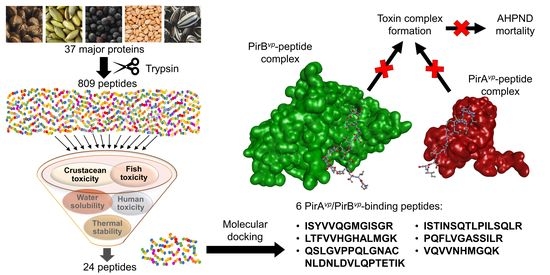
Targeting PirAvp and PirBvp Toxins of Vibrio parahaemolyticus with Oilseed Peptides: An In Silico Approach
Abstract
:1. Introduction
2. Results and Discussion
2.1. In Silico Tryptic Hydrolysis of Oilseed Meal Proteins
2.2. Toxicity of Peptides
2.3. Water Solubility and Thermal Stability of Peptides
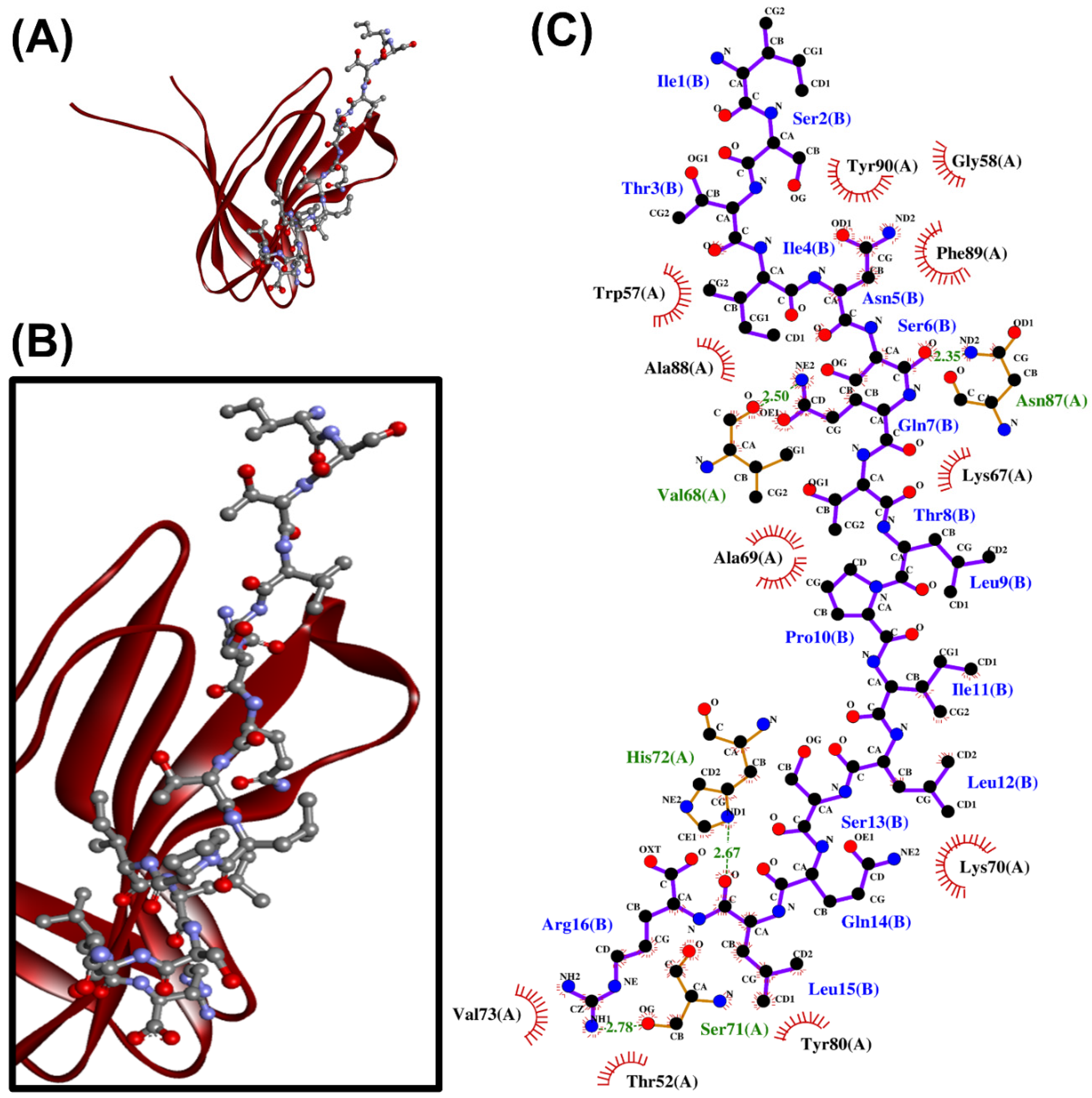
2.4. Molecular Docking Analysis
3. Materials and Methods
3.1. In Silico Hydrolysis of Oilseed Meal Proteins
3.2. Toxicity of Peptides
3.3. Water Solubility and Thermal Stability of Peptides
3.4. Molecular Docking Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tandel, G.M.; John, K.R.; Rosalind George, M.; Prince Jeyaseelan, M.J. Current status of viral diseases in Indian shrimp aquaculture. Acta Virol. 2017, 61, 131–137. [Google Scholar] [CrossRef]
- Kmmari, S.; Rathlavath, S.; Pillai, D.; Rajesh, G. Hepatopancreatic microsporidiasis (HPM) in shrimp culture: A review. Int. J. Curr. Microbiol. App. Sci. 2018, 7, 3208–3215. [Google Scholar] [CrossRef] [Green Version]
- Zorriehzahra, M.J.; Banaederakhshan, R. Early mortality syndrome (EMS) as new emerging threat in shrimp industry. Adv. Anim. Vet. Sci. 2015, 3, 64–72. [Google Scholar] [CrossRef] [Green Version]
- Santos, H.M.; Tsai, C.-Y.; Maquiling, K.R.A.; Tayo, L.L.; Mariatulqabtiah, A.R.; Lee, C.-W.; Chuang, K.P. Diagnosis and potential treatments for acute hepatopancreatic necrosis disease (AHPND): A review. Aquac. Int. 2020, 28, 169–185. [Google Scholar] [CrossRef]
- Dong, X.; Bi, D.; Wang, H.; Zou, P.; Xie, G.; Wan, X.; Yang, Q.; Zhu, Y.; Chen, M.; Guo, C.; et al. pirABvp-bearing Vibrio parahaemolyticus and Vibrio campbellii pathogens isolated from the same AHPND-affected pond possess highly similar pathogenic plasmids. Front. Microbiol. 2017, 8, 1859. [Google Scholar] [CrossRef]
- Han, J.E.; Mohney, L.L.; Tang, K.F.; Pantoja, C.R.; Lightner, D.V. Plasmid mediated tetracycline resistance of Vibrio parahaemolyticus associated with acute hepatopancreatic necrosis disease (AHPND) in shrimps. Aquac. Rep. 2015, 2, 17–21. [Google Scholar] [CrossRef] [Green Version]
- Okocha, R.C.; Olatoye, I.O.; Adedeji, O.B. Food safety impacts of antimicrobial use and their residues in aquaculture. Public Health Rev. 2018, 39, 21. [Google Scholar] [CrossRef] [PubMed]
- Han, J.E.; Tang, K.F.J.; Tran, L.H.; Lightner, D.V. Photorhabdus insect-related (Pir) toxin-like genes in a plasmid of Vibrio parahaemolyticus, the causative agent of acute hepatopancreatic necrosis disease (AHPND) of shrimp. Dis. Aquat. Org. 2015, 113, 33–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.-T.; Chen, I.-T.; Yang, Y.-T.; Ko, T.-P.; Huang, Y.-T.; Huang, J.-Y.; Huang, M.-F.; Lin, S.-J.; Chen, C.-Y.; Lin, S.-S.; et al. The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin. Proc. Natl. Acad. Sci. USA 2015, 112, 10798–10803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, L.; Nunan, L.; Redman, R.M.; Mohney, L.L.; Pantoja, C.R.; Fitzsimmons, K.; Lightner, D.V. Determination of the infectious nature of the agent of acute hepatopancreatic necrosis syndrome affecting penaeid shrimp. Dis. Aquat. Org. 2013, 105, 45–55. [Google Scholar] [CrossRef]
- Lin, S.-J.; Hsu, K.-C.; Wang, H.-C. Structural insights into the cytotoxic mechanism of Vibrio parahaemolyticus PirAvp and PirBvp toxins. Mar. Drugs 2017, 15, 373. [Google Scholar] [CrossRef] [Green Version]
- Singh, M.; Singh, P. Drugs and chemicals applied in aquaculture industry: A review of commercial availability, recommended dosage and mode of application. J. Entomol. Zool. Stud. 2018, 6, 903–907. [Google Scholar]
- Gómez-Jimenez, S.; Espinosa-Plascencia, A.; Valenzuela-Villa, F.; Bermúdez-Almada, M.D.C. Oxytetracycline (OTC) accumulation and elimination in hemolymph, muscle and hepatopancreas of white shrimp Litopenaeus vannamei following an OTC-feed therapeutic treatment. Aquaculture 2008, 274, 24–29. [Google Scholar] [CrossRef]
- Pinoargote, G.; Flores, G.; Cooper, K.; Ravishankar, S. Effects on survival and bacterial community composition of the aquaculture water and gastrointestinal tract of shrimp (Litopenaeus vannamei) exposed to probiotic treatments after an induced infection of acute hepatopancreatic necrosis disease. Aquac. Res. 2018, 49, 3270–3288. [Google Scholar] [CrossRef] [Green Version]
- Fuandila, N.N.; Widanarni, W.; Yuhana, M. Growth performance and immune response of prebiotic honey fed pacific white shrimp Litopenaeus vannamei to Vibrio parahaemolyticus infection. J. Appl. Aquac. 2020, 32, 221–235. [Google Scholar] [CrossRef]
- Jha, R.K.; Babikian, Y.H.; Babikian, H.Y.; Khoa, L.V.; Wisoyo, D.; Srisombat, S.; Jiaravanon, B. Efficacy of natural herbal formulation against acute hepatopancreatic necrosis disease (AHPND) causing Vibrio parahaemolyticus in Penaeus vannamei. Open J. Vet. Med. 2017, 2, 1–6. [Google Scholar] [CrossRef]
- Wang, G.; Yu, E.; Li, Z.; Yu, D.; Wang, H.; Gong, W. Effect of bioactive peptides (BPs) on the development of Pacific white shrimp (Litopenaeus vannamei Boone, 1931). J. Ocean Univ. China 2016, 15, 495–501. [Google Scholar] [CrossRef]
- Tang, C.-H.; Wang, X.-S.; Yang, X.-Q. Enzymatic hydrolysis of hemp (Cannabis sativa L.) protein isolate by various proteases and antioxidant properties of the resulting hydrolysates. Food Chem. 2009, 114, 1484–1490. [Google Scholar] [CrossRef]
- Zhou, C.; Yu, X.; Qin, X.; Ma, H.; Yagoub, A.E.A.; Hu, J. Hydrolysis of rapeseed meal protein under simulated duodenum digestion: Kinetic modeling and antioxidant activity. LWT 2016, 68, 523–531. [Google Scholar] [CrossRef]
- Lu, X.; Zhang, L.; Sun, Q.; Song, G.; Huang, J. Extraction, identification and structure-activity relationship of antioxidant peptides from sesame (Sesamum indicum L.) protein hydrolysate. Food Res. Int. 2019, 116, 707–716. [Google Scholar] [CrossRef]
- Vaghei, R.G.; Abolhasani, M.H.; Ghorbani, R.; Matinfar, A. Production of soybean meal-based feed and its effect on growth performance of western white shrimp. Iran. J. Fish. Sci. 2017, 16, 578–586. [Google Scholar]
- Shao, J.; Zhao, W.; Han, S.; Chen, Y.; Wang, B.; Wang, L. Partial replacement of fishmeal by fermented soybean meal in diets for juvenile white shrimp (Litopenaeus vannamei). Aquac. Nutr. 2019, 25, 145–153. [Google Scholar] [CrossRef] [Green Version]
- Cheng, A.-C.; Lin, H.-L.; Shiu, Y.-L.; Tyan, Y.-C.; Liu, C.-H. Isolation and characterization of antimicrobial peptides derived from Bacillus subtilis E20-fermented soybean meal and its use for preventing Vibrio infection in shrimp aquaculture. Fish. Shellfish Immunol. 2017, 67, 270–279. [Google Scholar] [CrossRef]
- Lin, S.-J.; Chen, Y.-F.; Hsu, K.-C.; Chen, Y.-L.; Ko, T.-P.; Lo, C.-F.; Wang, H.-C.; Wang, H.-C. Structural insights to the heterotetrameric interaction between the Vibrio parahaemolyticus PirAvp and PirBvp toxins and activation of the Cry-Like pore-forming domain. Toxins 2019, 11, 233. [Google Scholar] [CrossRef] [Green Version]
- Lazo, J.P.; Romaire, R.P.; Reigh, R.C. Evaluation of three in vitro enzyme assays for estimating protein digestibility in the pacific white shrimp Penaeus vannamei. J. World Aquac. Soc. 1998, 29, 441–450. [Google Scholar] [CrossRef]
- Duan, X.; Zhang, M.; Chen, F. Prediction and analysis of antimicrobial peptides from rapeseed protein using in silico approach. J. Food Biochem. 2021, 45, e13598. [Google Scholar] [CrossRef] [PubMed]
- Wong, F.-C.; Ong, J.-H.; Chai, T.-T. SARS-CoV-2 spike protein-, main protease- and papain-like-protease-targeting peptides from seed proteins following gastrointestinal digestion: An in silico study. Phytomed. Plus 2021, 1, 100016. [Google Scholar] [CrossRef]
- Jannathulla, R.; Dayal, J.S.; Ambasankar, K.; Yuvapushpa, R.; Kumar, J.A.; Muralidhar, M. Evaluation of fungal fermented rapeseed meal as a fishmeal substitute in the diet of Penaeus vannamei. J. Coast. Res. 2019, 86, 82–89. [Google Scholar] [CrossRef]
- Dayal, J.S.; Rajaram, V.; Ambasankar, K.; Ali, S.A. Sunflower oil cake as a replacement for fish meal in feeds of tiger shrimp, Penaeus monodon reared in tanks and in net cages. Indian J. Geo-Mar. Sci. 2011, 40, 460–470. [Google Scholar]
- Bae, J.; Hamidoghli, A.; Djaballah, M.S.; Maamri, S.; Hamdi, A.; Souffi, I.; Farris, N.W.; Bai, S.C. Effects of three different dietary plant protein sources as fishmeal replacers in juvenile whiteleg shrimp, Litopenaeus vannamei. Fish. Aquat. Sci. 2020, 23, 2. [Google Scholar] [CrossRef] [Green Version]
- Jimoh, W.; Aroyehun, H.T. Evaluation of cooked and mechanically defatted sesame (Sesamum indicum) seed meal as a replacer for soybean meal in the diet of African catfish (Clarias gariepinus). Turk. J. Fish. Aquat. Sci. 2011, 11, 185–190. [Google Scholar] [CrossRef]
- Omosowone, O.O.; Ogunrinde, A.M. Effect of partial or total supplementation of soybean meal with fluted pumpkin (Telfairia occidentalis) seed meal in the diet of hybrid Catfish (Heteroclarias) fingerlings. Food Sci. Nutr. 2018, 6, 1190–1195. [Google Scholar] [CrossRef]
- Simon, C.J.; Truong, H.; Habilay, N.; Hines, B. Feeding behaviour and bioavailability of essential amino acids in shrimp Penaeus monodon fed fresh and leached fishmeal and fishmeal-free diets. Animals 2021, 11, 847. [Google Scholar] [CrossRef]
- Sarma, R.; Wong, K.-Y.; Lynch, G.C.; Pettitt, B.M. Peptide solubility limits: Backbone and side-chain interactions. J. Phys. Chem. B. 2018, 122, 3528–3539. [Google Scholar] [CrossRef]
- Kvåle, A.; Yúfera, M.; Nygård, E.; Aursland, K.; Harboe, T.; Hamre, K. Leaching properties of three different micropaticulate diets and preference of the diets in cod (Gadus morhua L.) larvae. Aquaculture 2006, 251, 402–415. [Google Scholar] [CrossRef]
- Mo, W.Y.; Man, Y.B.; Wong, M.H. Use of food waste, fish waste and food processing waste for China’s aquaculture industry: Needs and challenge. Sci. Total Environ. 2018, 613–614, 635–643. [Google Scholar] [CrossRef]
- Osorio, D.; Rondón-Villarreal, P.; Torres, R. Peptides: A package for data mining of antimicrobial peptides. R J. 2015, 7, 4–14. [Google Scholar] [CrossRef]
- Delves-Broughton, J.; Weber, G. Nisin, natamycin and other commercial fermentates used in food biopreservation. In Protective Cultures, Antimicrobial Metabolites and Bacteriophages for Food and Beverage Biopreservation; Elsevier: Amsterdam, The Netherlands, 2011; pp. 63–99. [Google Scholar]
- Phiwsaiya, K.; Charoensapsri, W.; Taengphu, S.; Dong, H.T.; Sangsuriya, P.; Nguyen, G.T.T.; Pham, H.Q.; Amparyup, P.; Sritunyalucksana, K.; Taengchaiyaphum, S.; et al. A natural Vibrio parahaemolyticus ΔpirAVppirBVp+ mutant kills shrimp but produces neither Pir Vp toxins nor acute hepatopancreatic necrosis disease lesions. Appl. Environ. Microbiol. 2017, 83, e00680-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, R.; Pedrosa-Gerasmio, I.R.; Alenton, R.R.R.; Nozaki, R.; Kondo, H.; Hirono, I. Anti-PirA-like toxin immunoglobulin (IgY) in feeds passively immunizes shrimp against acute hepatopancreatic necrosis disease. J. Fish. Dis. 2019, 42, 1125–1132. [Google Scholar] [CrossRef]
- Almanza-Martínez, N.; Francisco Martínez Díaz, S.; Flores-Ramírez, G.; Zuñiga-Navarrete, F.; Gómez, I.; Cardona-Félix, C.S. An α-amylase-like protein interacts with PirB toxin from Vibrio parahaemolyticus in digestive tract tissue of white shrimp Litopenaeus vannamei. Aquac. Res. 2020, 51, 3910–3914. [Google Scholar] [CrossRef]
- Kotecka-Majchrzak, K.; Kasałka-Czarna, N.; Sumara, A.; Fornal, E.; Montowska, M. Multispecies identification of oilseed- and meat-specific proteins and heat-stable peptide markers in food products. Molecules 2021, 26, 1577. [Google Scholar] [CrossRef] [PubMed]
- Mamone, G.; Picariello, G.; Ramondo, A.; Nicolai, M.A.; Ferranti, P. Production, digestibility and allergenicity of hemp (Cannabis sativa L.) protein isolates. Food Res. Int. 2019, 115, 562–571. [Google Scholar] [CrossRef] [PubMed]
- Rezig, L.; Chibani, F.; Chouaibi, M.; Dalgalarrondo, M.; Hessini, K.; Guéguen, J.; Hamdi, S. Pumpkin (Cucurbita maxima) seed proteins: Sequential extraction processing and fraction characterization. J. Agric. Food Chem. 2013, 61, 7715–7721. [Google Scholar] [CrossRef]
- Perera, S.P.; McIntosh, T.C.; Wanasundara, J.P.D. Structural properties of cruciferin and napin of Brassica napus (Canola) show distinct responses to changes in pH and temperature. Plants 2016, 5, 36. [Google Scholar] [CrossRef] [Green Version]
- Wildermuth, S.R.; Young, E.E.; Were, L.M. Chlorogenic acid oxidation and its reaction with sunflower proteins to form green-colored complexes. Compr. Rev. Food Sci. Food Saf. 2016, 15, 829–843. [Google Scholar] [CrossRef]
- Onsaard, E. Sesame proteins. Int. Food Res. J. 2012, 19, 1287–1295. [Google Scholar]
- The UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [Green Version]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP-UWM database of bioactive peptides: Current opportunities. Int. J. Mol. Sci. 2019, 20, 5978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Yang, H.; Cai, Y.; Cao, Q.; Sun, L.; Wang, Z.; Li, W.; Liu, G.; Lee, P.W.; Tang, Y. In silico prediction of chemical aquatic toxicity for marine crustaceans via machine learning. Toxicol. Res. 2019, 8, 341–352. [Google Scholar] [CrossRef]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Raghava, G.P.S. In silico approach for predicting toxicity of peptides and proteins. PLoS ONE 2013, 8, e73957. [Google Scholar]
- Zhou, P.; Jin, B.; Li, H.; Huang, S.-Y. HPEPDOCK: A web server for blind peptide-protein docking based on a hierarchical algorithm. Nucleic Acids Res. 2018, 46, W443–W450. [Google Scholar] [CrossRef]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995, 8, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- Lamiable, A.; Thévenet, P.; Rey, J.; Vavrusa, M.; Derreumaux, P.; Tufféry, P. PEP-FOLD3: Faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016, 44, W449–W454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Y.; Maupetit, J.; Derreumaux, P.; Tufféry, P. Improved PEP-FOLD approach for peptide and miniprotein structure prediction. J. Chem. Theory Comput. 2014, 10, 4745–4758. [Google Scholar] [CrossRef]
- Thévenet, P.; Shen, Y.; Maupetit, J.; Guyon, F.; Derreumaux, P.; Tufféry, P. PEP-FOLD: An updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res. 2012, 40, W288–W293. [Google Scholar] [CrossRef] [Green Version]


| Source | Protein | UniProt Accession | Type * | Number of Residues | Number of Peptides |
|---|---|---|---|---|---|
| Hemp | Albumin | A0A219D1L6 | A | 119 | 18 |
| Edestin 1 | A0A090DLH8 | G | 488 | 62 | |
| Edestin 2 | A0A090DLI7 | G | 467 | 56 | |
| Edestin 3 | A0A219D3H6 | G | 468 | 54 | |
| 7S vicilin-like protein | A0A219D1T7 | G | 472 | 57 | |
| Pumpkin | 2S albumin large chain | Q39649 | A | 67 | 13 |
| 11S globulin delta chain | P13744 | G | 184 | 24 | |
| 11S globulin gamma chain | P13744 | G | 275 | 32 | |
| Rape | Cruciferin BnC1 subunit alpha | P33523 | G | 277 | 24 |
| Cruciferin BnC1 subunit beta | P33523 | G | 190 | 16 | |
| Cruciferin BnC2 subunit alpha | P33524 | G | 283 | 22 | |
| Cruciferin BnC2 subunit beta | P33524 | G | 190 | 19 | |
| Cruciferin CRU1 alpha chain | P33525 | G | 296 | 22 | |
| Cruciferin CRU1 beta chain | P33525 | G | 190 | 17 | |
| Cruciferin CRU4 alpha chain | P33522 | G | 254 | 20 | |
| Cruciferin CRU4 beta chain | P33522 | G | 189 | 17 | |
| Cruciferin subunit alpha | P11090 | G | 275 | 24 | |
| Cruciferin subunit beta | P11090 | G | 190 | 16 | |
| Napin-1A small chain | P24565 | A | 31 | 5 | |
| Napin-1A large chain | P24565 | A | 79 | 8 | |
| Napin-2 small chain | P01090 | A | 36 | 6 | |
| Napin-2 large chain | P01090 | A | 81 | 9 | |
| Napin-3 small chain | P80208 | A | 37 | 6 | |
| Napin-3 large chain | P80208 | A | 88 | 9 | |
| Napin-B small chain | P27740 | A | 36 | 6 | |
| Napin-B large chain | P27740 | A | 84 | 10 | |
| Napin embryo-specific small chain | P09893 | A | 38 | 6 | |
| Napin embryo-specific large chain | P09893 | A | 89 | 10 | |
| Sesame | 2S seed storage protein 1 small subunit | Q9XHP1 | A | 30 | 6 |
| 2S seed storage protein 1 large subunit | Q9XHP1 | A | 70 | 10 | |
| 11S globulin isoform 3 | Q2XSW7 | G | 468 | 54 | |
| 11S globulin isoform 4 | Q2XSW6 | G | 449 | 46 | |
| 11S globulin seed storage protein 2 acidic chain | Q9XHP0 | G | 256 | 31 | |
| 11S globulin seed storage protein 2 basic chain | Q9XHP0 | G | 182 | 18 | |
| Sunflower | 2S seed storage protein | P15461 | A | 134 | 15 |
| 11S globulin seed storage protein G3 acidic chain | P19084 | G | 285 | 22 | |
| 11S globulin seed storage protein G3 basic chain | P19084 | G | 188 | 19 |
| Source | Protein Type * | Total Number of Peptides | Number of Peptides Predicted as Non-Toxic (NT) and Toxic (T) against Different Organisms | |||||
|---|---|---|---|---|---|---|---|---|
| Crustacean | Fish | Human | ||||||
| NT | T | NT | T | NT | T | |||
| Hemp | G | 229 | 229 | 0 | 170 | 59 | 229 | 0 |
| A | 18 | 18 | 0 | 16 | 2 | 17 | 1 | |
| Pumpkin | G | 56 | 56 | 0 | 40 | 16 | 56 | 0 |
| A | 13 | 13 | 0 | 12 | 1 | 12 | 1 | |
| Rape | G | 197 | 197 | 0 | 115 | 82 | 194 | 0 |
| A | 75 | 75 | 0 | 61 | 14 | 70 | 5 | |
| Sesame | G | 149 | 149 | 0 | 109 | 40 | 149 | 0 |
| A | 16 | 16 | 0 | 15 | 1 | 14 | 2 | |
| Sunflower | G | 41 | 41 | 0 | 29 | 12 | 40 | 0 |
| A | 15 | 15 | 0 | 13 | 2 | 14 | 1 | |
| Total | 809 | 809 | 0 | 580 | 229 | 795 | 10 | |
| Source | Peptide | Aliphatic Index |
|---|---|---|
| Sesame | PI | 195 |
| GLIVMAR | 167 | |
| GHIITVAR | 146 | |
| ISTINSQTLPILSQLR | 146 | |
| IQVVGHK | 139 | |
| GVLYR | 136 | |
| GLQVISPPLQR | 133 | |
| VASA | 123 | |
| QEQFQCAGIVAMR | 68 | |
| MTFVR | 58 | |
| QTFHNIFR | 49 | |
| YWQSLQQHQQHR | 33 | |
| GQHQFGNVFR | 29 | |
| TGGYA | 20 | |
| HCMQWMR | 0 | |
| GSTWQQGQCR | 0 | |
| Hemp | PVV | 193 |
| GVLYK | 136 | |
| PQFLVGASSILR | 130 | |
| QGQIVTVPQNHAVVK | 110 | |
| ESVILPTSAASPPVK | 104 | |
| LGNLTSYQR | 87 | |
| VQVVNHMGQK | 87 | |
| ATA | 67 | |
| NIPSMCGMQPR | 35 | |
| TTWSWR | 0 | |
| WQSQCQFQR | 0 | |
| Sunflower | GHIVNVGQDLQIVR | 146 |
| VIQNLPNQCDLEVQQCTTCTG | 83 | |
| WVSFK | 58 | |
| GGWSN | 0 | |
| Rape | LTFVVHGHALMGK | 112 |
| QSLGVPPQLGNACNLDNLDVLQPTETIK | 108 | |
| ISYVVQGMGISGR | 107 | |
| TNANAQINTLAGR | 83 | |
| NLPNVCNMK | 76 | |
| TNANAMVSTLAGR | 75 | |
| CSGVSFVR | 73 | |
| ACQQWIR | 70 | |
| ACQQWLHK | 61 | |
| ATSQQFQWIEFK | 41 | |
| QQQGQQMQGQQMQQVISR | 38 | |
| VQGQHGPFQSTR | 24 | |
| QAMQSGGG | 13 | |
| QAMQSGSG | 13 | |
| QAMQPGGGSG | 10 | |
| TMPG | 0 | |
| TMPGPS | 0 | |
| TMPGPSY | 0 | |
| Lantibiotic nisin-A | ITSISLCTPGCKTGALMGCNMKTATCHCSIHVSK | 72 |
| Source | Peptide | Docking Score | Interaction with PirAvp Residues * | |||
|---|---|---|---|---|---|---|
| Hydrogen Bond | Hydrophobic Interaction | Salt Bridge | External Bond | |||
| Rape | LTFVVHGHALMGK | −194.881 | Asn87 | Trp57, Gly58, Pro60, Ala63, Ala69, Lys70, Tyr80, Gln83, Pro85, Asn87, Ala88, Phe89, Tyr90 | - | - |
| ISYVVQGMGISGR | −174.661 | Tyr80(2) | Trp57, Gly58, Ala59, Pro60, Val68, Ala69, Lys70, Ser71, His72, Tyr80, His81, Leu82, Gln83, Pro85, Asn87, Phe89, Tyr90 | - | - | |
| QSLGVPPQLGNACNLDNLDVLQPTETIK | −168.015 | Gln54, Trp57, Ser71(2), Tyr80, Asn87 | Thr52, Gln54, Trp57, Val68, Ala69, Lys70, Ser71, His72, Tyr80, His81, Pro85, Asn87, Phe89, Asn99 | - | Asn99(3) | |
| TNANAMVSTLAGR | −156.242 | Ala69, Ser71 | Pro35, Trp57, Val68, Ala69, Lys70, Ser71, His72, Asn87, Tyr80, Ala88, Tyr90 | - | - | |
| TNANAQINTLAGR | −151.751 | Ala69, His72, Tyr80, Asn87(2) | Trp57, Ala59, Pro60, Val68, Ala69, Lys70, Ser71, His72, Tyr80, Pro85, Asn87, Phe89, Tyr90 | - | - | |
| NLPNVCNMK | −147.564 | Lys29, Arg39(3), Arg84 | Tyr11, Lys29, Gly38, Arg39, Ser40, Arg84, Asp86, His111, Leu112, Glu113, His114, His115 | Asp86 | Arg84 | |
| CSGVSFVR | −146.934 | Asn87 | Trp57, Gly58, Ala59, Pro60, Ala69, Lys70, Pro85, Asp86, Asn87, Ala88, Phe89, Tyr90 | - | Asp86 | |
| Sesame | ISTINSQTLPILSQLR | −178.623 | Val68, Ser71, His72, Asn87 | Thr52, Trp57, Gly58, Lys67, Val68, Ala69, Lys70, Ser71, His72, Val73, Tyr80, Asn87, Ala88, Phe89, Tyr90 | - | - |
| GLQVISPPLQR | −171.949 | Asp10, Lys29, Arg39, Glu113 | Asp10, His13, Trp15, Asp27, Ser28, Lys29, Gly38, Arg39, Ser40, Arg84, Gln92, Tyr110, His111, Leu112, Glu113, His114, His115 | Asp10 | Arg84 | |
| GHIITVAR | −151.394 | Gly58, Ser71 | Trp57, Gly58, Ala59, Pro60, Val68, Ala69, Ser71, His72, Asn87, Phe89, Tyr90 | - | - | |
| GLIVMAR | −144.315 | Arg84(3) | Lys29, Val37, Gly38, Arg39, Gln83, Arg84, Pro85, Asp86, His111, Leu112, Glu113, His114, His115 | Asp86(3) | Glu113 | |
| Hemp | QGQIVTVPQNHAVVK | −177.911 | Tyr80, Asn87, Tyr90 | Trp57, Pro60, Ala69, Lys70, Ser71, His72, Tyr80, Leu82, Asn87, Ala88, Phe89, Tyr90 | - | Ala69 |
| PQFLVGASSILR | −175.973 | Trp57, Ser71, Asn87 | Trp57, Lys67, Val68, Lys70, Ser71, His72, Tyr80, His81, Leu82, Gln83, Asn87, Ala88, Phe89 | - | His81 | |
| LGNLTSYQR | −165.451 | Trp57, Ser71, Tyr80, Arg84, Asn87 | Trp57, Val68, Ala69, Lys70, Ser71, His72, Tyr80, Leu82, Gln83, Asp86, Asn87, Ala88, Tyr90 | Asp86(5) | - | |
| VQVVNHMGQK | −155.517 | Ala69, Ser71, Ala88 | Trp57, Ala59, Pro60, Val68, Ala69, Lys70, Asn87, Ala88, Phe89, Tyr90 | - | - | |
| ESVILPTSAASPPVK | −155.494 | Lys70, Asn87 | Pro35, Trp57, Gly58, Ala59, Pro60, Val68, Ala69, Lys70, Ser71, Tyr80, His81, Leu82, Pro85, Asn87, Ala88, Phe89, Tyr90 | - | - | |
| Sunflower | GHIVNVGQDLQIVR | −175.043 | Ala69, His72, Asn87 | Trp57, Pro60, Val68, Ala69, Lys70, His72, Tyr80, Asn87, Ala88, Tyr90 | Lys70 | - |
| VIQNLPNQCDLEVQQCTTCTG | −172.423 | Arg48, Ser71(2), Gln75, Arg76 | Val23, Arg48, Gly49, Glu50, Thr52, Gln54, Ser71, Gln75, Arg76, Ile97, Asn99, Gly100, Asn101 | Arg48(2) | Thr52 | |
| PirBvp | YNRVGRLKL | −174.899 | Trp57, Val68, Asn87, Ala88 | Trp57, Val68, Ala69, Lys70, Ser71, His72, Tyr80, Leu82, Pro85, Asn87, Ala88, Phe89 | - | - |
| WADNDSYNNANQD | −170.618 | His72, Asn87, Ala88 | Pro35, Ile53, Trp57, Ala69, Lys70, Ser71, His72, Tyr80, Leu82, Pro85, Asn87, Ala88, Phe89 | Lys70 | Leu82(2) | |
| FVVGENSGKPSVRLQL | −167.625 | Ser28, Gly38, Gln83, His111, Leu112 | Ser28, Lys29, His30, Thr31, Ile33, Glu36, Val37, Gly38, Arg39, Ser40, His81, Gln83, Arg84, His111, Leu112, Glu113 | Glu36 | Ser28, Ile33 | |
| YELFHPDEF | −159.953 | Ser71(3), Tyr80 | Pro35, Trp57, Lys67, Val68, Ala69, Lys70, Ser71, His72, Tyr80, Asn87, Ala88, Phe89, Tyr90 | - | Val68 | |
| DEIPQPLKPNM | −148.916 | Tyr80, Asn87 | Trp57, Gly58, Ala69, Lys70, Tyr80, Pro85, Asn87, Ala88, Phe89, Tyr90 | - | - | |
| MLADQEGSDKVAA | −142.536 | Lys70, Ser71, Asn87(3) | Trp57, Gly58, Ala59, Pro60, Val68, Ala69, Lys70, Ser71, His72, Val73, Asn87, Phe89, Tyr90 | - | Ser71 | |
| Source | Peptide | Docking Score | Interaction with PirBvp Residues * | ||
|---|---|---|---|---|---|
| Hydrogen Bond | Hydrophobic Interaction | External Bond | |||
| Rape | ISYVVQGMGISGR | −209.710 | Tyr35, Glu73, Tyr359 | Tyr35, Ala36, Ala39, Met40, Phe43, Ile48, Pro49, Asn60, Ile61, Pro64, Asp71, Ile72, Glu73, Gln78, Tyr359, Lys361, Gln400, GLn402, Phe429, Pro431, Thr436 | - |
| LTFVVHGHALMGK | −208.710 | Gly53, Asn60 | Tyr35, Ala39, Met40, Tyr50, Ala51, Gly52, Ser56, Thr57, Asn60, Asn114, Glu118, Tyr359, Phe429, Pro431, Phe434, Gly435, Thr436 | - | |
| QSLGVPPQLGNACNLDNLDVLQPTETIK | −201.070 | Asn28(2), Tyr35, Gly403 | Leu25, Asp27, Asn28, Tyr29, Tyr35, Thr66, Pro67, Pro70, Ile72, Asp230, Ala273, Val274, Tyr359, Asp364, Val397, Arg398, Gln400, Glu402, Gly403, His404, Phe429, Thr436 | Gln400(2) | |
| Sesame | ISTINSQTLPILSQLR | −198.924 | Arg21, Tyr29, Tyr359, Ser381, Arg398(2) | Arg21, Tyr29, Tyr35, Met40, Phe43, Ile48, Gln84, Asp85, Glu89, Tyr359, Lys361, Tyr362, Ser381, Arg398, Gln400, Glu402, Thr436 | - |
| Hemp | PQFLVGASSILR | −198.337 | Tyr359, Lys361, Phe434 | Tyr35, Ala36, Ala39, Met40, Ile48, Asn60, Ile61, Pro64, Asn65, Thr66, Ile72, Tyr359, Lys361, Tyr362, Ser381, Asp383, Gly403, Phe429, Phe434, Gly435, Thr436 | Tyr35 |
| VQVVNHMGQK | −196.671 | - | Tyr35, Ala39, Met40, Phe43, Ile48, Pro49, Tyr50, Ala51, Thr57, Asn60, Ile61, Asn65, Thr66, Pro67, Tyr359, Lys361, Gly403, His404, Phe429, Pro431, Thr436 | Asn65 | |
| PirAvp | TIQYQWGAPFMAGGWKVAKSHVVQRDET | −219.311 | Asn28, Asn65 | Asp27, Tyr29, Glu30, Val31, Tyr35, Met40, Ile48, Tyr50, Ala51, Thr57, Asn60, Pro64, Asn65, Thr66, Gln78, Asp81, Arg82, Gln84, Asp85, Tyr362, Ser396, Val397, Arg398, Glu402 | Val397 |
| WTVEPNGGVTEVDSKHTPIIPEVGRS | −191.468 | Tyr35, Ser396 | Asp27, Asn28, Tyr29, Val31, Tyr35, Ala36, Ile72, Gln78, Asp81, Asp364, Pro395, Ser396, Val397, Arg398, Phe429 | - | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ong, J.-H.; Wong, W.-L.; Wong, F.-C.; Chai, T.-T. Targeting PirAvp and PirBvp Toxins of Vibrio parahaemolyticus with Oilseed Peptides: An In Silico Approach. Antibiotics 2021, 10, 1211. https://doi.org/10.3390/antibiotics10101211
Ong J-H, Wong W-L, Wong F-C, Chai T-T. Targeting PirAvp and PirBvp Toxins of Vibrio parahaemolyticus with Oilseed Peptides: An In Silico Approach. Antibiotics. 2021; 10(10):1211. https://doi.org/10.3390/antibiotics10101211
Chicago/Turabian StyleOng, Joe-Hui, Wey-Lim Wong, Fai-Chu Wong, and Tsun-Thai Chai. 2021. "Targeting PirAvp and PirBvp Toxins of Vibrio parahaemolyticus with Oilseed Peptides: An In Silico Approach" Antibiotics 10, no. 10: 1211. https://doi.org/10.3390/antibiotics10101211
APA StyleOng, J.-H., Wong, W.-L., Wong, F.-C., & Chai, T.-T. (2021). Targeting PirAvp and PirBvp Toxins of Vibrio parahaemolyticus with Oilseed Peptides: An In Silico Approach. Antibiotics, 10(10), 1211. https://doi.org/10.3390/antibiotics10101211