Abstract
Resistances to extended-spectrum cephalosporins (ESC) and colistin are One Health issues since genes encoding these resistances can be transmitted between all sectors of the One Health concept, i.e., human, animal, and the environment. Among food-producing animals, sheep farming has long been overlooked. To fill in this knowledge gap, we looked for ESC- and colistin resistance in 21 faecal samples collected from sheep in one farm in the south of Portugal. ESC-resistant isolates were selected on MacConkey agar plates supplemented with cefotaxime. Susceptibility testing was performed by the disk-diffusion method according to CLSI, while colistin MIC was determined by broth microdilution. ESC- and colistin-resistance genes were identified by PCR, and the clonality of all isolates was assessed by XbaI-PFGE. The replicon content was determined by PCR according to the PCR-based replicon typing (PBRT) scheme. Sixty-two non-duplicate ESC-resistant E. coli isolates were identified, which all presented an extended-spectrum beta-lactamase (ESBL) phenotype, mostly due to the presence of CTX-M genes. One CTX-M-1-producing E. coli was concomitantly colistin-resistant and presented the plasmid-mediated mcr-1 gene. Nearly all isolates showed associated resistances to non-beta-lactam antibiotics, which could act as co-selectors, even in the absence of beta-lactam use. The results showed a high proportion of ESBL-producing E. coli in sheep faeces. Their dissemination was very dynamic, with the spread of successful clones between animals, but also a large diversity of clones and plasmids, sometimes residing in the same animal. This study highlights the need for global surveillance in all food-producing sectors, in order to avoid the dissemination of genes conferring resistance to last-resort antibiotics in human medicine.
1. Introduction
The discovery of antibiotics was one of the most important events in the history of human health, but their use and misuse has led to resistances to all known classes of molecules; antimicrobial resistance (AMR) has become a risk to global health [1,2]. The issue of AMR falls into the One Health concept, which postulates that the human, veterinary, and environmental sectors are interconnected [3,4,5]. Resistance to extended-spectrum cephalosporins (ESC) and colistin is of specific concern since these molecules, which are considered critically important antibiotics to human health, have now disseminated into both animals and the environment.
The localisation of resistance genes on plasmids largely favors their ability to be readily transmitted in an inter- or intra-species manner by horizontal transfer [6,7,8,9,10,11,12]. This is very often the case for extended-spectrum beta-lactamase (ESBL) determinants (mostly blaCTX-M genes) and the plasmid-borne colistin-resistance genes belonging to the mcr gene family. For example, the mcr-1 gene has been nearly exclusively found on plasmids from the incompatibility (Inc) groups IncX4, IncI2, and IncHI2 [13]. In contrast, blaCTX-M genes have been detected on a much larger variety of plasmids, IncF and IncI1 being among the most frequently identified ones [13].
Food-producing animals are an important reservoir of AMR, having a high impact on the whole ecosystem due to their place at the human–animal–environment interface. AMR can disseminate through the food chain, habitat sharing or the spreading of farm waste as landfill [6,14,15]. Sheep production, with an estimated 1 billion animals reared worldwide, is highly important in the livestock field because production is intended for both the food and textile industry [16]. The problem of AMR affects sheep production, just as it does all other farming systems. However, even though a few studies reported resistant bacteria in sheep, including in Portugal [17,18,19,20,21,22,23,24,25], this sector has been much less investigated than bovine, pig, or poultry farming.
In this context, and with a One Health perspective, our study aimed firstly at evaluating the presence of ESC- and colistin-resistant E. coli in healthy sheep from Portugal, and secondly at characterizing the collected isolates.
2. Results
2.1. ESBL-Producing Escherichia coli
2.1.1. Selection and Resistance Profile
ESC-resistant E. coli were identified in 19/21 animals (90.5%) on this farm. One to nine non-duplicate ESC-resistant bacteria were found per faecal sample; 62 isolates were obtained in total (Supplementary Materials Table S1). All identified bacteria presented the typical synergy of extended-spectrum-beta-lactamase (ESBL) production and were multi-drug resistant (MDR) according to the definition by Magiorakos et al. [26], i.e., resistance to three or more antibiotic categories, where beta-lactams are divided into six different categories. Only three isolates presented no resistance to non-beta-lactam antibiotics, while 11 isolates were resistant to only one non-beta-lactam antibiotic (tetracycline, n = 8; sulfamethoxazole-trimethoprim, n = 3). The most frequent associated resistances were to tetracycline (53/62, 85.5%) and sulfamethoxazole-trimethoprim (61.3%), followed by resistances to ciprofloxacin (27.4%), gentamicin (17.7%), nitrofurantoin (8.1%), and chloramphenicol (4.8%) (Figure 1). No resistance was observed for meropenem, tigecycline, and fosfomycin.
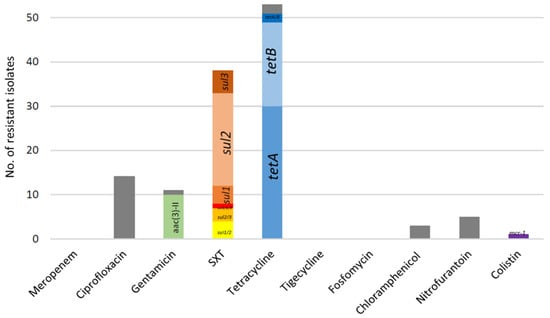
Figure 1.
Resistances associated to the 62 ESBL-producing E. coli from sheep from Portugal. In grey are resistance phenotypes for which the responsible gene was not looked for or not identified by PCR. Genes conferring low-level resistance to ciprofloxacin (qnr gene family and aac(6′)Ib-cr) were not included here since chromosomal mutations in the gyrA and parC genes were not looked for. SXT: sulfamethoxazole-trimethoprim.
2.1.2. Beta-Lactamases and Accessory Resistance Genes
The ESBL phenotype was largely due to the presence of blaCTX-M genes (93.5%), and only five blaSHV-12 genes were identified (including one concomitantly found with a blaCTX-M-32 gene). Among blaCTX-M genes, five variants were identified (Table 1), with a predominance of blaCTX-M-32 (41.9%) and blaCTX-M-15 (40.3%). A diversity of other resistance genes was detected (Figure 1 and Table S1), such as aac(6′)Ib-cr (35.5%), qnrS (3.2%), aac(3′)-II (16.1%), aph(3′)-III (4.8%), tetA (51.6%), tetB (33.9%), sul1 (19.3%), sul2 (43.5%), and sul3 (16.1%). The identified genes were coherent with the observed phenotypes.

Table 1.
Characteristics of all ESBL-producing isolates collected from sheep in Portugal.
2.1.3. Plasmid Content
Sixteen replicon types were present in the 62 ESBL-producing isolates, namely FII (n = 32) (including 3 FIA and 32 FIB), I1γ (n = 27), I1α (n = 13), HI1 (n = 3), X1 (n = 2), X2 (n = 19), X3 (n = 1), K (n = 2), L (n = 35), Y (n = 23), P (n = 2), B/O (n = 1), N (n = 8), and R (n = 1).
The 25 CTX-M-15-producing E. coli frequently carried the IncFIB (n = 21), IncY (n = 19), IncL (n = 19), IncFII (n = 18), and IncX2 (n = 17) replicon types, while the 26 CTX-M-32 producers (including the CTX-M-32 + SHV-12 producer) carried IncL (n = 15), IncI1γ (n = 13), IncI1α (n = 7), and IncFII (n = 7). The CTX-M-1- and SHV-12 producers mostly carried the IncI1γ replicon, which was present in three and five isolates, respectively.
2.1.4. Typing and Clonal Relation of ESBL-Producing E. coli
The phylogenetic analysis showed the absence of phylogroup B2 and the predominance of group A (n = 46), followed by B1 (n = 15) and D (n = 1). A high number of pulsed-field gel electrophoresis (PFGE) patterns (n = 33) was observed (Table 1 and Supplementary Materials Figure S1), highlighting a non-clonal population in this farm. Most PFGE profiles were singletons, but seven clusters (named F, H, L, S, AC, AE, and AF) encompassed clonal isolates identified in 2 to 14 different animals.
2.2. Detection of One mcr-1-Positive E. coli
The presence of the mcr-1 gene in the CTX-M-1-producing E. coli Ov11 was confirmed by Sanger sequencing. This isolate presented a colistin minimal inhibitory concentration (MIC) of 4 mg/mL, in coherence with the presence of the mcr-1 gene. In addition to colistin- and ESC resistance, Ov11 presented resistance to nitrofurantoin and an intermediate profile to ciprofloxacin and gentamicin. This intermediate profile can be explained by the presence of aac(6′)Ib-cr gene. Ov11 belonged to the phylogroup B1, had a unique PFGE pattern, and carried the IncI1γ and IncX1 plasmids.
3. Discussion
ESBL-producing Enterobacterales are a serious threat to human health according to the Centers for Disease Control and Prevention (CDC), with high impact on hospitalisations and healthcare costs [27]. ESBL-producing E. coli broke the barriers of the healthcare settings a long time ago and are now present in all compartments of the ecosystem, so that they are often sought as a marker of AMR burden [6,11,28]. While ESBL-producing E. coli have been recurrently reported in the poultry, pig, and bovine sectors, sheep farming has been under much less scrutiny. ESBL-producing E. coli have been reported in sheep in Chile, Pakistan, Switzerland, Tanzania, Turkey, and the United Kingdom [20,21,22,23,24,25], with proportions ranging from 1.5% to 11.1%. The presence of CTX-M-1- and CTX-M-32-producing E. coli in sheep was reported once from the centre of Portugal [19].
Here, we report the massive colonisation (90.5%) of sheep in one Portuguese farm. This is a surprisingly high proportion since animals were reared in an extensive farming system, being led to pastures each day. It was unfortunately not possible to trace the potential use of antibiotics. Of note, all but one E. coli belonged to the phylogroup A and B1, which are considered as commensals. Most of the animals presented more than one ESBL-producing E. coli strain, with up to nine different isolates in the same sheep. In certain cases, the presence of the same ESBL gene suggested its spread in the E. coli gut population of the sheep through horizontal gene transfer. However, different ESBL genes were also identified, proving that one animal can serve as a host for several ESC-resistance genes. PFGE analysis showed the dissemination of a few successful clones between animals, as particularly exemplified by the occurrence of the same PFGE pattern in 14 different animals. In parallel, a large diversity of ESBL-producing E. coli was also evidenced, suggesting plasmidic transmission among different genetic backgrounds. All these parameters undoubtedly contributed to the epidemiological success of ESBLs in that farm.
Fifty-nine (95.2%) isolates also displayed resistance phenotypes to non-beta-lactam antibiotics, so ESC resistance can be vertically and/or horizontally selected by the use of other classes of antibiotics. This is especially true for antibiotics such as tetracycline and sulfamethoxazole-trimethoprim, which are largely used in veterinary medicine, and which are also the ones for which we have identified here the greatest number of resistance phenotypes. The limitation of our study lies in the fact that only one farm was sampled, and our results are thus show the urgent need for a tighter survey in this sector.
As with the single report in Portugal, we identified blaCTX-M-1 and blaCTX-M-32 in sheep, but we also identified the blaCTX-M-15, blaCTX-M-14, blaCTX-M-98, and blaSHV-12 genes. CTX-M-15 is the most prevalent ESBL worldwide and particularly in humans in Europe [11,29,30]. Nevertheless, the presence of CTX-M-15 in food-producing animals is no longer an exception, even though CTX-M-1 is still more prevalent in animals [28]. In sheep, CTX-M-15 has been reported only in Switzerland, Tanzania, Turkey, and the United Kingdom [20,21,22,24]. The CTX-M-32 enzyme has emerged in the last years in the food-producing sector [19,28,31,32], and our study thus confirms its importance in livestock.
Among the 25 CTX-M-15-producing E. coli isolates in our study, 22 harboured IncF plasmids. IncF plasmids are known to be very successful vectors of the blaCTX-M-15 genes (an important example is their broad occurrence in ST131 clones) and have disseminated worldwide in the human, animal, and environmental sectors [13,33]. On the contrary, the blaCTX-M-32 genes were often found on IncN plasmids, which were identified in only 5/26 of our isolates [34,35]. However, in this study, the characterisation of plasmids remains to be performed and only poor information on the ESBL-carrying plasmid can be inferred from the replicon content.
The plasmid-mediated colistin-resistance gene mcr-1 has first been reported in food-producing animals worldwide [10,36]. Colistin is considered a last-resort option for human infections caused by carbapenem-resistant Gram-negative microorganisms, and is, for this reason, classified by the World Health Organisation (WHO), together with other antibiotics such as last-generation cephalosporins and fluoroquinolones, as a critically important antibiotic for human medicine [12,37]. Fortunately, mcr-1 genes are still rare in both hospital settings and the community worldwide, thus reinforcing the importance of their surveillance to avoid any emergence in humans. The mcr-1 gene has been described from various livestock sources (cattle, pigs, poultry, fish, milk, cheese, and eggs) as well as from pets and wild animals, [7,9,36,38,39,40,41,42,43], but our study is its first description in sheep. The Ov11 strain did not contain the main replicons (IncX4, IncHI2, and IncI2) usually described as carrying the mcr-1 gene, so a chromosomal location cannot be excluded. The surveillance and monitoring of mcr-1 remains essential in a One Health approach of AMR, to avoid its spread in a sector that presents high proportions of ESBL-producing E. coli but still very few resistances to colistin [14].
The importance of food-producing animals in AMR spread cannot be overlooked, since their role as a reservoir has been recurrently demonstrated [5,28]. In line with the results presented here, it is essential and urgent to implement holistic approaches to understand the drivers, dynamics, and epidemiology of AMR in all food-producing systems [4,44].
4. Materials and Methods
4.1. Sampling Scheme
In 2016, 21 faecal samples were collected from one farm in southern Portugal. Animals were reared in an extensive farming system and were led each day to the pastures. Fresh animal faeces were collected in sterile containers and transported at 4 °C to the laboratory, where they were stored at −20 °C for further characterisation.
4.2. Bacterial Isolation and Identification
Samples were thawed and 2 g of each sample of faeces was inoculated into 40 mL of either Tryptic Soy Broth (TSB, Liofilchem, Roseto degli Abruzzi, Italy) or TSB containing a cefotaxime 30 µg disk (Liofilchem, Roseto degli Abruzzi, Italy) and incubated for 24 h at 37 °C. A volume of 100 µL of each enrichment step was plated on MacConkey Agar (MAC, Liofilchem, Roseto degli Abruzzi, Italy) supplemented with cefotaxime (2 µg/mL). One colony per morphology was picked from selective media for further analysis. Samples were plated on antibiotic-free MAC as a growth control. One colony per morphology was picked and plated onto CHROMagar Orientation (CHROMagar, Paris, France) for presumptive identification, and identification was further confirmed by API 20E galleries (Biomérieux, Marcy l’Etoile, France). All isolates were stored in TSB + 20% glycerol for further characterisation.
4.3. Antimicrobial Susceptibility Testing and Identification
A total of 18 antibiotics were used in the susceptibility test, by disk diffusion, with amoxicillin, amoxicillin and clavulanic acid, cefoxitin, cefotaxime, ceftazidime, ceftiofur, cefepime, ceftaroline, aztreonam, meropenem, ciprofloxacin, gentamicin, sulfamethoxazole-trimethoprim, tigecycline, tetracycline, fosfomycin, chloramphenicol, and nitrofurantoin (Liofilchem, Roseto degli Abruzzi, Italy). The interpretation was performed according to CLSI [45]. Tigecycline resistance was interpreted according to EUCAST [46]. E. coli ATCC 25922 was used as quality control. In accordance with EUCAST standards, the minimal inhibitory concentration (MIC) for colistin was determined by the broth microdilution method [46]. MDR profiles were determined according to standard criteria [26]. ESBL detection was performed using the double-disk synergy test (DDST) [45].
4.4. Antimicrobial Resistance Genes
The genes mcr-1 to -5 were screened by PCR for all resistant isolates obtained [47]. ESBL-producing isolates were screened for the presence of blaTEM, blaSHV, blaOXA, and blaCTX-M (groups 1, 2, 8, 9, and 25) by PCR [48,49]. Isolates with positive amplifications were subjected to Sanger sequencing (Genewiz, Bishop’s Stortford, UK). PCR assays were performed using Super Hot Master Mix (Bioron, Römerberg, Germany); the primers used are listed in Supplementary Materials Table S2.
PCR assays were also performed to determine the presence of other accessory resistance genes: qnrA, qnrB, qnrS, aac(6′)-Ib-cr, aac(3)-II, aac(3)-IV, ant(2″)-I, aph(3′)-I, aph(3′)-II, aph(3′)-III, sul1, sul2, sul3, tetA, and tetB (Table S2).
4.5. E. coli Characterisation and Plasmid Content
Phylogenetic groups were detected by PCR as previously described [50]. Pulsed-field gel electrophoresis (PFGE) was performed on all ESBL-producing E. coli isolates using the restriction enzyme XbaI (Bioron, Römerberg, Germany) [51]. DNA fingerprints were analyzed and the dendrogram of patterns was made using the Dice correlation coefficient, with tolerance and optimisation set at 0.5% and 1%, respectively (BioNumerics, Sint-Martens-Latem, Belgium). Replicon types were detected using the PBRT 2.0 kit (Diatheva, Fano, Italy).
5. Conclusions
We showed the massive colonisation of sheep by ESBL-producing E. coli in one farm in the south of Portugal. Clonal dissemination was evidenced, as well as the circulation of a wide variety of ESBL-producing E. coli clones, highlighting a very dynamic situation. The mcr-1 gene was also identified in this farm, and for the first time in sheep. Our results proved once more the essential role of monitoring AMR dissemination in all sectors of the One Health continuum, in order to avoid the selection and emergence of resistant pathogens, which is of strategic importance for human health.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antibiotics10111403/s1, Figure S1: Clonal relation of 62 ESBL-producing E. coli from sheep from Portugal. Table S1: Characterization of 62 ESBL-producing E. coli from sheep from Portugal. Table S2: List of primers used in the standard PCRs assays. References [52,53,54,55,56,57,58,59,60,61,62,63,64,65] are cited in the supplementary materials.
Author Contributions
Conceptualisation, J.D.P. and H.M.N.F.; methodology, J.D.P., M.H. and H.M.N.F.; investigation, J.D.P.; resources, M.H., J.-Y.M. and H.M.N.F.; writing—original draft preparation, J.D.P.; writing—review and editing, M.H., J.-Y.M. and H.M.N.F.; supervision, H.M.N.F.; project administration, H.M.N.F.; funding acquisition, M.H., J.-Y.M. and H.M.N.F. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Applied Molecular Biosciences Unit—UCIBIO, which is financed by national funds from FCT (UIDP/04378/2020 and UIDB/04378/2020) and by the French Agency for Food, Environmental and Occupational Health and Safety (ANSES). JDP was supported by a scholarship (9152/0-13) from Coordination for the Improvement of Higher Education Personnel (CAPES); from the project EcoARUn: POCI-01-0145-FEDER-030310 funded by FEDER, through COMPETE2020-Programa Operacional Competitividade e Internacionalização (POCI), and by national funds (OE), through FCT/MCTES and by strategic funding to CESAM (UIDP/50017/2020+UIDB/50017/2020), through national funds. HMNF and JDP thank AgriFood XXI I&D&I project (NORTE-01-0145-FEDER-000041) co-financed by European Regional Development Fund, through the NORTE 2020 (Programa Operacional Regional do Norte 2014/2020).
Acknowledgments
Thank you to the Faculty of Pharmacy of the University of Porto (FFUP) and the Portuguese government for the infrastructure and resources employed in this work. Thank you also to microbiology laboratory team of FFUP and ANSES for collaboration and support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Naylor, N.R.; Atun, R.; Zhu, N.; Kulasabanathan, K.; Silva, S.; Chatterjee, A.; Knight, G.M.; Robotham, J.V. Estimating the burden of antimicrobial resistance: A systematic literature review. Antimicrob. Resist. Infect. Control. 2018, 7, 58. [Google Scholar] [CrossRef] [PubMed]
- Dadgostar, P. Antimicrobial Resistance: Implications and Costs. Infect. Drug Resist. 2019, 12, 3903–3910. [Google Scholar] [CrossRef]
- Iramiot, J.S.; Kajumbula, H.; Bazira, J.; Kansiime, C.; Asiimwe, B.B. Antimicrobial resistance at the human–animal interface in the Pastoralist Communities of Kasese District, South Western Uganda. Sci. Rep. 2020, 10, 14737. [Google Scholar] [CrossRef] [PubMed]
- Booton, R.D.; Meeyai, A.; Alhusein, N.; Buller, H.; Feil, E.; Lambert, H.; Mongkolsuk, S.; Pitchforth, E.; Reyher, K.K.; Sakcamduang, W.; et al. One Health drivers of antibacterial resistance: Quantifying the relative impacts of human, animal and environmental use and transmission. One Health 2021, 12, 100220. [Google Scholar] [CrossRef] [PubMed]
- Torres, R.T.; Carvalho, J.; Fernandes, J.; Palmeira, J.D.; Cunha, M.V.; Fonseca, C. Mapping the scientific knowledge of antimicrobial resistance in food-producing animals. One Health 2021, 13, 100324. [Google Scholar] [CrossRef]
- Palmeira, J.D.; Haenni, M.; Metayer, V.; Madec, J.Y.; Ferreira, H.M.N. Epidemic spread of IncI1/pST113 plasmid carrying the Extended-Spectrum Beta-Lactamase (ESBL) bla(CTX-M-8) gene in Escherichia coli of Brazilian cattle. Vet. Microbiol. 2020, 243, 108629. [Google Scholar] [CrossRef]
- Palmeira, J.D.; Ferreira, H.; Madec, J.Y.; Haenni, M. Draft genome of a ST443 mcr-1- and bla(CTX-M-2)-carrying Escherichia coli from cattle in Brazil. J. Glob. Antimicrob. Resist. 2018, 13, 269–270. [Google Scholar] [CrossRef]
- Dantas Palmeira, J.; Ferreira, H.; Madec, J.Y.; Haenni, M. Pandemic Escherichia coli ST648 isolate harbouring fosA3 and bla(CTX-M-8) on an IncI1/ST113 plasmid: A new successful combination for the spread of fosfomycin resistance? J. Glob. Antimicrob. Resist. 2018, 15, 254–255. [Google Scholar] [CrossRef]
- Torres, R.T.; Cunha, M.V.; Araujo, D.; Ferreira, H.; Fonseca, C.; Palmeira, J.D. Emergence of colistin resistance genes (mcr-1) in Escherichia coli among widely distributed wild ungulates. Environ. Pollut. 2021, 291, 118136. [Google Scholar] [CrossRef]
- Wang, R.; van Dorp, L.; Shaw, L.P.; Bradley, P.; Wang, Q.; Wang, X.; Jin, L.; Zhang, Q.; Liu, Y.; Rieux, A.; et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 2018, 9, 1179. [Google Scholar] [CrossRef]
- Canton, R.; Gonzalez-Alba, J.M.; Galán, J.C. CTX-M Enzymes: Origin and Diffusion. Front. Microbiol. 2012, 3, 110. [Google Scholar] [CrossRef] [PubMed]
- WHO. Critically Important Antimicrobials for Human Medicine, 6th ed.; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- Madec, J.-Y.; Haenni, M. Antimicrobial resistance plasmid reservoir in food and food-producing animals. Plasmid 2018, 99, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Baquero, F.; Coque, T.M.; Martínez, J.-L.; Aracil-Gisbert, S.; Lanza, V.F. Gene Transmission in the One Health Microbiosphere and the Channels of Antimicrobial Resistance. Front. Microbiol. 2019, 10, 2892. [Google Scholar] [CrossRef] [PubMed]
- Woolhouse, M.; Ward, M.; van Bunnik, B.; Farrar, J. Antimicrobial resistance in humans, livestock and the wider environment. Philos Trans. R. Soc. Lond B Biol. Sci. 2015, 370, 20140083. [Google Scholar] [CrossRef]
- Morris, S.T. 2—Overview of sheep production systems. In Advances in Sheep Welfare; Ferguson, D.M., Lee, C., Fisher, A., Eds.; Woodhead Publishing: Sawston, UK, 2017; pp. 19–35. [Google Scholar] [CrossRef]
- Silva, N.; Phythian, C.J.; Currie, C.; Tassi, R.; Ballingall, K.T.; Magro, G.; McNeilly, T.N.; Zadoks, R.N. Antimicrobial resistance in ovine bacteria: A sheep in wolf’s clothing? PLoS ONE 2020, 15, e0238708. [Google Scholar] [CrossRef] [PubMed]
- Scott, L.; Menzies, P.; Reid-Smith, R.J.; Avery, B.P.; McEwen, S.A.; Moon, C.S.; Berke, O. Antimicrobial resistance in fecal generic Escherichia coli and Salmonella spp. obtained from Ontario sheep flocks and associations between antimicrobial use and resistance. Can. J. Vet. Res. 2012, 76, 109–119. [Google Scholar] [PubMed]
- Ramos, S.; Igrejas, G.; Silva, N.; Jones-Dias, D.; Capelo-Martinez, J.-L.; Caniça, M.; Poeta, P. First report of CTX-M producing Escherichia coli, including the new ST2526, isolated from beef cattle and sheep in Portugal. Food Control. 2013, 31, 208–210. [Google Scholar] [CrossRef]
- Seni, J.; Falgenhauer, L.; Simeo, N.; Mirambo, M.M.; Imirzalioglu, C.; Matee, M.; Rweyemamu, M.; Chakraborty, T.; Mshana, S.E. Multiple ESBL-Producing Escherichia coli Sequence Types Carrying Quinolone and Aminoglycoside Resistance Genes Circulating in Companion and Domestic Farm Animals in Mwanza, Tanzania, Harbor Commonly Occurring Plasmids. Front. Microbiol. 2016, 7, 142. [Google Scholar] [CrossRef]
- Geser, N.; Stephan, R.; Hächler, H. Occurrence and characteristics of extended-spectrum β-lactamase (ESBL) producing Enterobacteriaceae in food producing animals, minced meat and raw milk. Bmc Vet. Res. 2012, 8, 21. [Google Scholar] [CrossRef]
- Snow, L.C.; Wearing, H.; Stephenson, B.; Teale, C.J.; Coldham, N.G. Investigation of the presence of ESBL-producing Escherichia coli in the North Wales and West Midlands areas of the UK in 2007 to 2008 using scanning surveillance. Vet. Rec. 2011, 169, 656. [Google Scholar] [CrossRef]
- Benavides, J.A.; Salgado-Caxito, M.; Opazo-Capurro, A.; González Muñoz, P.; Piñeiro, A.; Otto Medina, M.; Rivas, L.; Munita, J.; Millán, J. ESBL-Producing Escherichia coli Carrying CTX-M Genes Circulating among Livestock, Dogs, and Wild Mammals in Small-Scale Farms of Central Chile. Antibiotics 2021, 10, 510. [Google Scholar] [CrossRef]
- Pehlivanoglu, F.; Turutoglu, H.; Ozturk, D.; Yardimci, H. Molecular Characterization of ESBL-Producing Escherichia coli Isolated from Healthy Cattle and Sheep. Acta Vet. 2016, 66, 520–533. [Google Scholar] [CrossRef]
- Mehmood, A.S.Q. Phenotypic and molecular characterization of esblproducing the enterobacteriaceae from animal fecal samples in southern punjab, pakistan. Sci. Int. 2021, 33, 45–48. [Google Scholar]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States; CDC: Atlanta, GA, USA, 2019.
- Dantas Palmeira, J.; Ferreira, H.M.N. Extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae in cattle production—A threat around the world. Heliyon 2020, 6, e03206. [Google Scholar] [CrossRef] [PubMed]
- Valsdottir, F.; Elfarsdottir Jelle, A.; Gudlaugsson, O.; Hilmarsdottir, I. Long-lasting outbreak due to CTX-M-15-producing Klebsiella pneumoniae ST336 in a rehabilitation ward: Report and literature review. J. Hosp. Infect. 2017, 97, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Pasanen, T.; Jalava, J.; Horsma, J.; Salo, E.; Pakarinen, M.; Tarkka, E.; Vaara, M.; Tissari, P. An outbreak of CTX-M-15-producing Escherichia coli, Enterobacter cloacae, and Klebsiella in a children’s hospital in Finland. Scand. J. Infect. Dis. 2014, 46, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Cottell, J.L.; Kanwar, N.; Castillo-Courtade, L.; Chalmers, G.; Scott, H.M.; Norby, B.; Loneragan, G.H.; Boerlin, P. blaCTX-M-32 on an IncN plasmid in Escherichia coli from beef cattle in the United States. Antimicrob. Agents Chemother. 2013, 57, 1096–1097. [Google Scholar] [CrossRef]
- Tamang, M.D.; Nam, H.-M.; Gurung, M.; Jang, G.-C.; Kim, S.-R.; Jung, S.-C.; Park, Y.H.; Lim, S.-K. Molecular Characterization of CTX-M β-Lactamase and Associated Addiction Systems in Escherichia coli Circulating among Cattle, Farm Workers, and the Farm Environment. Appl. Environ. Microbiol. 2013, 79, 3898–3905. [Google Scholar] [CrossRef]
- Lahlaoui, H.; De Luca, F.; Maradel, S.; Ben-Haj-Khalifa, A.; Ben Hamouda, H.; Kheder, M.; Ben Moussa, M.; Rossillini, G.-M.; Docquier, J.-D. Occurrence of conjugative IncF-type plasmids harboring the blaCTX-M-15 gene in Enterobacteriaceae isolates from newborns in Tunisia. Pediatr. Res. 2015, 77, 107–110. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.-H.; Hu, Z.-Q. Epidemiology and genetics of CTX-M extended-spectrum β-lactamases in Gram-negative bacteria. Crit. Rev. Microbiol. 2013, 39, 79–101. [Google Scholar] [CrossRef]
- Novais, Â.; Cantón, R.; Moreira, R.; Peixe, L.; Baquero, F.; Coque, T.M. Emergence and Dissemination of Enterobacteriaceae Isolates Producing CTX-M-1-Like Enzymes in Spain Are Associated with IncFII (CTX-M-15) and Broad-Host-Range (CTX-M-1, -3, and -32) Plasmids. Antimicrob. Agents Chemother. 2007, 51, 796–799. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Zając, M.; Sztromwasser, P.; Bortolaia, V.; Leekitcharoenphon, P.; Cavaco, L.M.; Ziȩtek-Barszcz, A.; Hendriksen, R.S.; Wasyl, D. Occurrence and Characterization of mcr-1-Positive Escherichia coli Isolated from Food-Producing Animals in Poland, 2011–2016. Front. Microbiol. 2019, 10, 1753. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.; Urmi, U.L.; Rana, M.; Sultana, F.; Jahan, N.; Hossain, B.; Iqbal, S.; Hossain, M.M.; Mosaddek, A.S.M.; Nahar, S. High abundance of the colistin resistance gene mcr-1 in chicken gut-bacteria in Bangladesh. Sci. Rep. 2020, 10, 17292. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Fanning, S.; Gan, X.; Liu, C.; Nguyen, S.; Wang, M.; Wang, W.; Jiang, T.; Xu, J.; Li, F. Salmonella harbouring the mcr-1 gene isolated from food in China between 2012 and 2016. J. Antimicrob. Chemother. 2019, 74, 826–828. [Google Scholar] [CrossRef]
- Lv, L.; Cao, Y.; Yu, P.; Huang, R.; Wang, J.; Wen, Q.; Zhi, C.; Zhang, Q.; Liu, J.-H. Detection of mcr-1 Gene among Escherichia coli Isolates from Farmed Fish and Characterization of mcr-1-Bearing IncP Plasmids. Antimicrob. Agents Chemother. 2018, 62, e02378-17. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Ali, T.; Gao, J.; Ur Rahman, S.; Yu, D.; Barkema, H.W.; Huo, W.; Xu, S.; Shi, Y.; Kastelic, J.P.; et al. Co-Occurrence of Plasmid-Mediated Colistin Resistance (mcr-1) and Extended-Spectrum beta-Lactamase Encoding Genes in Escherichia coli from Bovine Mastitic Milk in China. Microb. Drug Resist. 2020, 26, 685–696. [Google Scholar] [CrossRef]
- Nagy, Á.; Székelyhidi, R.; Hanczné Lakatos, E.; Kapcsándi, V. Review on the occurrence of the mcr-1 gene causing colistin resistance in cow’s milk and dairy products. Heliyon 2021, 7, e06800. [Google Scholar] [CrossRef]
- Lei, L.; Wang, Y.; Schwarz, S.; Walsh, T.R.; Ou, Y.; Wu, Y.; Li, M.; Shen, Z. mcr-1 in Enterobacteriaceae from Companion Animals, Beijing, China, 2012–2016. Emerg. Infect. Dis. 2017, 23, 710–711. [Google Scholar] [CrossRef]
- FAO. Drivers, Dynamics and Epidemiology of Antimicrobial Resistance in Animal Production; FAO: Rome, Italy, 2016. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 27th ed.; CLSI: Annapolis Junction, MD, USA, 2017. [Google Scholar]
- EUCAST. Breakpoint Tables for Interpretation of MICs and Zone Diameters; EUCAST: Vaxjo, Sweden, 2021. [Google Scholar]
- Rebelo, A.R.; Bortolaia, V.; Kjeldgaard, J.S.; Pedersen, S.K.; Leekitcharoenphon, P.; Hansen, I.M.; Guerra, B.; Malorny, B.; Borowiak, M.; Hammerl, J.A.; et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Eurosurveillance 2018, 23, 17-00672. [Google Scholar] [CrossRef] [PubMed]
- Woodford, N.; Fagan, E.J.; Ellington, M.J. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J. Antimicrob. Chemother. 2005, 57, 154–155. [Google Scholar] [CrossRef] [PubMed]
- Dallenne, C.; Da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef]
- Saidani, M.; Messadi, L.; Chaouechi, A.; Tabib, I.; Saras, E.; Soudani, A.; Daaloul-Jedidi, M.; Mamlouk, A.; Ben Chehida, F.; Chakroun, C.; et al. High Genetic Diversity of Enterobacteriaceae Clones and Plasmids Disseminating Resistance to Extended-Spectrum Cephalosporins and Colistin in Healthy Chicken in Tunisia. Microb. Drug Resist. 2019, 25, 1507–1513. [Google Scholar] [CrossRef] [PubMed]
- Cavaco, L.M.; Frimodt-Møller, N.; Hasman, H.; Guardabassi, L.; Nielsen, L.; Aarestrup, F.M. Prevalence of quinolone resistance mechanisms and associations to minimum inhibitory concentrations in quinolone-resistant Escherichia coli isolated from humans and swine in Denmark. Microb. Drug Resist. 2008, 14, 163–169. [Google Scholar] [CrossRef]
- Cattoir, V.; Weill, F.X.; Poirel, L.; Fabre, L.; Soussy, C.J.; Nordmann, P. Prevalence of qnr genes in Salmonella in France. J. Antimicrob. Chemother. 2007, 59, 751–754. [Google Scholar] [CrossRef]
- Park, C.H.; Robicsek, A.; Jacoby, G.A.; Sahm, D.; Hooper, D.C. Prevalence in the United States of aac(6’)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob. Agents Chemother. 2006, 50, 3953–3955. [Google Scholar] [CrossRef]
- Vliegenthart, J.S.; Ketelaar-van Gaalen, P.A.; van de Klundert, J.A. Nucleotide sequence of the aacC2 gene, a gentamicin resistance determinant involved in a hospital epidemic of multiply resistant members of the family Enterobacteriaceae. Antimicrob. Agents Chemother. 1989, 33, 1153–1159. [Google Scholar] [CrossRef]
- Bräu, B.; Pilz, U.; Piepersberg, W. Genes for gentamicin-(3)-N-acetyltransferases III and IV: I. Nucleotide sequence of the AAC(3)-IV gene and possible involvement of an IS140 element in its expression. Mol. Gen. Genet. 1984, 193, 179–187. [Google Scholar] [CrossRef]
- Cameron, F.H.; Groot Obbink, D.J.; Ackerman, V.P.; Hall, R.M. Nucleotide sequence of the AAD(2″) aminoglycoside adenylyltransferase determinant aadB. Evolutionary relationship of this region with those surrounding aadA in R538-1 and dhfrII in R388. Nucleic. Acids Res. 1986, 14, 8625–8635. [Google Scholar] [CrossRef] [PubMed]
- Melano, R.; Corso, A.; Petroni, A.; Centrón, D.; Orman, B.; Pereyra, A.; Moreno, N.; Galas, M. Multiple antibiotic-resistance mechanisms including a novel combination of extended-spectrum beta-lactamases in a Klebsiella pneumoniae clinical strain isolated in Argentina. J. Antimicrob. Chemother. 2003, 52, 36–42. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Aarestrup, F.M.; Lertworapreecha, M.; Evans, M.C.; Bangtrakulnonth, A.; Chalermchaikit, T.; Hendriksen, R.S.; Wegener, H.C. Antimicrobial susceptibility and occurrence of resistance genes among Salmonella enterica serovar Weltevreden from different countries. J. Antimicrob. Chemother. 2003, 52, 715–718. [Google Scholar] [CrossRef] [PubMed]
- Oka, A.; Sugisaki, H.; Takanami, M. Nucleotide sequence of the kanamycin resistance transposon Tn903. J. Mol. Biol. 1981, 147, 217–226. [Google Scholar] [CrossRef]
- Waters, S.H.; Rogowsky, P.; Grinsted, J.; Altenbuchner, J.; Schmitt, R. The tetracycline resistance determinants of RP1 and Tn1721: Nucleotide sequence analysis. Nucleic. Acids Res. 1983, 11, 6089–6105. [Google Scholar] [CrossRef]
- Sengeløv, G.; Agersø, Y.; Halling-Sørensen, B.; Baloda, S.B.; Andersen, J.S.; Jensen, L.B. Bacterial antibiotic resistance levels in Danish farmland as a result of treatment with pig manure slurry. Environ. Int. 2003, 28, 587–595. [Google Scholar] [CrossRef]
- Rahmani, M.; Peighambari, S.M.; Svendsen, C.A.; Cavaco, L.M.; Agersø, Y.; Hendriksen, R.S. Molecular clonality and antimicrobial resistance in Salmonella entericaserovars Enteritidis and Infantis from broilers in three Northern regions of Iran. BMC Vet. Res. 2013, 9, 66. [Google Scholar] [CrossRef]
- Perreten, V.; Boerlin, P. A new sulfonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob. Agents Chemother. 2003, 47, 1169–1172. [Google Scholar] [CrossRef]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).