Bacteriophages with Potential to Inactivate Aeromonas hydrophila in Cockles: In Vitro and In Vivo Preliminary Studies
Abstract
1. Introduction
2. Results
2.1. Phage Isolation and Enrichment
2.2. Virion Morphology
2.3. Phage Host Range Determination and Efficiency of Plating (EOP) Analysis
2.4. One-Step Growth Curve Analysis
2.5. Bacterial Killing Curves
2.5.1. Bacterial Killing Curves Using Single Phage Suspensions
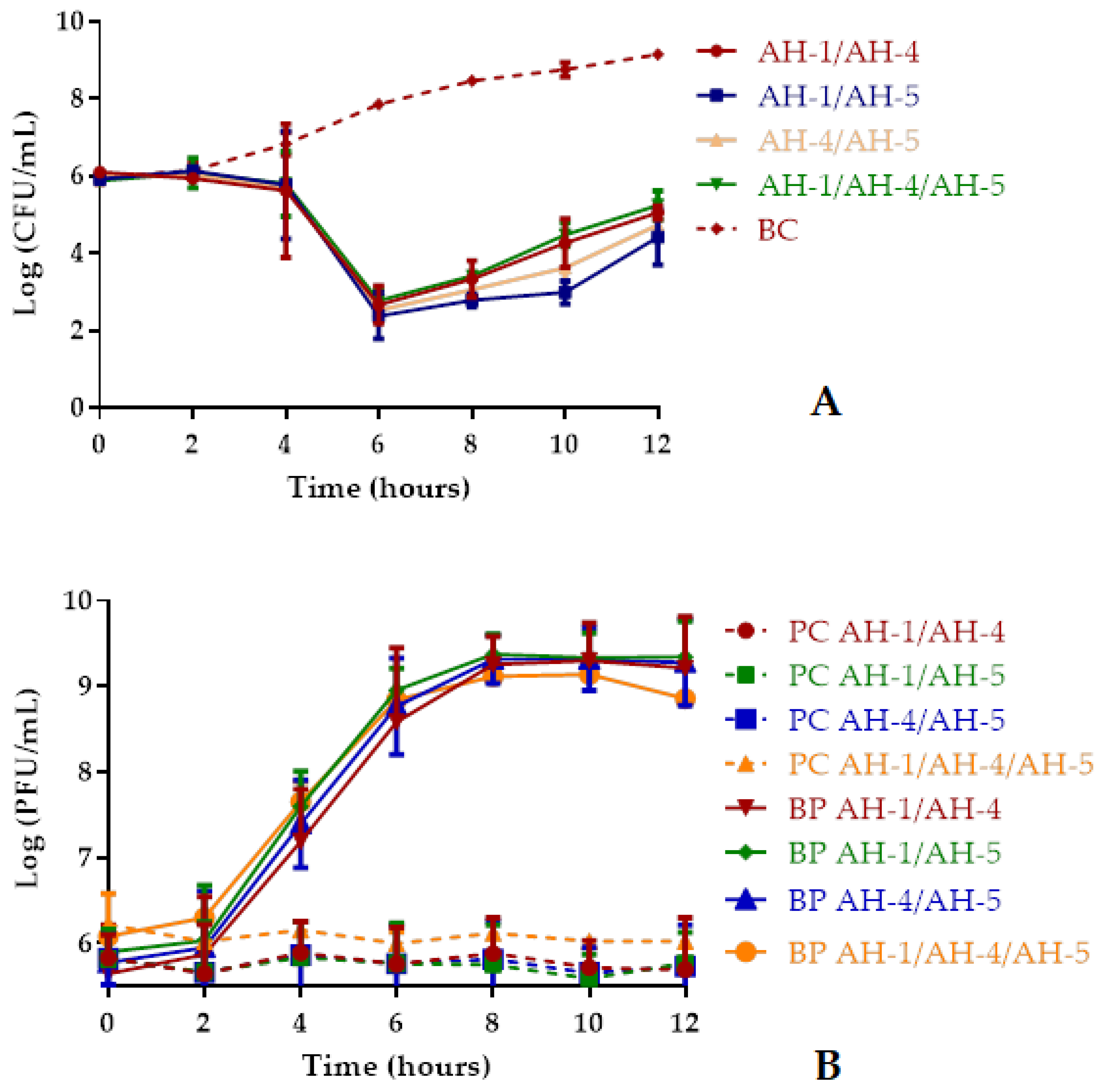
2.5.2. Bacterial Killing Curves Using Phage Cocktails
2.6. Determination of the Rate of Emergence of Phage-Resistant Mutants
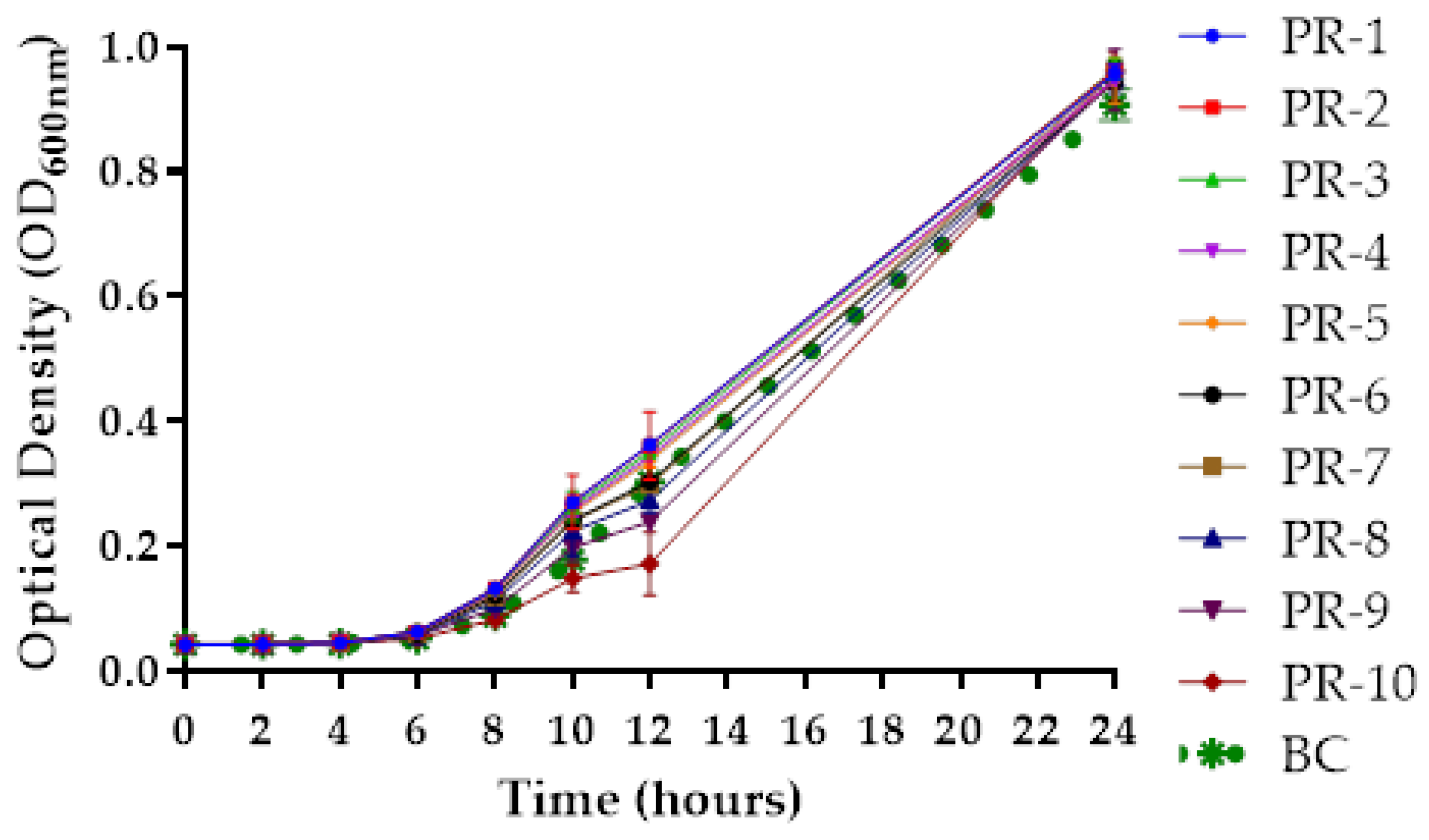
2.7. Growth Curve of Phage-Resistant A. hydrophila Strains
2.8. Bacterial Killing Curves at Different MOIs
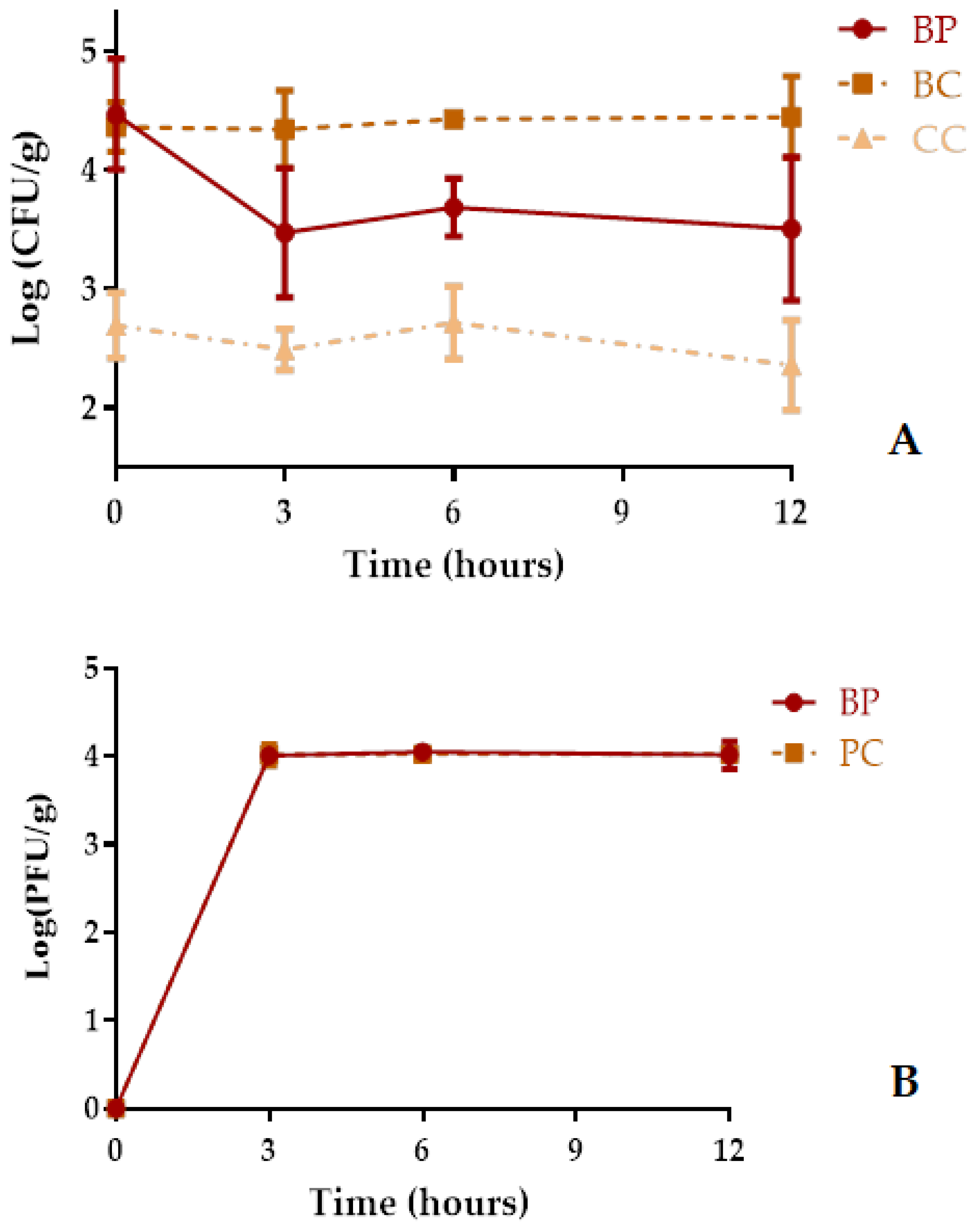
2.9. Phage Application during Cerastoderma edule Depuration in a Static System
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Phage Isolation and Purification
4.3. Electron Microscope Examination
4.4. Phage Host Range and Efficiency of Plating (EOP) Analysis
4.5. One Step Growth Assays
4.6. Bacterial Kill Curves Using Single-Phage Suspensions and Phage Cocktails
4.7. Rate of Emergence of Bacterial Mutants Resistant to Phages
4.8. Growth Curve of Phage-Resistant A. hydrophila Strains
4.9. Bacterial Kill Curves at Different MOIs
4.10. Phage Application during Cerastoderma edule Depuration in Static Seawater
4.10.1. Collection of C. edule Samples
4.10.2. Depuration of Artificially Contaminated Cockles in the Presence of Phage AH-1 in a Static System
4.11. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oliveira, J.; Cunha, A.; Castilho, F.; Romalde, J.; Pereira, M. Microbial contamination and purification of bivalve shellfish: Crucial aspects in monitoring and future perspectives—A mini-review. Food Control. 2011, 22, 805–816. [Google Scholar] [CrossRef]
- FAO. The State of World Fisheries and Aquaculture 2020. Sustainability in Action; FAO: Rome, Italy, 2020. [Google Scholar]
- Pereira, C.; Costa, P.; Duarte, J.; Balcão, V.M.; Almeida, A. Phage therapy as a potential approach in the biocontrol of patho-genic bacteria associated with shellfish consumption. Int. J. Food Microbiol. 2021, 338, 108995. [Google Scholar] [CrossRef] [PubMed]
- Zannella, C.; Mosca, F.; Mariani, F.; Franci, G.; Folliero, V.; Galdiero, M.; Tiscar, P.G.; Galdiero, M. Microbial Diseases of Bivalve Mollusks: Infections, Immunology and Antimicrobial Defense. Mar. Drugs 2017, 15, 182. [Google Scholar] [CrossRef] [PubMed]
- Fernández, E.; Alvarez-Salgado, X.A.; Beiras, R.; Ovejero, A.; Méndez, G. Coexistence of urban uses and shellfish production in an upwelling-driven, highly productive marine environment: The case of the Ría de Vigo (Galicia, Spain). Reg. Stud. Mar. Sci. 2016, 8, 362–370. [Google Scholar] [CrossRef]
- Butt, A.A.; Aldridge, K.E.; Sanders, C. V Infections related to the ingestion of seafood Part I: Viral and bacterial infections. Lancet Infect. Dis. 2004, 4, 201–212. [Google Scholar] [CrossRef]
- De Silva, B.C.J.; Hossain, S.; Dahanayake, P.S.; Lee, D.-W.; Wickramanayake, M.V.K.S.; Heo, G.-J. Multi-drug resistant mes-ophilic aeromonads isolated from marketed scallops (Patinopecten yessoensis) harboring resistance genes. Fish. Aquat. Life 2020, 28, 1–10. [Google Scholar] [CrossRef]
- Silva, A.C.M.d.M.; Nascimento, D.L.; Machado, R.Z.; Costa, F.N. Caracterização de Aeromonas spp isoladas de amostras de ostras e água por método microbiológico e molecular. Ciência Anim. Bras. 2014, 15, 362–368. [Google Scholar] [CrossRef]
- Chittick, B.; Stoskopf, M.; Law, M.; Overstreet, R.; Levine, J. Evaluation of potential health risks to Eastern Elliptio (Elliptio complanata) (Mollusca: Bivalvia: Unionida: Unionidae) and implications for sympatric endangered freshwater mussel species. J. Aquat. Ecosyst. Stress Recover. 2001, 9, 35–42. [Google Scholar] [CrossRef]
- Ribeiro, E.B.; Noleto, K.S.; de Oliveira, S.R.S.; de Jesus, W.B.; de Sousa Serra, I.M.R.; da Silva de Almeida, Z.; de Oliveira Mota Andrade, T.; de Araújo Soares, R.; Antonio, Í.G.; Santos, D.M.S.; et al. Biomarkers (glutathione S-transferase and catalase) and microorganisms in soft tissues of Crassostrea rhizophorae to assess contamination of seafood in Brazil. Mar. Pollut. Bull. 2020, 158, 111348. [Google Scholar] [CrossRef]
- Krovacek, K.; Dumontet, S.; Eriksson, E.; Baloda, S.B. Isolation, and virulence profiles, of Aeromonas hydrophila implicated in an outbreak of food poisoning in Sweden. Microbiol. Immunol. 1995, 39, 655–661. [Google Scholar] [CrossRef]
- Grim, C.J.; Kozlova, E.V.; Ponnusamy, D.; Fitts, E.C.; Sha, J.; Kirtley, M.L.; van Lier, C.J.; Tiner, B.L.; Erova, T.E.; Joseph, S.J.; et al. Functional genomic characterization of virulence factors from necrotizing fasciitis-causing strains of Aeromonas hydrophila. Appl. Environ. Microbiol. 2014, 80, 4162–4183. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Albores, A.; Lopez-Santamarina, A.; Rodriguez, J.A.; Ibarra, I.S.; Mondragón, A.D.C.; Miranda, J.M.; Lamas, A.; Cepeda, A. Complementary methods to improve the depuration of bivalves: A review. Foods 2020, 9, 129. [Google Scholar] [CrossRef] [PubMed]
- Polo, D.; Álvarez, C.; Díez, J.; Darriba, S.; Longa, Á.; Romalde, J.L. Viral elimination during commercial depuration of shellfish. Food Control 2014, 43, 206–212. [Google Scholar] [CrossRef]
- Anjana, S.; Abhilash, S.; Varghese, B.; Sabu, S.; Sunooj, K.; Xavier, K.M. Performance evaluation of ultra violet assisted vertical re-circulating depuration system on microbial, heavy metal reduction and composition of black clam (Villorita cyprinoides). LWT 2021, 138, 110628. [Google Scholar] [CrossRef]
- Lee, R.; Lovatelli, A.; Ababouch, L. Bivalve Depuration: Fundamental and Practical Aspects; FAO Fisheries Technical Paper; FAO: Rome, Italy, 2008; ISBN 978-92-5-106006-3. [Google Scholar]
- Groubert, T.N.; Oliver, J.D. Interaction of Vibrio vulnificus and the Eastern Oyster, Crassostrea virginica. J. Food Prot. 1994, 57, 224–228. [Google Scholar] [CrossRef]
- Martínez, O.; Rodríguez-Calleja, J.M.; Santos, J.; Otero, A.; García-López, M.L. Foodborne and indicator bacteria in farmed molluscan shellfish before and after depuration. J. Food Prot. 2009, 72, 1443–1449. [Google Scholar] [CrossRef]
- Rong, R.; Lin, H.; Wang, J.; Khan, M.N.; Li, M. Reductions of Vibrio parahaemolyticus in oysters after bacteriophage application during depuration. Aquaculture 2014, 418-419, 171–176. [Google Scholar] [CrossRef]
- Tamplin, M.L.; Capers, G.M. Persistence of Vibrio vulnificus in tissues of Gulf Coast oysters, Crassostrea virginica, exposed to seawater disinfected with UV light. Appl. Environ. Microbiol. 1992, 58, 1506–1510. [Google Scholar] [CrossRef]
- Vezzulli, L.; Stagnaro, L.; Grande, C.; Tassistro, G.; Canesi, L.; Pruzzo, C. Comparative 16SrDNA Gene-based microbiota profiles of the Pacific oyster (Crassostrea gigas) and the Mediterranean mussel (Mytilus galloprovincialis) from a shellfish farm (Ligurian Sea, Italy). Microb. Ecol. 2017, 75, 495–504. [Google Scholar] [CrossRef]
- WHO; Lees, D.; Younger, A.; Dore, B. World Health Organization (WHO) Depuration and Relaying; Rees, G., Pond, K., Kay, D., Bartram, J., Santo Domingo, J., Eds.; IWA Publishing: London, UK, 2010; ISBN 9781843392255. [Google Scholar]
- Clokie, M.R.J.; Millard, A.D.; Letarov, A.V.; Heaphy, S. Phages in nature. Bacteriophage 2011, 1, 31–45. [Google Scholar] [CrossRef]
- Galarce, N.E.; Bravo, J.L.; Robeson, J.P.; Borie, C.F. Bacteriophage cocktail reduces Salmonella enterica serovar Enteritidis counts in raw and smoked salmon tissues. Rev. Argent. Microbiol. 2014, 46, 333–337. [Google Scholar] [CrossRef]
- Almeida, A.; Cunha, Â.; Gomes, N.C.; Alves, E.; Costa, L.; Faustino, M.A. Phage therapy and photodynamic therapy: Low environmental impact approaches to inactivate microorganisms in fish farming plants. Mar. Drugs 2009, 7, 268–313. [Google Scholar] [CrossRef]
- Jun, J.W.; Kim, H.J.; Kil Yun, S.; Chai, J.Y.; Park, S.C. Eating oysters without risk of vibriosis: Application of a bacteriophage against Vibrio parahaemolyticus in oysters. Int. J. Food Microbiol. 2014, 188, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.; Moreirinha, C.; Rocha, R.J.; Calado, R.; Romalde, J.L.; Nunes, M.L.; Almeida, A. Application of bacteriophages during depuration reduces the load of Salmonella typhimurium in cockles. Food Res. Int. 2016, 90, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.; Moreirinha, C.; Teles, L.; Rocha, R.J.; Calado, R.; Romalde, J.L.; Nunes, M.L.; Almeida, A. Application of phage therapy during bivalve depuration improves Escherichia coli decontamination. Food Microbiol. 2017, 61, 102–112. [Google Scholar] [CrossRef]
- Akmal, M.; Rahimi-Midani, A.; Hafeez-Ur-Rehman, M.; Hussain, A.; Choi, T.-J. Isolation, characterization, and application of a bacteriophage infecting the fish pathogen Aeromonas hydrophila. Pathogens 2020, 9, 215. [Google Scholar] [CrossRef]
- Aberoum, A.; Jooyandeh, H. A Review on occurrence and characterization of the Aeromonas species from marine fishes. World J. Fish Mar. Sci. 2010, 2, 519–523. [Google Scholar]
- ChiHsin, H.; ChongYi, L.; JongKang, L.; ChanShing, L. Control of the eel (Anguilla japonica) pathogens, Aeromonas hydrophila and Edwardsiella tarda, by bacteriophages. J. Fish. Soc. Taiwan 2000, 27, 21–31. [Google Scholar]
- Wu, J.-L.; Lin, H.-M.; Jan, L.; Hsu, Y.-L.; Chang, L.-H. Biological control of fish bacterial pathogen, Aeromonas hydrophila, by Bbcteriophage AH 1. Fish Pathol. 1981, 15, 271–276. [Google Scholar] [CrossRef]
- Jun, J.W.; Kim, J.H.; Shin, S.P.; Han, J.E.; Chai, J.Y.; Park, S.C. Protective effects of the Aeromonas phages pAh1-C and pAh6-C against mass mortality of the cyprinid loach (Misgurnus anguillicaudatus) caused by Aeromonas hydrophila. Aquaculture 2013, 416–417, 289–295. [Google Scholar] [CrossRef]
- Hassan, S.W.; Ali, S.M.; Almisherfi, M.M. Isolation and molecular characterization of some marine Aeromonas phages: Protective effects for Nile tilapia infected with Aeromonas hydrophila. J. Pure Appl. Microbiol. 2018, 12, 1175–1185. [Google Scholar] [CrossRef]
- Hoang, H.A.; Xuan, T.T.T.; Nga, L.P.; Oanh, D.T.H. Selection of phages to control Aeromonas hydrophila—An infectious agent in striped catfish. Biocontrol Sci. 2019, 24, 23–28. [Google Scholar] [CrossRef] [PubMed]
- El-araby, D.; El-didamony, G.; Th, M.M. New approach to use phage therapy against Aeromonas hydrophila induced motile Aeromonas septicemia in Nile tilapia. Mar. Sci. Res. Dev. 2016, 6, 6–11. [Google Scholar]
- Chow, M.S.; Rouf, M.A. Isolation and partial characterization of two Aeromonas hydrophila bacteriophages. Appl. Environ. Microbiol. 1983, 45, 1670–1676. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.B.; Lin, N.T.; Tseng, Y.H.; Weng, S.F. Genomic characterization of the novel Aeromonas hydrophila phage ahp1 suggests the derivation of a new subgroup from phiKMV-Like family. PLoS ONE 2016, 11, e0162060. [Google Scholar] [CrossRef] [PubMed]
- Kabwe, M.; Brown, T.; Speirs, L.; Ku, H.; Leach, M.; Chan, H.T.; Petrovski, S.; Lock, P.; Tucci, J. Novel bacteriophages capable of disrupting biofilms from clinical strains of Aeromonas hydrophila. Front. Microbiol. 2020, 11, 194. [Google Scholar] [CrossRef]
- Le, T.S.; Nguyen, T.H.; Vo, H.P.; Doan, V.C.; Nguyen, H.L.; Tran, M.T.; Tran, T.T.; Southgate, P.C.; İpek Kurtböke, D. Protective effects of bacteriophages against Aeromonas hydrophila species causing Motile Aeromonas Septicemia (MAS) in striped catfish. Antibiotics 2018, 7, 16. [Google Scholar] [CrossRef]
- Schulz, P.; Robak, S.; Dastych, J.; Siwicki, A.K. Influence of bacteriophages cocktail on European eel (Anguilla anguilla) im-munity and survival after experimental challenge. Fish Shellfish Immunol. 2019, 84, 28–37. [Google Scholar] [CrossRef]
- Schulz, P.; Pajdak-Czaus, J.; Robak, S.; Dastych, J.; Siwicki, A.K. Bacteriophage-based cocktail modulates selected immuno-logical parameters and post-challenge survival of rainbow trout (Oncorhynchus mykiss). J. Fish Dis. 2019, 42, 1151–1160. [Google Scholar] [PubMed]
- Liu, J.; Gao, S.; Dong, Y.; Lu, C.; Liu, Y. Isolation and characterization of bacteriophages against virulent Aeromonas hydrophila. BMC Microbiol. 2020, 20, 1–13. [Google Scholar] [CrossRef]
- Pereira, C.; Moreirinha, C.; Lewicka, M.; Almeida, P.; Clemente, C.; Cunha, Â.; Delgadillo, I.; Romalde, J.; Nunes, M.L.; Al-meida, A. Bacteriophages with potential to inactivate Salmonella typhimurium: Use of single phage suspensions and phage cocktails. Virus Res. 2016, 220, 179–192. [Google Scholar] [CrossRef]
- Costa, P.; Pereira, C.; Gomes, A.T.P.C.; Almeida, A. Efficiency ofsingle phage suspensions and phage cocktail in the inactivation of Escherichia coli and Salmonella typhimurium: An in vitro preliminary study. Microorganisms 2019, 7, 94. [Google Scholar] [CrossRef]
- Pereira, C.; Silva, Y.J.; Santos, A.L.; Cunha, Â.; Gomes, N.C.M.; Almeida, A. Bacteriophages with potential for inactivation of fish pathogenic bacteria: Survival, host specificity and effect on bacterial community structure. Mar. Drugs 2011, 9, 2236–2255. [Google Scholar] [CrossRef]
- Mirzaei, K.M.; Nilsson, A.S. Isolation of phages for phage therapy: A comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS ONE 2015, 10, e0118557. [Google Scholar] [CrossRef]
- Børsheim, K. Native marine bacteriophages. FEMS Microbiol. Ecol. 1993, 102, 141–159. [Google Scholar] [CrossRef]
- Kellogg, C.; Rose, J.; Jiang, S.; Thurmond, J.; Paul, J. Genetic diversity of related vibriophages isolated from marine environments around Florida and Hawaii, USA. Mar. Ecol. Prog. Ser. 1995, 120, 89–98. [Google Scholar] [CrossRef]
- Wichels, A.; Biel, S.S.; Gelderblom, H.R.; Brinkhoff, T.; Muyzer, G.; Schütt, C. Bacteriophage diversity in the North Sea. Appl. Environ. Microbiol. 1998, 64, 4128–4133. [Google Scholar] [CrossRef]
- Mateus, L.; Costa, L.; Silva, Y.; Pereira, C.; Cunha, A.; Almeida, A. Efficiency of phage cocktails in the inactivation of Vibrio in aquaculture. Aquaculture 2014, 424-425, 167–173. [Google Scholar] [CrossRef]
- Abedon, S.T.; Culler, R.R. Optimizing bacteriophage plaque fecundity. J. Biol. 2007, 249, 582–592. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T. Lysis from without. Bacteriophage 2011, 1, 46–49. [Google Scholar] [CrossRef]
- Drulis-Kawa, Z.; Majkowska-Skrobek, G.; Maciejewska, B.; Delattre, A.-S.; Lavigne, R. Learning from bacteriophages - Advantages and limitations of phage and phage-encoded protein applications. Curr. Protein Pept. Sci. 2012, 13, 699–722. [Google Scholar] [CrossRef]
- Bull, J.J.; Gill, J.J. The habits of highly effective phages: Population dynamics as a framework for identifying therapeutic phages. Front. Microbiol. 2014, 5, 618. [Google Scholar] [CrossRef]
- Duarte, J.; Pereira, C.; Moreirinha, C.; Salvio, R.; Lopes, A.; Wang, D.; Almeida, A. New insights on phage efficacy to control Aeromonas salmonicida in aquaculture systems: An in vitro preliminary study. Aquaculture 2018, 495, 970–982. [Google Scholar] [CrossRef]
- Filippov, A.; Sergueev, K.V.; He, Y.; Huang, X.Z.; Gnade, B.T.; Mueller, A.J.; Fernandez-Prada, C.; Nikolich, M.P. Bacterio-phage-resistant mutants in Yersinia pestis: Identification of phage receptors and attenuation for mice. PLoS ONE 2011, 6, 1–11. [Google Scholar] [CrossRef]
- Pereira, S.; Pereira, C.; Santos, L.; Klumpp, J.; Almeida, A. Potential of phage cocktails in the inactivation of Enterobacter cloacae —An in vitro study in a buffer solution and in urine samples. Virus Res. 2016, 211, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.; Moreirinha, C.; Lewicka, M.; Almeida, P.; Clemente, C.; Romalde, J.L.; Nunes, M.L.; Almeida, A. Characterization and in vitro evaluation of new bacteriophages for the biocontrol of Escherichia coli. Virus Res. 2017, 227, 171–182. [Google Scholar] [CrossRef] [PubMed]
- O’Flynn, G.; Ross, R.P.; Fitzgerald, G.F.; Coffey, A. Evaluation of a cocktail of three bacteriophages for biocontrol of Escherichia coli O157:H7. Appl. Environ. Microbiol. 2004, 70, 3417–3424. [Google Scholar] [CrossRef]
- Tanji, Y.; Shimada, T.; Yoichi, M.; Miyanaga, K.; Hori, K.; Unno, H. Toward rational control of Escherichia coli O157:H7 by a phage cocktail. Appl. Microbiol. Biotechnol. 2004, 64, 270–274. [Google Scholar] [CrossRef]
- Capparelli, R.; Nocerino, N.; Iannaccone, M.; Ercolini, D.; Parlato, M.; Chiara, M.; Iannelli, D. Bacteriophage therapy of Sal-monella enterica: A fresh appraisal of bacteriophage therapy. J. Infect. Dis. 2010, 201, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Capparelli, R.; Nocerino, N.; Lanzetta, R.; Silipo, A.; Amoresano, A.; Giangrande, C.; Becker, K.; Blaiotta, G.; Evidente, A.; Cimmino, A.; et al. Bacteriophage-resistant Staphylococcus aureus mutant confers broad immunity against staphylococcal infection in mice. PLoS ONE 2010, 5, e11720. [Google Scholar] [CrossRef] [PubMed]
- León, M.; Kokkari, C.; García, K.; Castillo, D.; Katharios, P.; Bastías, R. Diversification of Vibrio anguillarum driven by the bacteriophage CHOED. Front. Microbiol. 2019, 10, 1396. [Google Scholar] [CrossRef] [PubMed]
- Arisaka, F.; Kanamaru, S.; Leiman, P.; Rossmann, M.G. The tail lysozyme complex of bacteriophage T4. Int. J. Biochem. Cell Biol. 2003, 35, 16–21. [Google Scholar] [CrossRef]
- Lopes, A.; Pereira, C.; Almeida, A. Sequential combined effect of phages and antibiotics on the inactivation of E. coli. Microorganisms 2018, 6, 125. [Google Scholar] [CrossRef]
- Christiansen, R.H.; Madsen, L.; Dalsgaard, I.; Castillo, D.; Kalatzis, P.G.; Middelboe, M. Effect of bacteriophages on the growth of Flavobacterium psychrophilum and development of phage-resistant strains. Microb. Ecol. 2016, 71, 845–859. [Google Scholar] [CrossRef]
- Kim, J.H.; Son, J.S.; Choi, Y.J.; Choresca, C.H.; Shin, S.P.; Han, J.E.; Jun, J.W.; Kang, D.H.; Oh, C.; Heo, S.J.; et al. Isolation and characterization of a lytic Myoviridae bacteriophage PAS-1 with broad infectivity in Aeromonas salmonicida. Curr. Microbiol. 2012, 64, 418–426. [Google Scholar] [CrossRef]
- Silva, Y.; Moreirinha, C.; Pereira, C.; Costa, L.; Rocha, R.J.M.; Cunha, Â.; Gomes, N.C.M.; Calado, R.; Almeida, A. Biological control of Aeromonas salmonicida infection in juvenile Senegalese sole (Solea senegalensis) with phage AS-A. Aquaculture 2016, 450, 225–233. [Google Scholar] [CrossRef]
- Silva, Y.J.; Costa, L.; Pereira, C.; Mateus, C.; Cunha, Â.; Calado, R.; Gomes, N.; Pardo, M.A.; Hernandez, I.; Almeida, A. Phage therapy as an approach to prevent Vibrio anguillarum infections in fish larvae production. PLoS ONE 2014, 9, e114197. [Google Scholar] [CrossRef] [PubMed]
- Alves, E.; Carvalho, C.M.B.; Tomé, J.P.C.; Faustino, M.A.F.; Neves, M.G.P.M.S.; Tomé, A.C.; Cavaleiro, J.A.S.; Cunha, Â.; Mendo, S.; Almeida, A. Photodynamic inactivation of recombinant bioluminescent Escherichia coli by cationic porphyrins under artificial and solar irradiation. J. Ind. Microbiol. Biotechnol. 2008, 35, 1447–1454. [Google Scholar] [CrossRef]
- Tacão, M.; Correia, A.; Henriques, I.S. Low prevalence of carbapenem-resistant bacteria in river water: Resistance is mostly related to intrinsic mechanisms. Microb. Drug Resist. 2015, 21, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.H. Bacteriophages; Interscience Publishers: New York, NY, USA, 1959. [Google Scholar]
- Kutter, E. Phage host range and efficiency of plating. In Bacteriophage: Methods and Protocols; Humana Press: Totowa, NJ, USA, 2009; pp. 141–149. ISBN 978-1-58829-682-5. [Google Scholar]
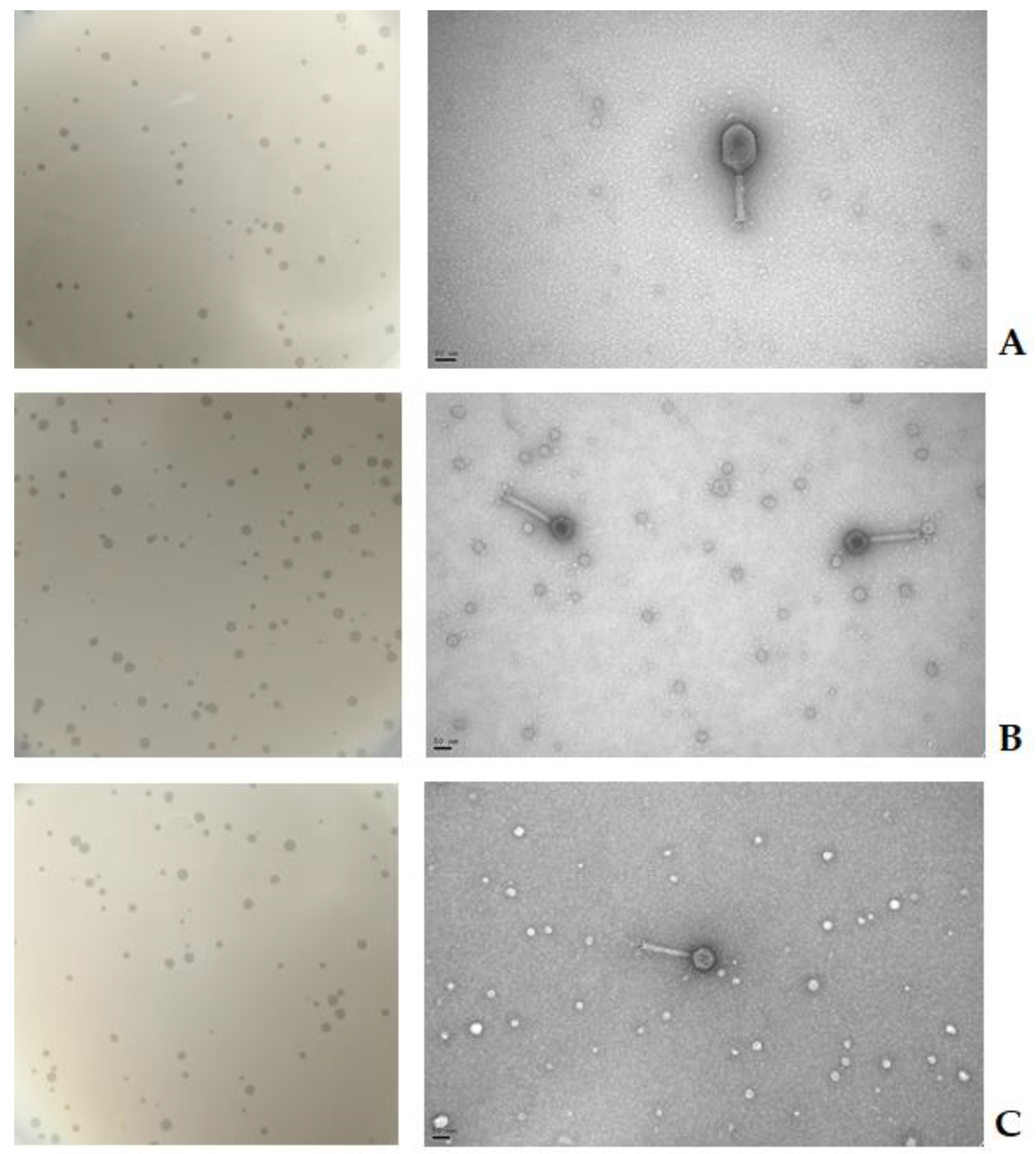



| Species | Infectivity of Phage | Efficacy of Plating (%) | ||||
| AH-1 | AH-4 | AH-5 | AH-1 | AH-4 | AH-5 | |
| Aeromonas hydrophila ATCC 7966 | + | + | + | 100 | 100 | 100 |
| Aeromonas hydrophila 839 | + | + | + | 83.3 ± 3.52 | 91.14 ± 5.73 | 76.19 ± 2.61 |
| Aeromonas caviae 838 | − | − | − | 0 | 0 | 0 |
| Aeromonas salmonicida CECT 894 | − | − | − | 0 | 0 | 0 |
| Aeromonas IR13 | − | − | − | 0 | 0 | 0 |
| Bioluminescent Escherichia coli | − | + | - | 0 | 3.31 ± 0.50 | 0 |
| Escherichia coli ATCC 25922 | − | − | − | 0 | 0 | 0 |
| Escherichia coli ATCC 13706 | − | − | − | 0 | 0 | 0 |
| Listeria innocua NCTC 11288 | − | − | − | 0 | 0 | 0 |
| Listeria monocytogenes NCTC 1194 | − | − | − | 0 | 0 | 0 |
| Photobacterium damselae damselae DSM 7482 | − | − | − | 0 | 0 | 0 |
| Salmonella typhimurium ATCC 13311 | + | + | − | 56.10 ± 2.32 | 10.17 ± 0.35 | 0 |
| Salmonella typhimurium ATCC 14028 | − | − | − | 0 | 0 | 0 |
| Salmonella typhimurium WG49 | − | − | − | 0 | 0 | 0 |
| Shigella flexneri DSM 4782 | − | − | − | 0 | 0 | 0 |
| Staphylococcus aureus ATCC 6538 | − | − | − | 0 | 0 | 0 |
| Vibrio parahaemolyticus DSM 27657 | − | − | − | 0 | 0 | 0 |
| Vibrio anguillarum DSM 21597 | − | − | − | 0 | 0 | 0 |
| Aliivibrio fischeri ATCC 49387 | − | − | − | 0 | 0 | 0 |
| Phages/Phage Cocktails | Control Sample (CFU/mL) | Sample Treated with Phages | Frequency of Mutants |
| AH-1 | 1.24 ± 0.45 × 109 | 3.85 ± 2.77 × 104 | 3.10 × 10−3 |
| AH-4 | 2.41 ± 0.93 × 109 | 2.75 ± 1.20 × 104 | 1.14 × 10−3 |
| AH-5 | 3.27 ± 0.11 × 109 | 1.64 ± 0.35 × 104 | 5.02 × 10−4 |
| AH-1/AH-4 | 6.43 ± 0.12 × 109 | 5.32 ± 2.46 × 104 | 8.26 × 10−4 |
| AH-1/AH-5 | 8.15 ± 0.36 × 109 | 5.22 ± 3.12 × 104 | 6.40 × 10−4 |
| AH-4/AH-5 | 2.17 ± 0.58 × 109 | 1.55 ± 1.15 × 104 | 7.13 × 10−4 |
| AH-1/AH-4/AH-5 | 9.23 ± 2.08 × 109 | 5.53 ± 3.24 × 104 | 5.99 × 10−4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duarte, J.; Pereira, C.; Costa, P.; Almeida, A. Bacteriophages with Potential to Inactivate Aeromonas hydrophila in Cockles: In Vitro and In Vivo Preliminary Studies. Antibiotics 2021, 10, 710. https://doi.org/10.3390/antibiotics10060710
Duarte J, Pereira C, Costa P, Almeida A. Bacteriophages with Potential to Inactivate Aeromonas hydrophila in Cockles: In Vitro and In Vivo Preliminary Studies. Antibiotics. 2021; 10(6):710. https://doi.org/10.3390/antibiotics10060710
Chicago/Turabian StyleDuarte, João, Carla Pereira, Pedro Costa, and Adelaide Almeida. 2021. "Bacteriophages with Potential to Inactivate Aeromonas hydrophila in Cockles: In Vitro and In Vivo Preliminary Studies" Antibiotics 10, no. 6: 710. https://doi.org/10.3390/antibiotics10060710
APA StyleDuarte, J., Pereira, C., Costa, P., & Almeida, A. (2021). Bacteriophages with Potential to Inactivate Aeromonas hydrophila in Cockles: In Vitro and In Vivo Preliminary Studies. Antibiotics, 10(6), 710. https://doi.org/10.3390/antibiotics10060710









