrossRef]
- Vivas, A.; IJspeert, C.; Pan, J.Y.; Vermeul, K.; van den Berg, A.; Passier, R.; Keller, S.S.; van der Meer, A.D. Generation and Culture of Cardiac Microtissues in a Microfluidic Chip with a Reversible Open Top Enables Electrical Pacing, Dynamic Drug Dosing and Endothelial Cell Co-Culture. Adv. Mater. Technol. 2022, 7, 2101355. [Google Scholar] [CrossRef]
- Yoo, J.; Jung, Y.; Char, K.; Jang, Y. Advances in cell coculture membranes recapitulating in vivo microenvironments. Trends Biotechnol. 2023, 41, 214–227. [Google Scholar] [CrossRef]
- Liu, Y.; Kongsuphol, P.; Chiam, S.Y.; Zhang, Q.X.; Gourikutty, S.B.N.; Saha, S.; Biswas, S.K.; Ramadan, Q. Adipose-on-a-chip: A dynamic microphysiological in vitro model of the human adipose for immune-metabolic analysis in type II diabetes. Lab Chip 2019, 19, 241–253. [Google Scholar] [CrossRef]
- Liu, H.; Jie, M.; He, Z.; Li, H.F.; Lin, J.M. Study of antioxidant effects on malignant glioma cells by constructing a tumor-microvascular structure on microchip. Anal. Chim. Acta 2017, 978, 1–9. [Google Scholar] [CrossRef]
- Sato, K.; Kikuchi, S.; Yoshida, E.; Ishii, R.; Sasaki, N.; Tsunoda, K.; Sato, K. Patterned Co-culture of Live Cells on a Microchip by Photocrosslinking with Benzophenone. Anal. Sci. 2016, 32, 113–116. [Google Scholar] [CrossRef]
- Sun, W.; Chen, Y.; Wang, Y.; Luo, P.; Zhang, M.; Zhang, H.; Hu, P. Interaction study of cancer cells and fibroblasts on a spatially confined oxygen gradient microfluidic chip to investigate the tumor microenvironment. Analyst 2018, 143, 5431–5437. [Google Scholar] [CrossRef]
- Chen, H.J.; Miller, P.; Shuler, M.L. A pumpless body-on-a-chip model using a primary culture of human intestinal cells and a 3D culture of liver cells. Lab Chip 2018, 18, 2036–2046. [Google Scholar] [CrossRef]
- Hattersley, S.M.; Sylvester, D.C.; Dyer, C.E.; Stafford, N.D.; Haswell, S.J.; Greenman, J. A microfluidic system for testing the responses of head and neck squamous cell carcinoma tissue biopsies to treatment with chemotherapy drugs. Ann. Biomed. Eng. 2012, 40, 1277–1288. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, S.; Fan, F.; Xu, N.; Meng, X.L.; Zhang, Y.; Lin, J.M. Neurotoxicity mechanism of aconitine in HT22 cells studied by microfluidic chip-mass spectrometry. J. Pharm. Anal. 2023, 13, 88–98. [Google Scholar] [CrossRef]
- Jeong, S.; Kim, S.; Buonocore, J.; Park, J.; Welsh, C.J.; Li, J.; Han, A. A Three-Dimensional Arrayed Microfluidic Blood-Brain Barrier Model With Integrated Electrical Sensor Array. IEEE Trans. Biomed. Eng. 2018, 65, 431–439. [Google Scholar] [CrossRef]
- Yeste, J.; García-Ramírez, M.; Illa, X.; Guimerà, A.; Hernández, C.; Simó, R.; Villa, R. A compartmentalized microfluidic chip with crisscross microgrooves and electrophysiological electrodes for modeling the blood-retinal barrier. Lab Chip 2017, 18, 95–105. [Google Scholar] [CrossRef]
- Esch, M.B.; Ueno, H.; Applegate, D.R.; Shuler, M.L. Modular, pumpless body-on-a-chip platform for the co-culture of GI tract epithelium and 3D primary liver tissue. Lab Chip 2016, 16, 2719–2729. [Google Scholar] [CrossRef]
- Li, X.; Fan, B.; Cao, S.; Chen, D.; Zhao, X.; Men, D.; Yue, W.; Wang, J.; Chen, J. A microfluidic flow cytometer enabling absolute quantification of single-cell intracellular proteins. Lab Chip 2017, 17, 3129–3137. [Google Scholar] [CrossRef]
- Lee, J.M.; Seo, H.I.; Bae, J.H.; Chung, B.G. Hydrogel microfluidic co-culture device for photothermal therapy and cancer migration. Electrophoresis 2017, 38, 1318–1324. [Google Scholar] [CrossRef]
- Wang, X.; Li, X.; Ding, J.; Long, X.; Zhang, H.; Zhang, X.; Jiang, X.; Xu, T. 3D bioprinted glioma microenvironment for glioma vascularization. J. Biomed. Mater. Res. A 2021, 109, 915–925. [Google Scholar] [CrossRef]
- Ko, J.; Ahn, J.; Kim, S.; Lee, Y.; Lee, J.; Park, D.; Jeon, N.L. Tumor spheroid-on-a-chip: A standardized microfluidic culture platform for investigating tumor angiogenesis. Lab Chip 2019, 19, 2822–2833. [Google Scholar] [CrossRef]
- Moshksayan, K.; Kashaninejad, N.; Warkiani, M.E.; Lock, J.G.; Moghadas, H.; Firoozabadi, B.; Saidi, M.S.; Nguyen, N.-T. Spheroids-on-a-chip: Recent advances and design considerations in microfluidic platforms for spheroid formation and culture. Sens. Actuators B Chem. 2018, 263, 151–176. [Google Scholar] [CrossRef]
- Ma, Y.; Pan, J.Z.; Zhao, S.P.; Lou, Q.; Zhu, Y.; Fang, Q. Microdroplet chain array for cell migration assays. Lab Chip 2016, 16, 4658–4665. [Google Scholar] [CrossRef]
- Lin, L.; He, Z.; Jie, M.; Lin, J.M.; Zhang, J. 3D microfluidic tumor models for biomimetic engineering of glioma niche and detection of cell morphology, migration and phenotype change. Talanta 2021, 234, 122702. [Google Scholar] [CrossRef]
- Song, H.; Wang, Y.; Rosano, J.M.; Prabhakarpandian, B.; Garson, C.; Pant, K.; Lai, E. A microfluidic impedance flow cytometer for identification of differentiation state of stem cells. Lab Chip 2013, 13, 2300–2310. [Google Scholar] [CrossRef]
- Tan, H.Y.; Trier, S.; Rahbek, U.L.; Dufva, M.; Kutter, J.P.; Andresen, T.L. A multi-chamber microfluidic intestinal barrier model using Caco-2 cells for drug transport studies. PLoS ONE 2018, 13, e0197101. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, X.; Chen, J.; Luo, Y.; Chen, Z.; Lin, D.; Zhang, J.; Liu, D. Gel-Free Single-Cell Culture Arrays on a Microfluidic Chip for Highly Efficient Expansion and Recovery of Colon Cancer Stem Cells. ACS Biomater. Sci. Eng. 2022, 8, 3623–3632. [Google Scholar] [CrossRef]
- Tehranirokh, M.; Kouzani, A.Z.; Francis, P.S.; Kanwar, J.R. Microfluidic devices for cell cultivation and proliferation. Biomicrofluidics 2013, 7, 051502. [Google Scholar] [CrossRef]
- Sacchi, M.; Bansal, R.; Rouwkema, J. Bioengineered 3D Models to Recapitulate Tissue Fibrosis. Trends Biotechnol. 2020, 38, 623–636. [Google Scholar] [CrossRef]
- van Grunsven, L.A. 3D in vitro models of liver fibrosis. Adv. Drug Deliv. Rev. 2017, 121, 133–146. [Google Scholar] [CrossRef]
- Jang, K.J.; Otieno, M.A.; Ronxhi, J.; Lim, H.K.; Ewart, L.; Kodella, K.R.; Petropolis, D.B.; Kulkarni, G.; Rubins, J.E.; Conegliano, D.; et al. Reproducing human and cross-species drug toxicities using a Liver-Chip. Sci. Transl. Med. 2019, 11, eaax5516. [Google Scholar] [CrossRef]
- Materne, E.M.; Ramme, A.P.; Terrasso, A.P.; Serra, M.; Alves, P.M.; Brito, C.; Sakharov, D.A.; Tonevitsky, A.G.; Lauster, R.; Marx, U. A multi-organ chip co-culture of neurospheres and liver equivalents for long-term substance testing. J. Biotechnol. 2015, 205, 36–46. [Google Scholar] [CrossRef]
- Caruso, G.; Musso, N.; Grasso, M.; Costantino, A.; Lazzarino, G.; Tascedda, F.; Gulisano, M.; Lunte, S.M.; Caraci, F. Microfluidics as a Novel Tool for Biological and Toxicological Assays in Drug Discovery Processes: Focus on Microchip Electrophoresis. Micromachines 2020, 11, 593. [Google Scholar] [CrossRef]
- Jeong, S.Y.; Lee, J.H.; Shin, Y.; Chung, S.; Kuh, H.J. Co-Culture of Tumor Spheroids and Fibroblasts in a Collagen Matrix-Incorporated Microfluidic Chip Mimics Reciprocal Activation in Solid Tumor Microenvironment. PLoS ONE 2016, 11, e0159013. [Google Scholar] [CrossRef]
- Ibrahim, L.I.; Hajal, C.; Offeddu, G.S.; Gillrie, M.R.; Kamm, R.D. Omentum-on-a-chip: A multicellular, vascularized microfluidic model of the human peritoneum for the study of ovarian cancer metastases. Biomaterials 2022, 288, 121728. [Google Scholar] [CrossRef]
- Oh, S.; Ryu, H.; Tahk, D.; Ko, J.; Chung, Y.; Lee, H.K.; Jeon, N.L. “Open-top” microfluidic device for in vitro three-dimensional capillary beds. Lab Chip 2017, 17, 3405–3414. [Google Scholar] [CrossRef]
- Wang, S.; Mao, S.; Li, M.; Li, H.F.; Lin, J.M. Near-physiological microenvironment simulation on chip to evaluate drug resistance of different loci in tumour mass. Talanta 2019, 191, 67–73. [Google Scholar] [CrossRef]
- Guimarães, C.F.; Cruz-Moreira, D.; Caballero, D.; Pirraco, R.P.; Gasperini, L.; Kundu, S.C.; Reis, R.L. Shining a Light on Cancer-Photonics in Microfluidic Tumor Modeling and Biosensing. Adv. Healthc. Mater. 2023, 12, e2201442. [Google Scholar] [CrossRef]
- Kim, S.; Lee, H.; Chung, M.; Jeon, N.L. Engineering of functional, perfusable 3D microvascular networks on a chip. Lab Chip 2013, 13, 1489–1500. [Google Scholar] [CrossRef]
- Liang, Y.; Yoon, J.Y. In situ sensors for blood-brain barrier (BBB) on a chip. Sens. Actuators Rep. 2021, 3, 100031. [Google Scholar] [CrossRef]
- Benz, F.; Liebner, S. Structure and Function of the Blood–Brain Barrier (BBB). Handb. Exp. Pharmacol. 2020, 273, 3–31. [Google Scholar]
- Park, J.; Koito, H.; Li, J.; Han, A. Microfluidic compartmentalized co-culture platform for CNS axon myelination research. Biomed. Microdevices 2009, 11, 1145–1153. [Google Scholar] [CrossRef] [PubMed]
- Dinh, N.D.; Chiang, Y.Y.; Hardelauf, H.; Baumann, J.; Jackson, E.; Waide, S.; Sisnaiske, J.; Frimat, J.-P.; van Thriel, C.; Janasek, D.; et al. Microfluidic construction of minimalistic neuronal co-cultures. Lab Chip 2013, 13, 1402–1412. [Google Scholar] [CrossRef] [PubMed]
- Booth, R.; Kim, H. Characterization of a microfluidic in vitro model of the blood-brain barrier (μBBB). Lab Chip 2012, 12, 1784–1792. [Google Scholar] [CrossRef] [PubMed]
- Leiby, K.L.; Raredon, M.S.B.; Niklason, L.E. Bioengineering the Blood-gas Barrier. Compr. Physiol. 2020, 10, 415–452. [Google Scholar] [PubMed]
- Sellgren, K.L.; Butala, E.J.; Gilmour, B.P.; Randell, S.H.; Grego, S. A biomimetic multicellular model of the airways using primary human cells. Lab Chip 2014, 14, 3349–3358. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting organ-level lung functions on a chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef] [PubMed]
- Benam, K.H.; Villenave, R.; Lucchesi, C.; Varone, A.; Hubeau, C.; Lee, H.H.; E Alves, S.; Salmon, M.; Ferrante, T.C.; Weaver, J.C.; et al. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat. Methods 2016, 13, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Wiedenmann, S.; Breunig, M.; Merkle, J.; von Toerne, C.; Georgiev, T.; Moussus, M.; Schulte, L.; Seufferlein, T.; Sterr, M.; Lickert, H.; et al. Single-cell-resolved differentiation of human induced pluripotent stem cells into pancreatic duct-like organoids on a microwell chip. Nat. Biomed. Eng. 2021, 5, 897–913. [Google Scholar] [CrossRef]
- Kim, S.K.; Kim, Y.H.; Park, S.; Cho, S.W. Organoid engineering with microfluidics and biomaterials for liver, lung disease, and cancer modeling. Acta Biomater. 2021, 132, 37–51. [Google Scholar] [CrossRef]
- Sin, A.; Chin, K.C.; Jamil, M.F.; Kostov, Y.; Rao, G.; Shuler, M.L. The design and fabrication of three-chamber microscale cell culture analog devices with integrated dissolved oxygen sensors. Biotechnol. Prog. 2004, 20, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, E.; Kaazempur-Mofrad, M.; Borenstein, J. Concept and computational design for a bioartificial nephron-on-a-chip. Int. J. Artif. Organs 2008, 31, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.A.; Kang, E.; Ju, J.; Kim, D.S.; Lee, S.H. Spheroid-based three-dimensional liver-on-a-chip to investigate hepatocyte-hepatic stellate cell interactions and flow effects. Lab Chip 2013, 13, 3529–3537. [Google Scholar] [CrossRef] [PubMed]
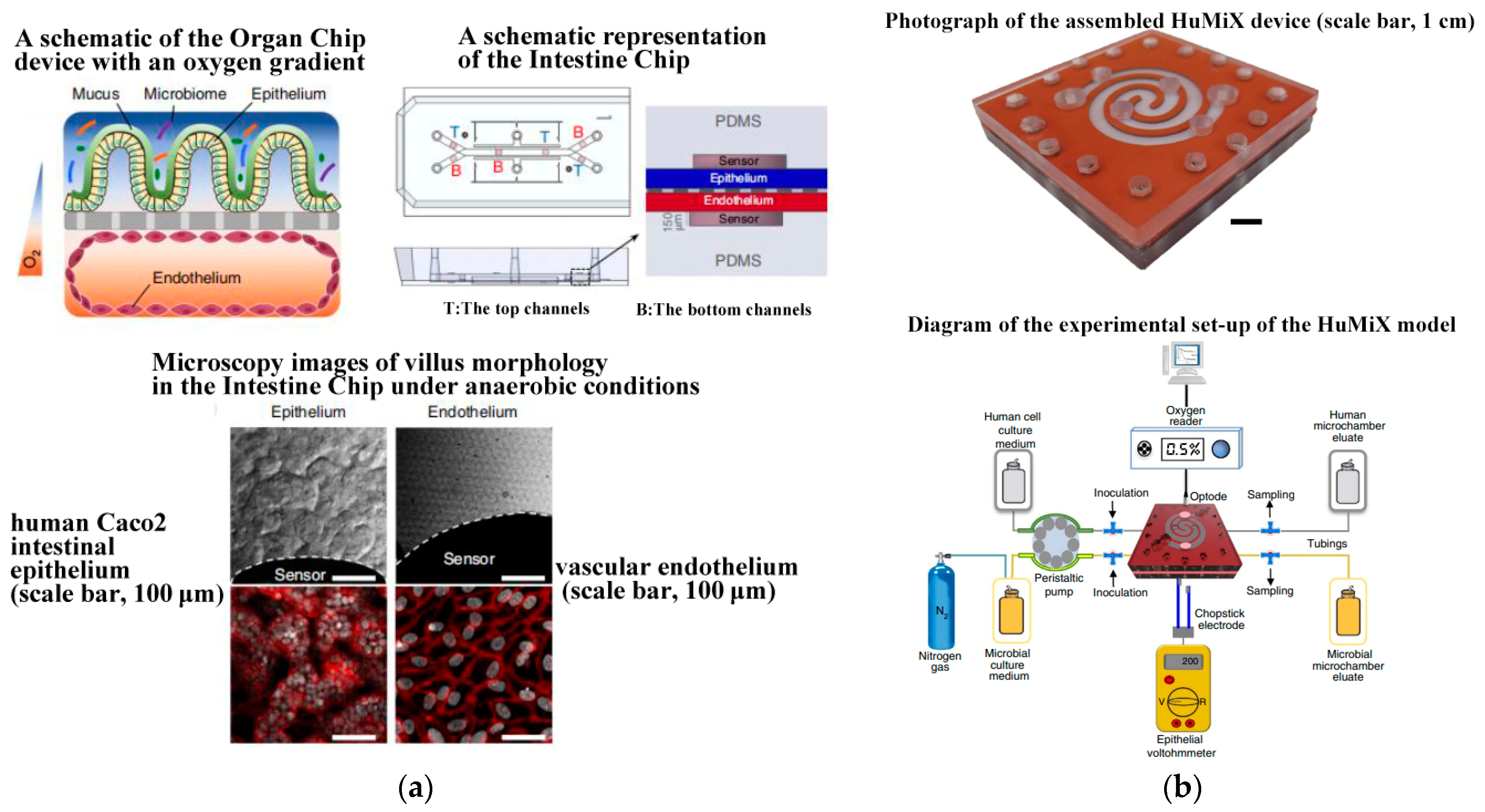
- Jalili-Firoozinezhad, S.; Gazzaniga, F.S.; Calamari, E.L.; Camacho, D.M.; Fadel, C.W.; Bein, A.; Swenor, B.; Nestor, B.; Cronce, M.J.; Tovaglieri, A.; et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng. 2019, 3, 520–531. [Google Scholar] [CrossRef]
- Puschhof, J.; Pleguezuelos-Manzano, C.; Clevers, H. Organoids and organs-on-chips: Insights into human gut-microbe interactions. Cell Host Microbe 2021, 29, 867–878. [Google Scholar] [CrossRef] [PubMed]
- Glaser, D.E.; Curtis, M.B.; Sariano, P.A.; Rollins, Z.A.; Shergill, B.S.; Anand, A.; Deely, A.M.; Shirure, V.S.; Anderson, L.; Lowen, J.M.; et al. Organ-on-a-chip model of vascularized human bone marrow niches. Biomaterials 2022, 280, 121245–121289. [Google Scholar] [CrossRef] [PubMed]
- Jung, D.J.; Shin, T.H.; Kim, M.; Sung, C.O.; Jang, S.J.; Jeong, G.S. A one-stop microfluidic-based lung cancer organoid culture platform for testing drug sensitivity. Lab Chip 2019, 19, 2854–2865. [Google Scholar] [CrossRef] [PubMed]
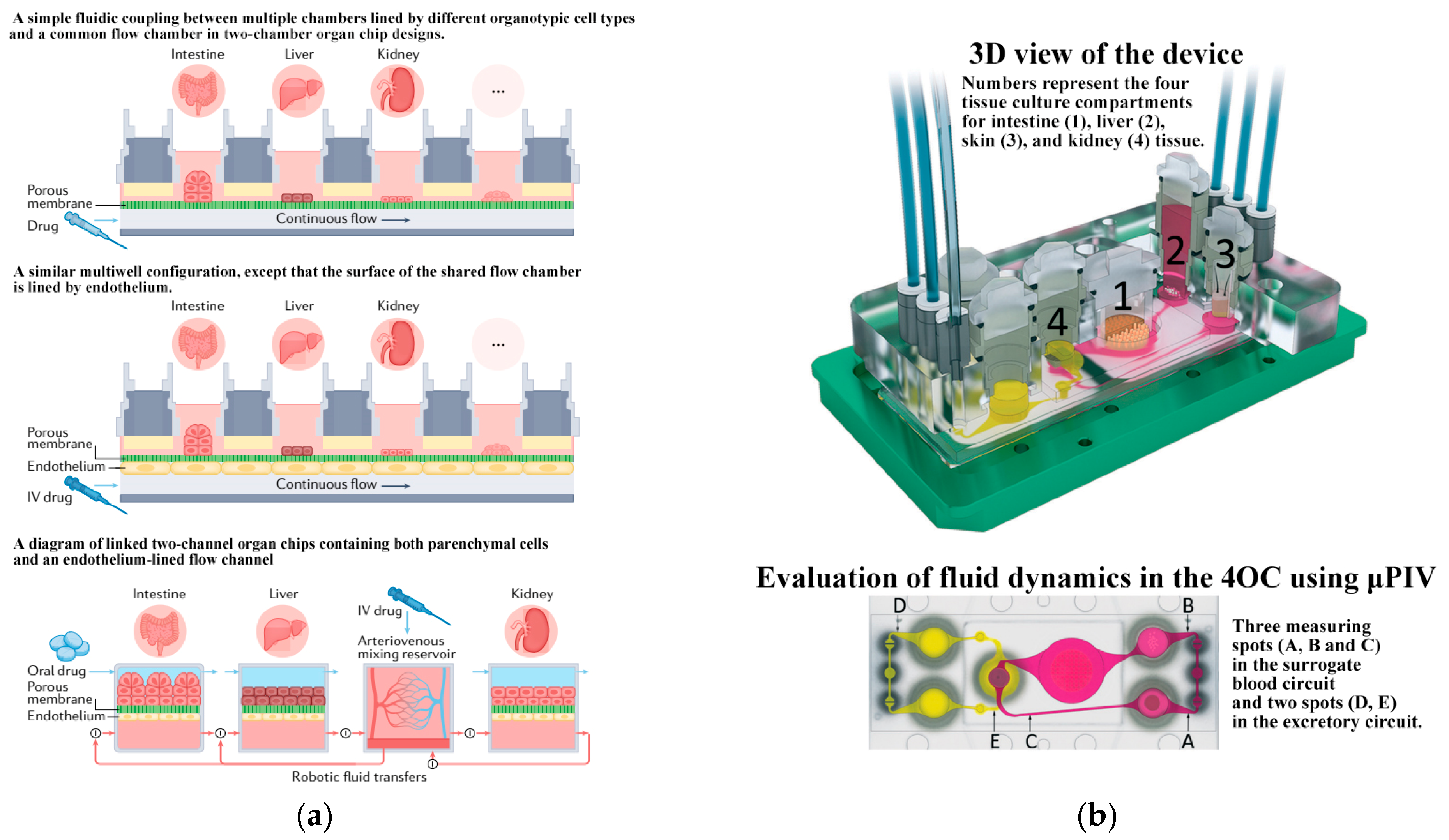
- Novak, R.; Ingram, M.; Marquez, S.; Das, D.; Delahanty, A.; Herland, A.; Maoz, B.M.; Jeanty, S.S.F.; Somayaji, M.R.; Burt, M.; et al. Robotic fluidic coupling and interrogation of multiple vascularized organ chips. Nat. Biomed. Eng. 2020, 4, 407–420. [Google Scholar] [CrossRef]
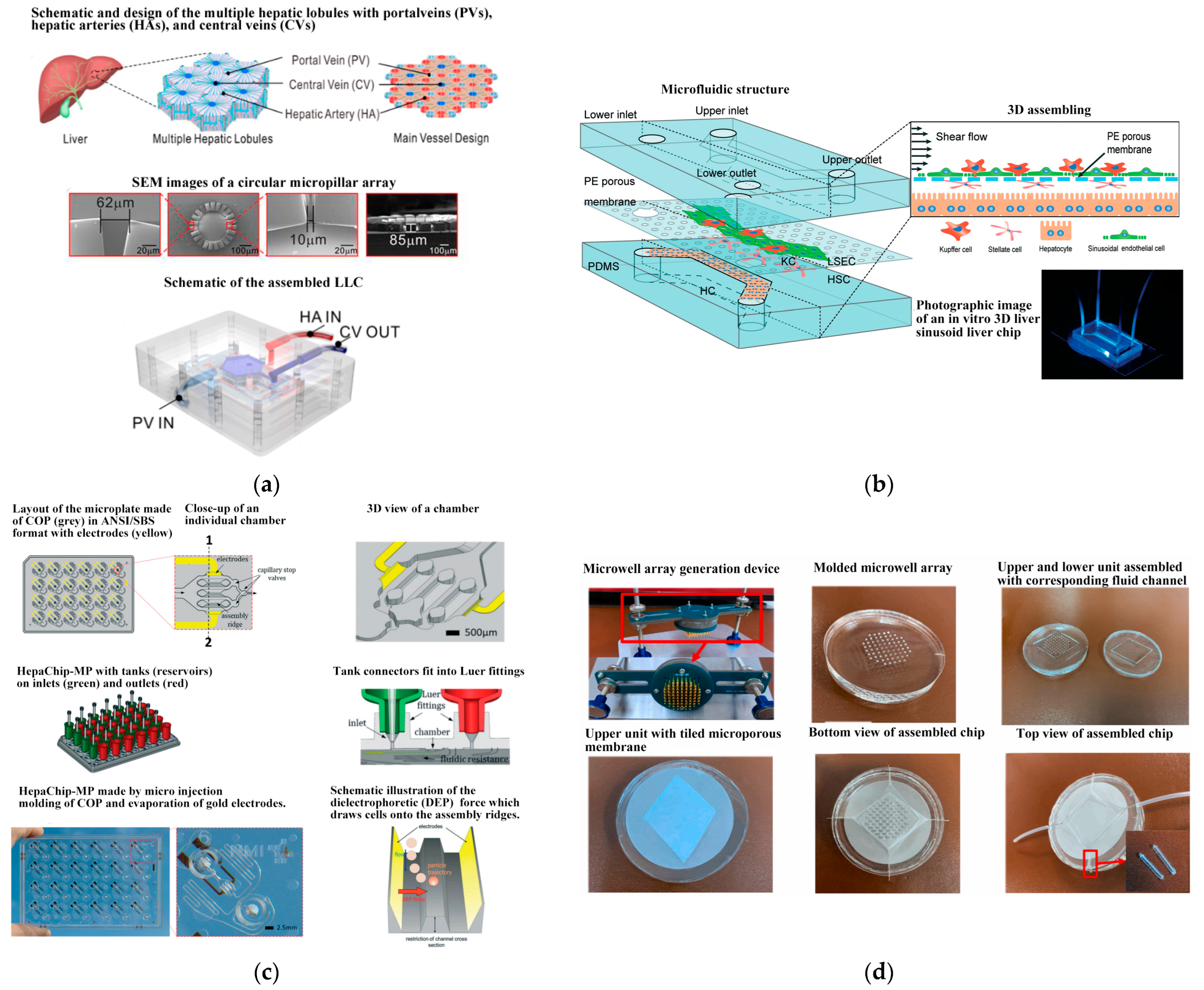
- Kang, Y.B.; Sodunke, T.R.; Lamontagne, J.; Cirillo, J.; Rajiv, C.; Bouchard, M.J.; Noh, M. Liver sinusoid on a chip: Long-term layered co-culture of primary rat hepatocytes and endothelial cells in microfluidic platforms. Biotechnol. Bioeng. 2015, 112, 2571–2582. [Google Scholar] [CrossRef]
- Ya, S.; Ding, W.; Li, S.; Du, K.; Zhang, Y.; Li, C.; Liu, J.; Li, F.; Li, P.; Luo, T.; et al. On-Chip Construction of Liver Lobules with Self-Assembled Perfusable Hepatic Sinusoid Networks. ACS Appl. Mater. Interfaces 2021, 13, 32640–32652. [Google Scholar] [CrossRef]
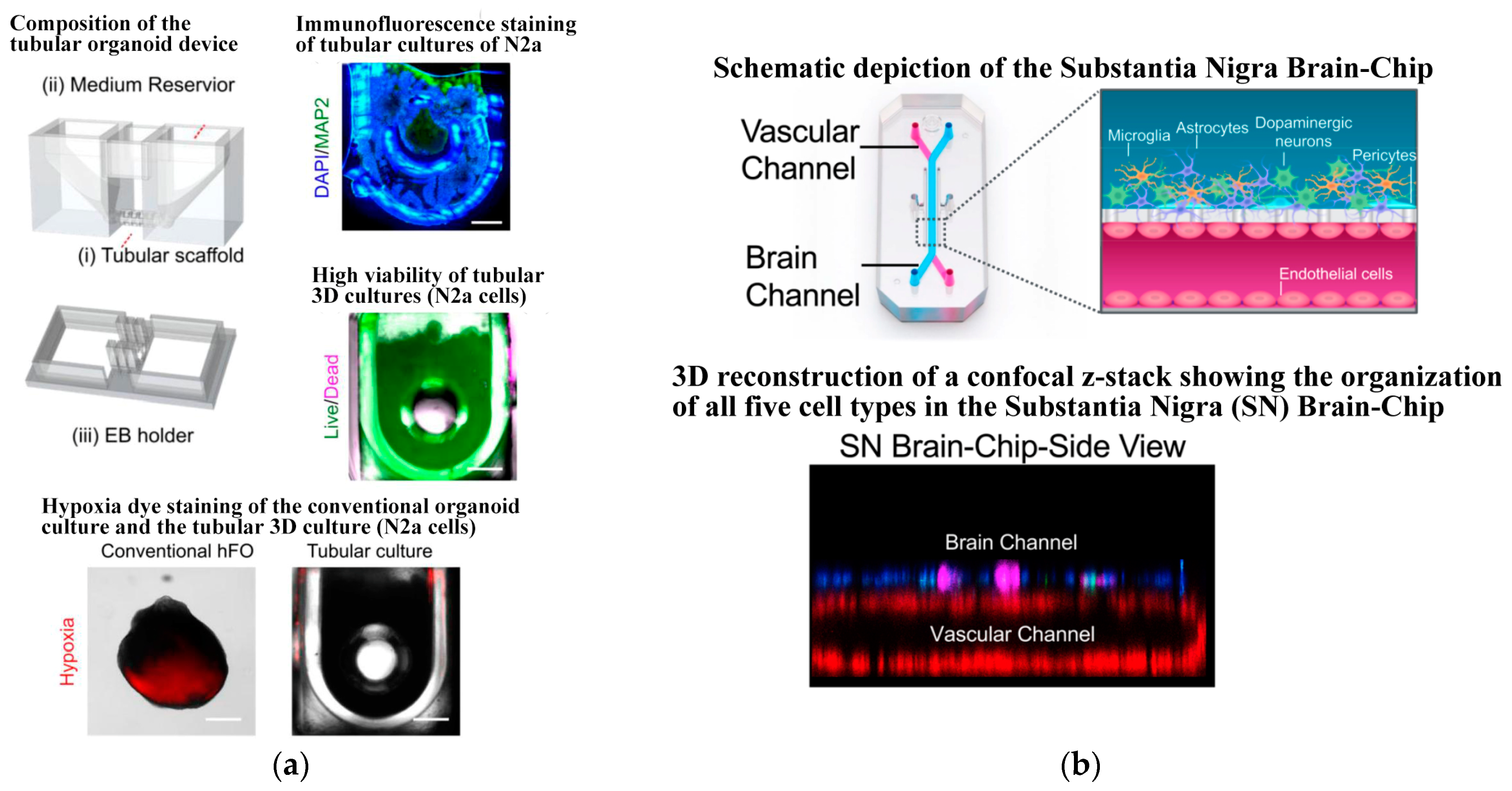
- Ao, Z.; Cai, H.; Wu, Z.; Song, S.; Karahan, H.; Kim, B.; Lu, H.-C.; Kim, J.; Mackie, K.; Guo, F. Tubular human brain organoids to model microglia-mediated neuroinflammation. Lab Chip 2021, 21, 2751–2762. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Li, N.; Yang, H.; Luo, C.; Gong, Y.; Tong, C.; Gao, Y.; Lü, S.; Long, M. Mimicking liver sinusoidal structures and functions using a 3D-configured microfluidic chip. Lab Chip 2017, 17, 782–794. [Google Scholar] [CrossRef]
- Busche, M.; Tomilova, O.; Schütte, J.; Werner, S.; Beer, M.; Groll, N.; Hagmeyer, B.; Pawlak, M.; Jones, P.D.; Schmees, C.; et al. HepaChip-MP–a twenty-four chamber microplate for a continuously perfused liver coculture model. Lab Chip 2020, 20, 2911–2926. [Google Scholar] [CrossRef]
- Zheng, Y.B.; Ma, L.D.; Wu, J.L.; Wang, Y.M.; Meng, X.S.; Hu, P.; Liang, Q.L.; Xie, Y.Y.; Luo, G. Design and fabrication of an integrated 3D dynamic multicellular liver-on-a-chip and its application in hepatotoxicity screening. Talanta 2022, 241, 123262. [Google Scholar] [CrossRef]
- Shah, P.; Fritz, J.V.; Glaab, E.; Desai, M.S.; Greenhalgh, K.; Frachet, A.; Niegowska, M.; Estes, M.; Jäger, C.; Seguin-Devaux, C.; et al. A microfluidics-based in vitro model of the gastrointestinal human–microbe interface. Nat. Commun. 2016, 7, 11535. [Google Scholar] [CrossRef] [PubMed]
- Pediaditakis, I.; Kodella, K.R.; Manatakis, D.V.; Le, C.Y.; Hinojosa, C.D.; Tien-Street, W.; Manolakos, E.S.; Vekrellis, K.; Hamilton, G.A.; Ewart, L.; et al. Modeling alpha-synuclein pathology in a human brain-chip to assess blood-brain barrier disruption. Nat. Commun. 2021, 12, 5907–6023. [Google Scholar] [CrossRef] [PubMed]
- Chou, D.B.; Frismantas, V.; Milton, Y.; David, R.; Pop-Damkov, P.; Ferguson, D.; MacDonald, A.; Bölükbaşı, V.; Joyce, C.E.; Teixeira, L.S.M.; et al. On-chip recapitulation of clinical bone marrow toxicities and patient-specific pathophysiology. Nat. Biomed. Eng. 2020, 4, 394–406. [Google Scholar] [CrossRef]
- Ingber, D.E. Human organs-on-chips for disease modelling, drug development and personalized medicine. Nat. Rev. Genet. 2022, 23, 467–491. [Google Scholar] [CrossRef]
- Maschmeyer, I.; Lorenz, A.K.; Schimek, K.; Hasenberg, T.; Ramme, A.P.; Hübner, J.; Lindner, M.; Drewell, C.; Bauer, S.; Thomas, A.; et al. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip 2015, 15, 2688–2699. [Google Scholar] [CrossRef]
- Smith, V.M.; Nguyen, H.; Rumsey, J.W.; Long, C.J.; Shuler, M.L.; Hickman, J.J. A Functional Human-on-a-Chip autoimmune disease model of myasthenia gravis for development of therapeutics. Front. Cell Dev. Biol. 2021, 9, 745897. [Google Scholar] [CrossRef]
- Wang, Y.I.; Carmona, C.; Hickman, J.J.; Shuler, M.L. Multiorgan microphysiological systems for drug development: Strategies, advances, and challenges. Adv. Healthc. Mater. 2018, 7, 1701000. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Li, C.; Cao, J.; Cui, Z.; Du, J.; Fu, Z.; Yang, H.; Chen, P. Organ-on-a-chip meets artificial intelligence in drug evaluation. Theranostics 2023, 13, 4526. [Google Scholar] [CrossRef] [PubMed]
- Scheinpflug, J.; Pfeiffenberger, M.; Damerau, A.; Schwarz, F.; Textor, M.; Lang, A.; Schulze, F. Journey into bone models: A review. Genes 2018, 9, 247. [Google Scholar] [CrossRef] [PubMed]



| Detection Classification | Detection Target | Detection Method or Marking | Ref. |
|---|---|---|---|
| System or barrier permeability assessment | The permeability of molecules | Glucose | [78] |
| Rhodamine | [79] | ||
| Cell viability | Colorimetry | [80,81] | |
| Staining of live/dead cells | [82] | ||
| Lactic dehydrogenase (LDH) Activity assay | [83,84] | ||
| Electrophysiological activity | Transendothelial electrical resistance (TEER) | [85,86,87] | |
| Immunofluorescence of cellular marker substances | Actin | [88] | |
| Green fluorescence protein (GFP) | [45,89,90] | ||
| Formation of spherical bodies | Electron microscope | [91,92,93] | |
| Intercellular interaction | Cell migration | Mass spectrometry analysis, qPCR, Immunofluorescence | [80,81,88,89,94] |
| Cell differentiation | [95,96,97,98] | ||
| Cellular fibrosis | [70,99,100,101] | ||
| Cytotoxicity testing | [102,103] |
| Application | Co-Culture Type | Ref. | |
|---|---|---|---|
| Angiogenesis | Endothelial cells, Glioma cells | [79] | |
| HUVECs, LFs | [109] | ||
| Endothelial cells, Adipocytes, Mesothelial cells, Tumor cells | [105] | ||
| Blood–brain Barrier | Astrocytes, Endothelial cells | [110,114] | |
| Blood–gas Barrier | Human alveolar epithelial cells, Human pulmonary microvascular endothelial cells | [117] | |
| HAECs, primary human lung microvascular endothelial cells | [118] | ||
| Other Organoid Chips | liver chips | liver cells, endothelial cells | [130,132,133,134] |
| intestinal chips | intestinal epithelium, microorganisms | [125,135] | |
| bone chips | CD34+cells, bone-marrow-derived stromal cells, human vascular endothelium | [137] | |
| brain chips | glial cells and immune cells | [131] | |
| dopaminergic neurons, astrocytes, microglia, pericytes, and microvascular brain endothelial cells | [136] | ||
| multi-organ chip | intestine, liver, skin, and kidney tissue | [139] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; He, W.; Song, Y.; Zhang, X.; Sun, J.; Zhou, Z. Advances of 3D Cell Co-Culture Technology Based on Microfluidic Chips. Biosensors 2024, 14, 336. https://doi.org/10.3390/bios14070336
Li C, He W, Song Y, Zhang X, Sun J, Zhou Z. Advances of 3D Cell Co-Culture Technology Based on Microfluidic Chips. Biosensors. 2024; 14(7):336. https://doi.org/10.3390/bios14070336
Chicago/Turabian StyleLi, Can, Wei He, Yihua Song, Xia Zhang, Jianfei Sun, and Zuojian Zhou. 2024. "Advances of 3D Cell Co-Culture Technology Based on Microfluidic Chips" Biosensors 14, no. 7: 336. https://doi.org/10.3390/bios14070336
APA StyleLi, C., He, W., Song, Y., Zhang, X., Sun, J., & Zhou, Z. (2024). Advances of 3D Cell Co-Culture Technology Based on Microfluidic Chips. Biosensors, 14(7), 336. https://doi.org/10.3390/bios14070336






