Screening of Novel Antimicrobial Diastereomers of Azithromycin–Thiosemicarbazone Conjugates: A Combined LC-SPE/Cryo NMR, MS/MS and Molecular Modeling Approach
Abstract
1. Introduction
2. Results and Discussion
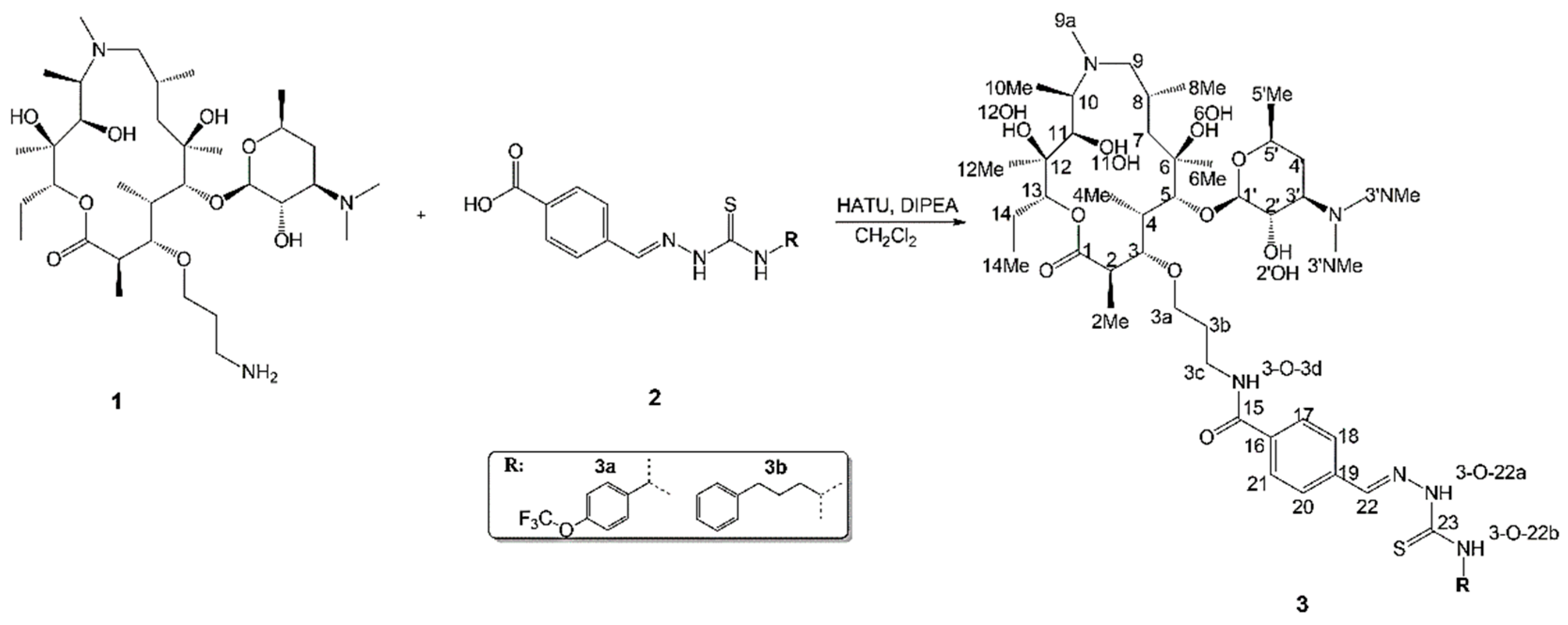
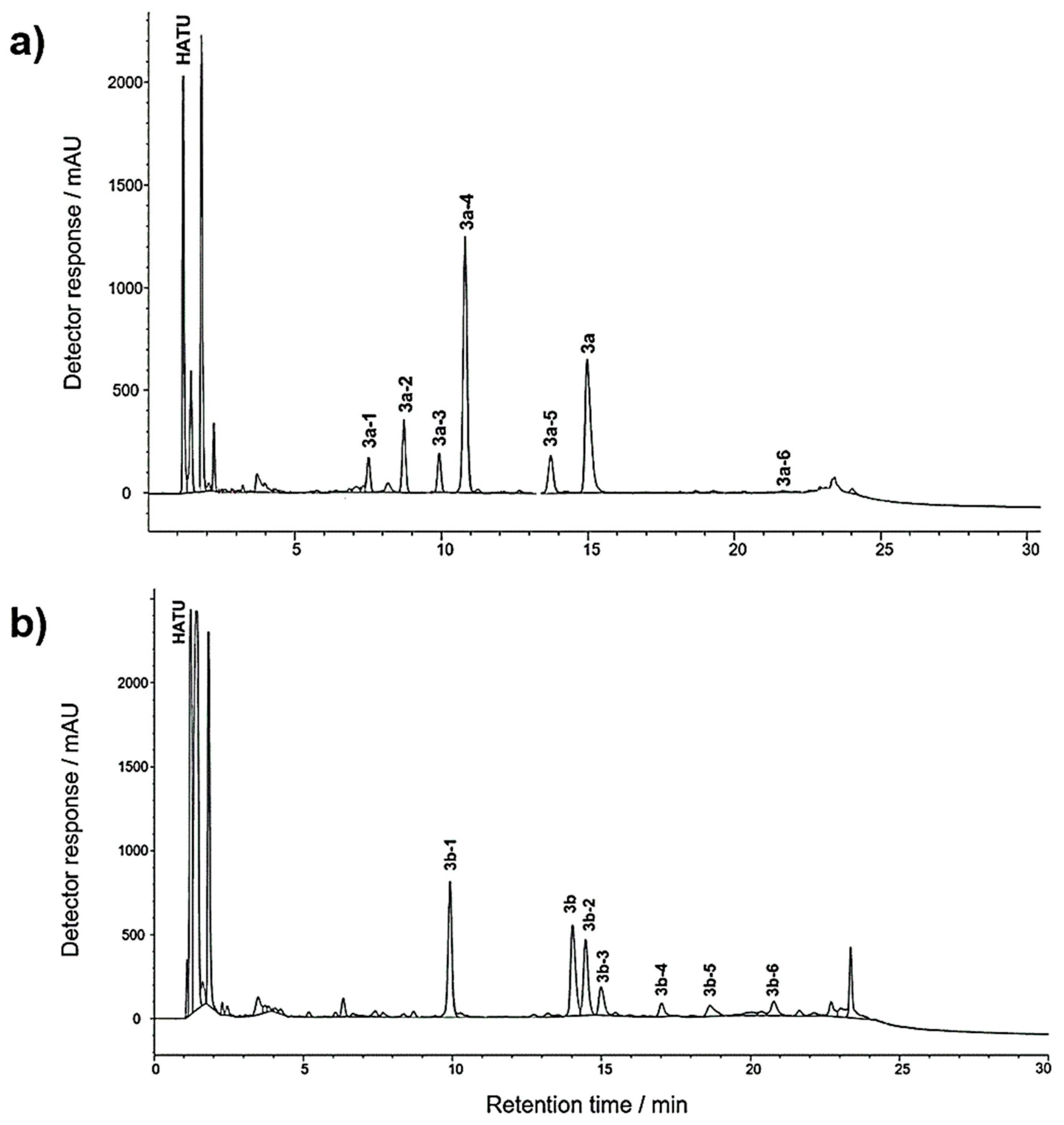
2.1. SPE Preconcentration
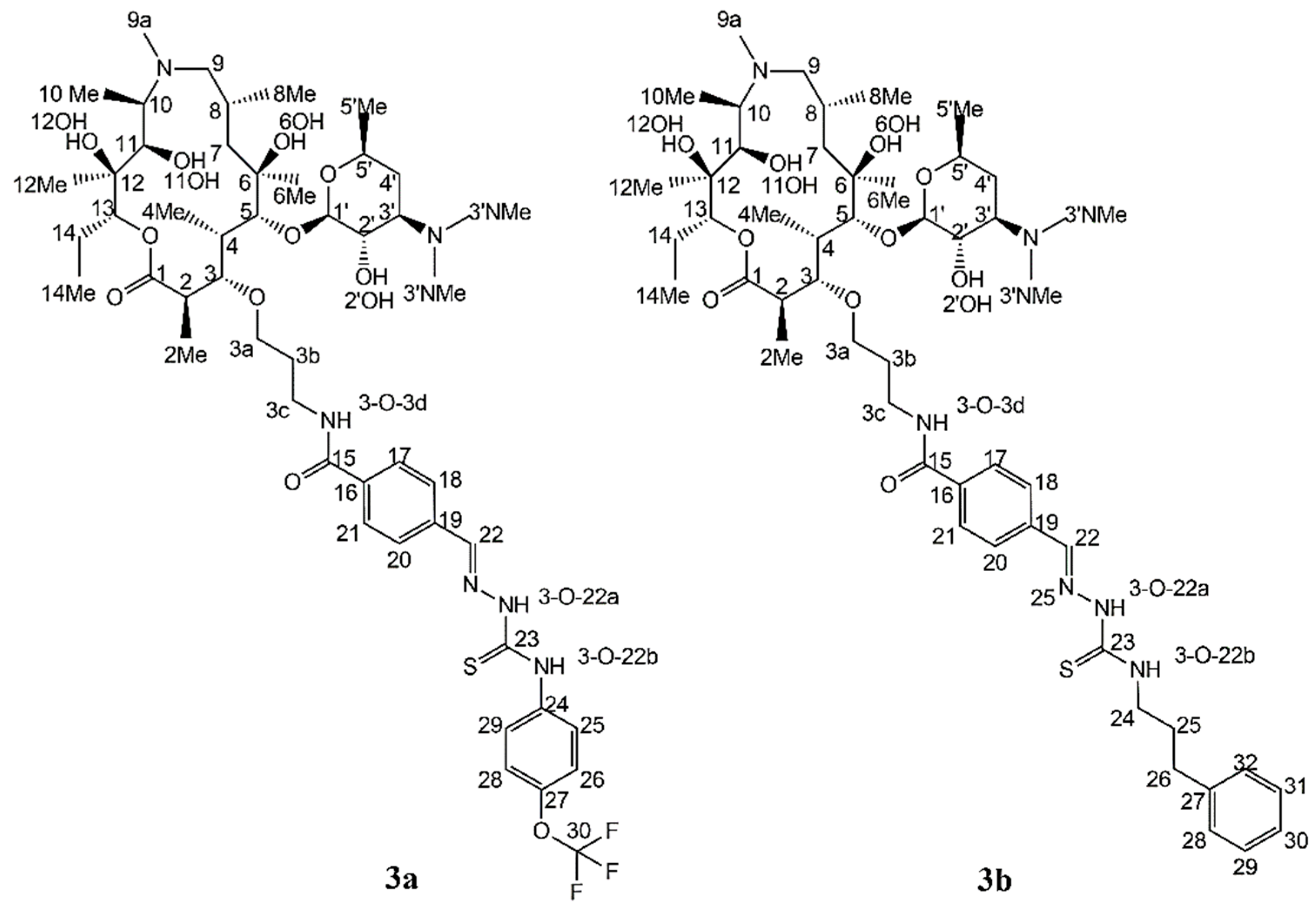
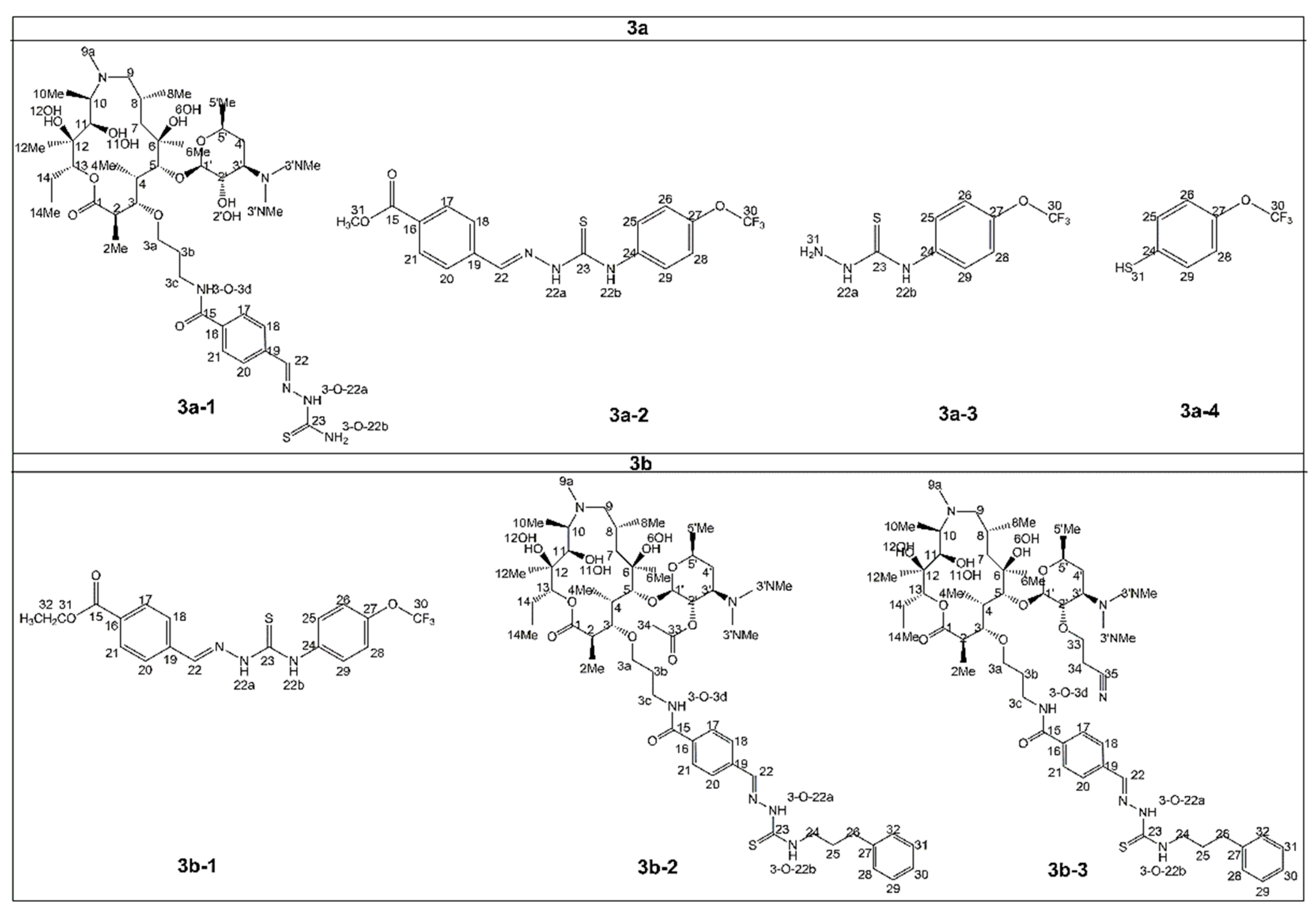
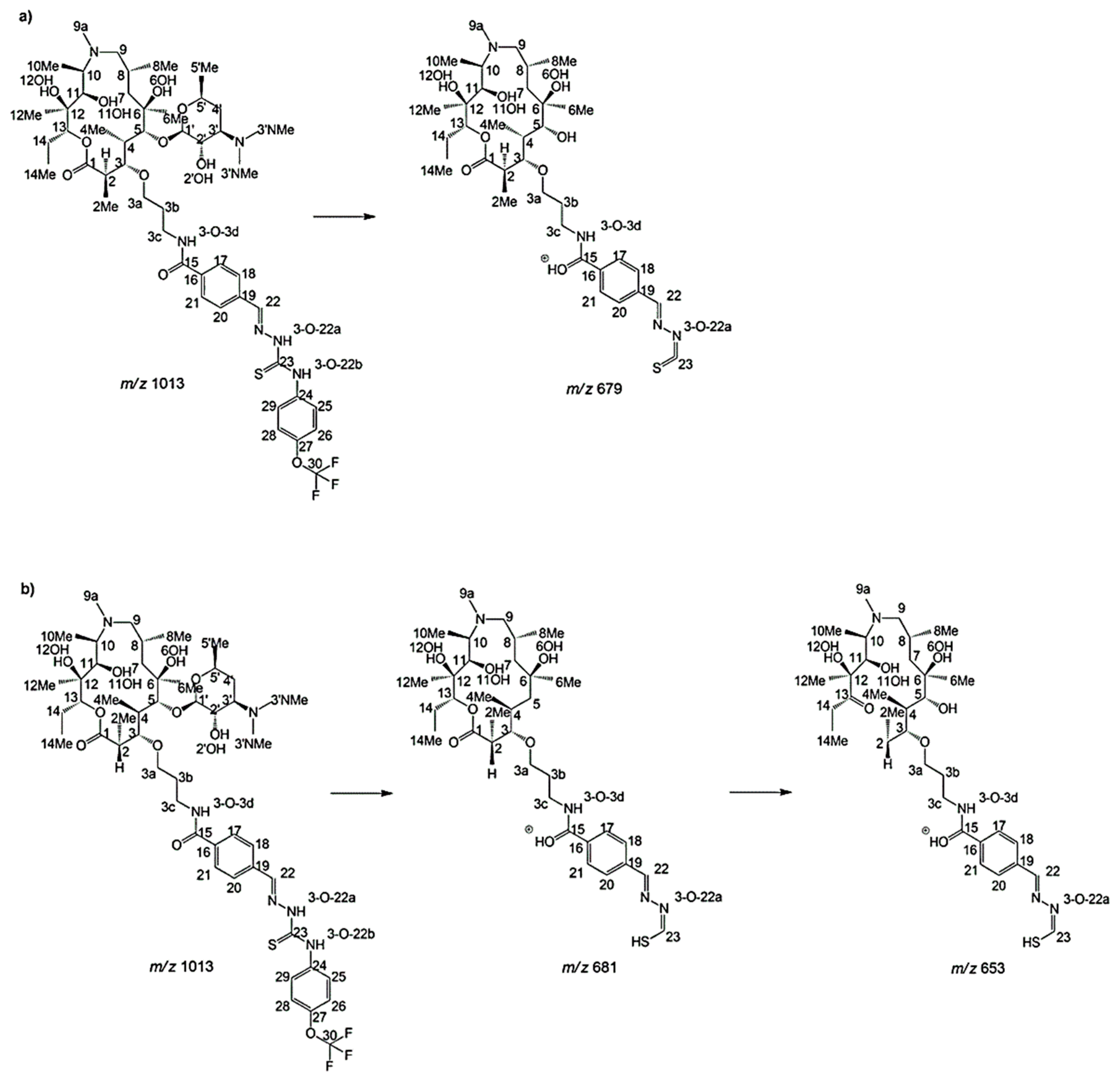
2.2. Structure of Identified Reaction Components Not Related to Isomers
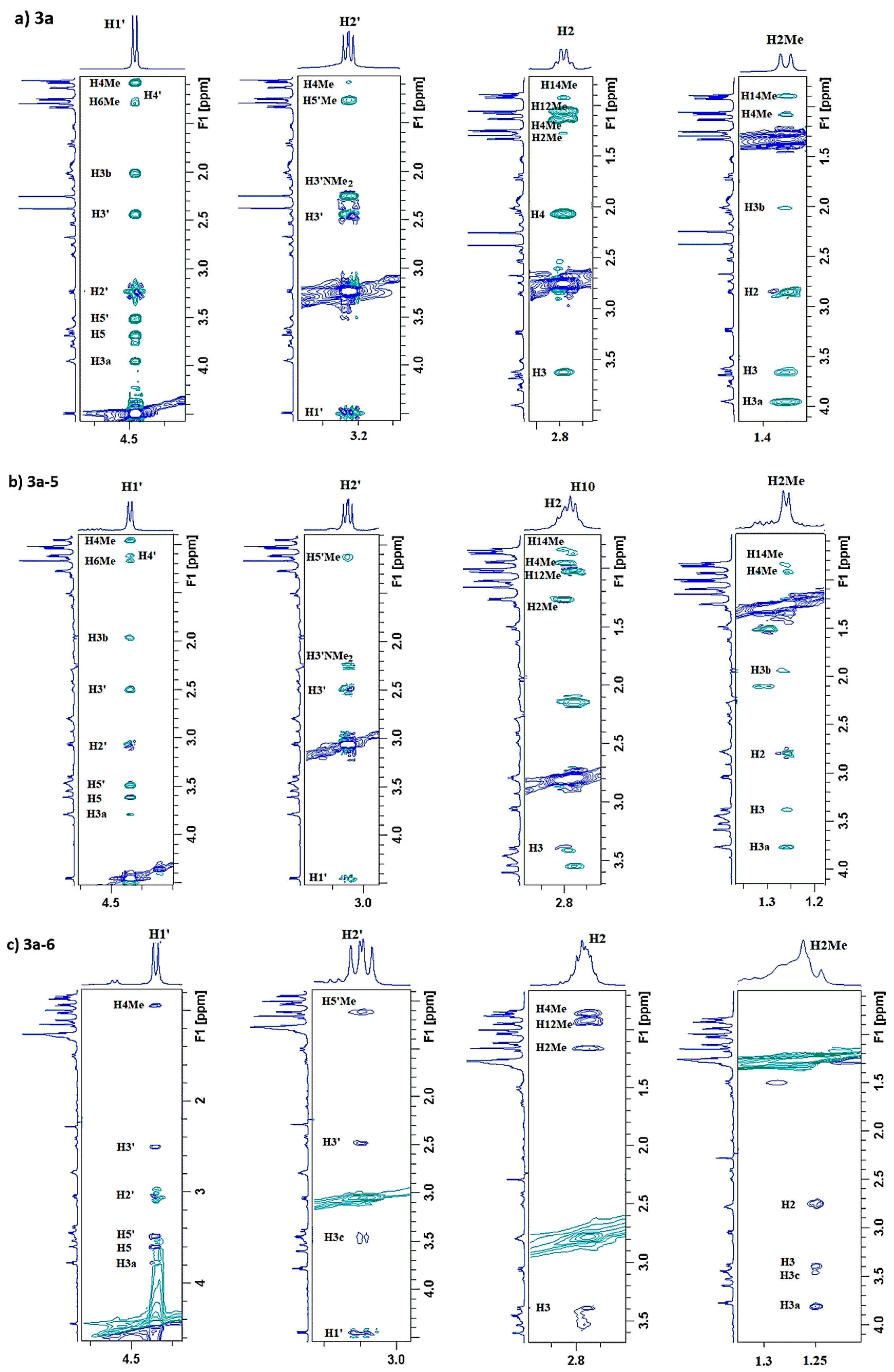
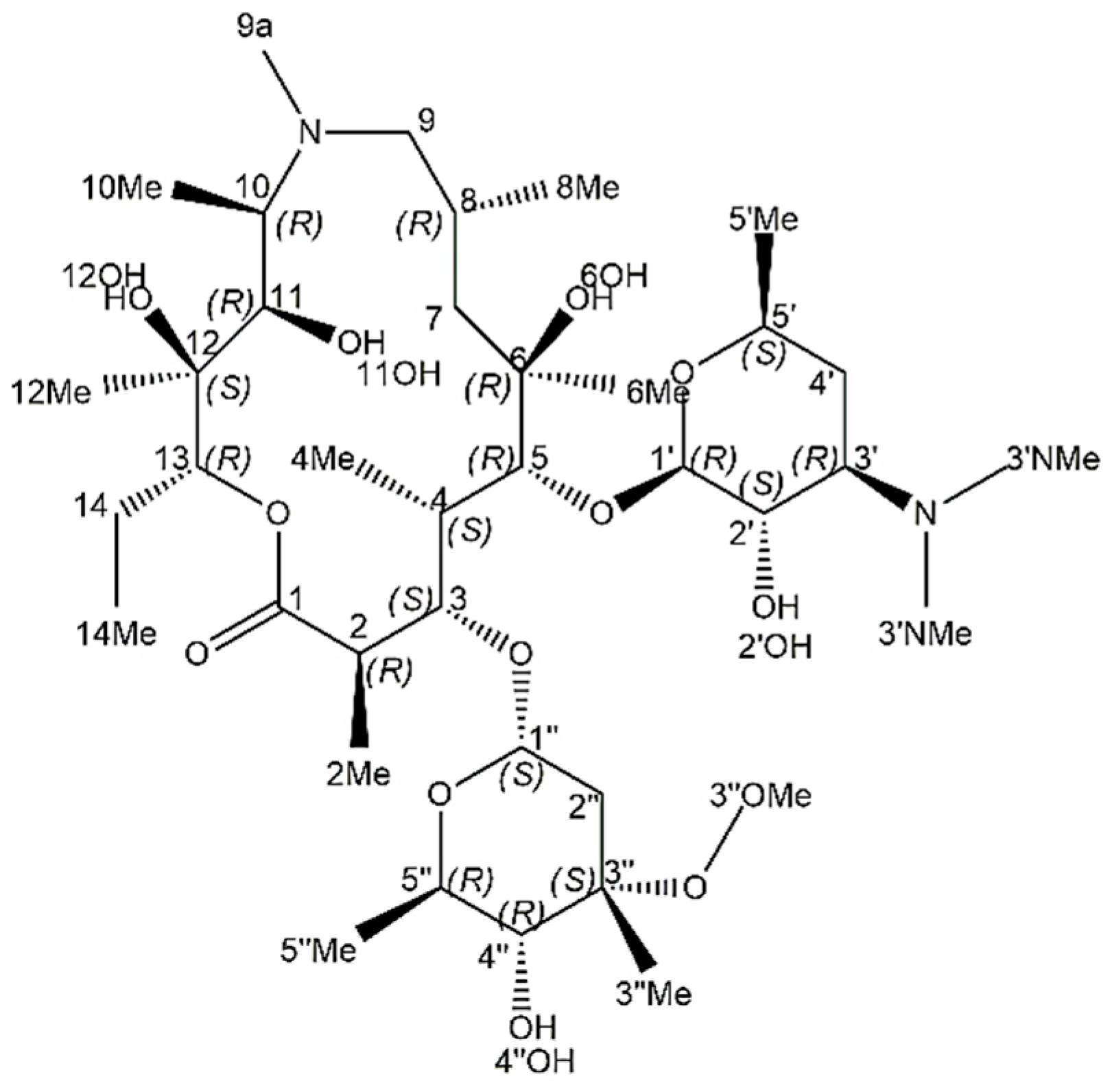
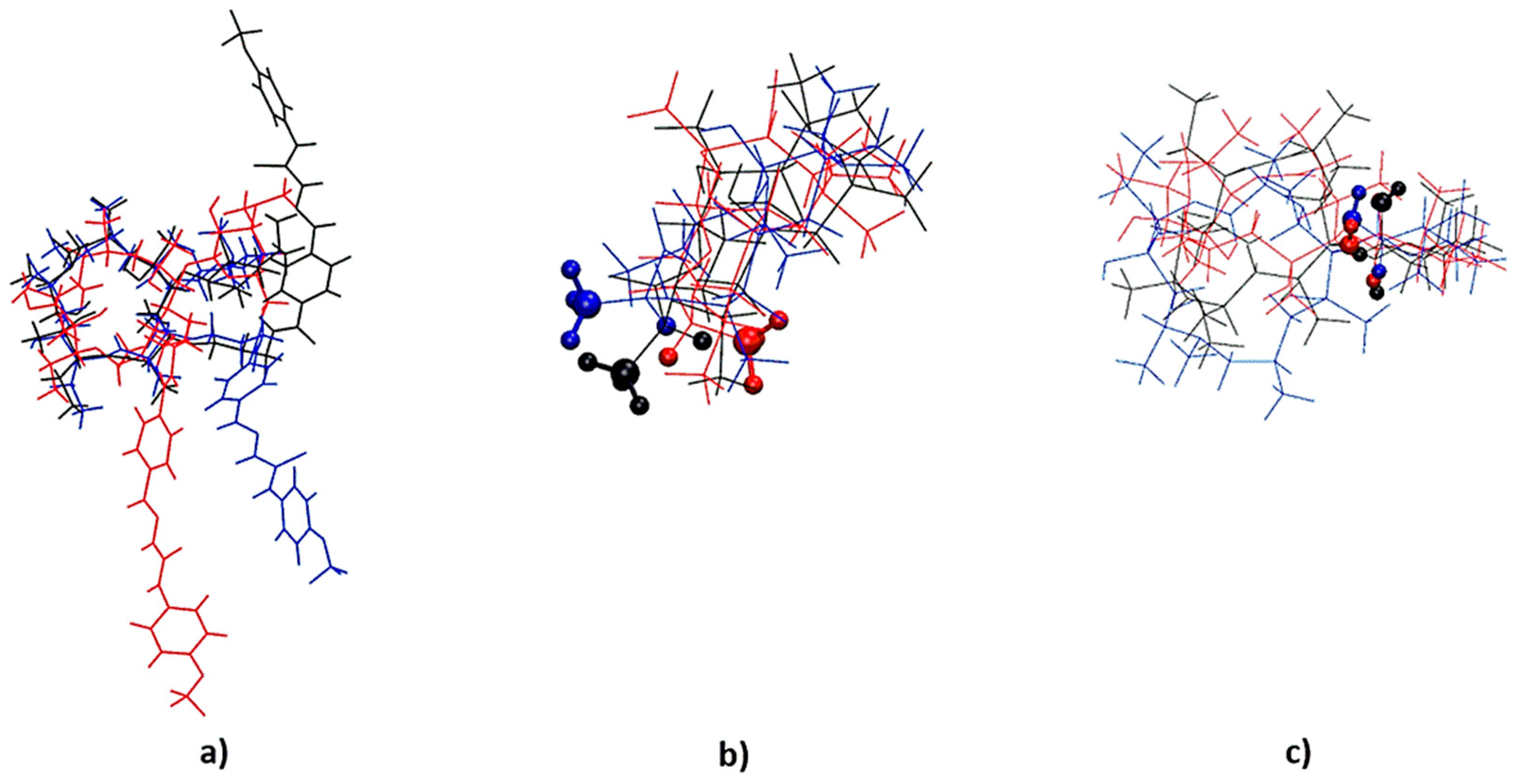
2.3. Structure of Stereoisomers
2.4. Antibacterial Evaluation
3. Materials and Methods
3.1. Materials and Reagents
3.2. Sample Preparation
3.3. Liquid Chromatography
3.4. On-Line HPLC-SPE
3.5. NMR Spectroscopy
3.6. MS Analysis
3.7. Molecular Dynamics Simulations
3.8. Antibacterial Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arsic, B.; Novak, P.; Rimoli, M.G.; Barber, J.; Kragol, G.; Sodano, F. Macrolides Properties, Synthesys and Applications; De Gruyter: Berlin, Germany, 2018. [Google Scholar]
- Parnham, M.J.; Eraković Faber, V.; Gimarellos-Bourboulis, E.J.; Perletti, G.; Verleden, G.M.; Vos, R. Azithromycin: Mechanism of action and their relevance for clinical applications. Pharm. Therap. 2014, 143, 225–245. [Google Scholar] [CrossRef]
- Schlünzen, F.; Zarivach, R.; Harm, J.; Bashan, A.; Tocilj, A.; Albrecht, R.; Yonath, A.; Franceschi, F. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 2001, 413, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Hansen, J.L.; Ippolito, J.A.; Ban, N.; Nissen, P.; Moore, P.B.; Steitz, T.A. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol. Cell 2002, 10, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Dunkle, J.A.; Xiong, L.; Mankin, A.S.; Cate, J.H.D. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc. Natl. Acad. Sci. USA 2010, 107, 17152–17157. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.N. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol. 2014, 12, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Grgičević, I.; Mikulandra, I.; Bukvić, M.; Banjanac, M.; Habinovec, I.; Bertoša, B.; Novak, P. Discovery of Macrozones, new antimicrobial thiosemicarbazone-based azithromycin conjugates: Design, synthesis and in vitro biological evaluation. Int. J. Antimicrob. Agents 2020, 56, 106147. [Google Scholar] [CrossRef]
- Mikulandra, I.; Jednačak, T.; Bertoša, B.; Parlov-Vuković, J.; Kušec, I.; Novak, P. Interactions of aminopropyl-azithromycin derivatives, precursors in the synthesis of bioactive macrozones, with E. coli ribosome: NMR and molecular docking studies. Materials 2021, 14, 5561. [Google Scholar] [CrossRef]
- Singh, S.; Handa, T.; Narayanam, M.; Sahu, A.; Junwal, M.; Shah, R.P. A critical review on the use of modern sophisticated hyphenated tools in the characterization of impurities and degradation products. J. Pharm. Biomed. Anal. 2012, 69, 148–173. [Google Scholar] [CrossRef]
- Görög, S. Critical review of reports on impurity and degradation product profiling in the last decade. Trends Anal. Chem. 2018, 101, 2–16. [Google Scholar] [CrossRef]
- Sturm, S.; Seger, C. Liquid chromatography–nuclear magnetic resonance coupling as alternative to liquid chromatography–mass spectrometry hyphenations: Curious option or powerful and complementary routine tool? J. Chromatogr. A 2012, 1259, 50–61. [Google Scholar] [CrossRef]
- Grosso, C.; Jäger, A.K.; Staerk, D. Coupling of a high-resolution monoamine oxidase-A inhibitor assay and HPLC-SPE-NMR for advanced bioactivity profiling of plant extracts. Phytochem. Anal. 2013, 2, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Novak, P.; Tepeš, P.; Cindrić, M.; Ilijaš, M.; Dragojević, S.; Mihaljević, K. Combined use of liquid chromatography-nuclear magnetic resonance spectroscopy and liquid chromatography-mass spectrometry for the characterisation of an acarbose degradation product. J. Chromatogr. A 2004, 1033, 299–303. [Google Scholar] [CrossRef] [PubMed]
- Novak, P.; Tepeš, P.; Ilijaš, M.; Fistrić, I.; Bratoš, I.; Avdagić, A.; Gabelica Marković, V.; Dumić, M. LC-NMR and LC-MS identification of an impurity in a novel antifungal drug icofungipen. J. Pharm. Biomed. Anal. 2009, 50, 68–72. [Google Scholar] [CrossRef] [PubMed]
- Novak, P.; Tepeš, P.; Fistrić, I.; Bratoš, I.; Gabelica, V. The Application of LC-NMR and LC-MS for the Separation and Rapid Structure elucidation of an Unknown Impurity in drug 5-Aminosalicylic Acid. J. Pharm. Biomed. Anal. 2006, 40, 1268–1272. [Google Scholar] [CrossRef] [PubMed]
- Habinovec, I.; Mikulandra, I.; Sekula, L.E.; Gašperov, J.; Kazazić, S.; Novak, P. Rapid structure determination of bioactive 4″-tetrahydrofurfuryl macrozone reaction mixture components by LC-SPE/cryo NMR and MS. Molecules 2021, 26, 6316. [Google Scholar] [CrossRef]
- Harča, M.; Habinovec, I.; Meštrović, E.; Biljan, I.; Novak, P. Rapid Identification of Unknown Impurities in 3-Bromo-5-(trifluoromethyl)aniline by LC-SPE/NMR. Croat. Chem. Acta 2016, 89, 543–547. [Google Scholar] [CrossRef]
- Seger, C.; Godejohann, M.; Spraul, M.; Stuppner, H.; Hadacek, F. Reaction product analysis by high-performance liquid chromatography-solid-phase extraction-nuclear magnetic resonance Application to the absolute configuration determination of naturally occurring polyyne alcohols. J. Chromatogr. A 2006, 1136, 82–88. [Google Scholar] [CrossRef]
- Novak, P.; Barber, J.; Čikoš, A.; Arsić, B.; Plavec, J.; Lazarevski, G.; Tepeš, P.; Košutić-Hulita, N. Free and bound state structures of 6-O-methyl homoerythromycins and epitope mapping of their interactions with ribosomes. Bioorg. Med. Chem. 2009, 17, 5857–5867. [Google Scholar] [CrossRef]
- Henninger, T.C.; Xu, X.; Abbanat, D.; Baum, E.Z.; Foleno, B.D.; Hilliard, J.J.; Bush, K.; Hlasta, D.J.; Macielag, M.J. Synthesis and antibacterial activity of C-6 carbamate ketolides, a novel series of orally active ketolide antibiotics. Bioorg. Med. Chem. Lett. 2004, 14, 4495–4499. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Hoover, W.G. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A 1985, 31, 1695–1697. [Google Scholar] [CrossRef] [PubMed]
- Martyna, G.J.; Tuckerman, M.E.; Tobias, D.J.; Klein, M.L. Explicit reversible integrators for extended systems dynamics. Mol. Phys. 1996, 87, 1117–1157. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]

| Atom | |||
|---|---|---|---|
| Isomer | C-2 | C-1′ | C-2′ |
| 3a | R | R | S |
| 3a-5 | R | R | R |
| 3a-6 | S | S | R |
| 3b | R | R | S |
| 3b-6 | R | S | R |
| Compound | S. pneumoniae B0652 | S. pneumoniae B0326 | S. pneumoniae B0633 | S. pyogenes B0542 | S. pyogenes B0545 | S. pyogenes B0544 | S. aureus ATCC 29213 | S. aureus B0331 | S. aureus B0330 | E. faecalis ATCC 29212 | M. catarrhalis ATCC 23246 | E. coli ATCC 25922 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| * eryS | M | cMLS | eryS | M | cMLS | eryS | M | cMLS | ||||
| MIC (μg/mL) | ||||||||||||
| 3a | 8 | 8 | 64 | 2 | 8 | 32 | 8 | 16 | 32 | 8 | 4 | >64 |
| 3a-5 | 16 | 32 | 64 | 16 | 16 | 32 | 16 | 16 | 8 | 16 | 2 | >64 |
| 3a-6 | 4 | 16 | 64 | 4 | 16 | 32 | 16 | 8 | 16 | 64 | 8 | >64 |
| 3b | 2 | 16 | 64 | 4 | 16 | 64 | 8 | 8 | 16 | 32 | 4 | >64 |
| 3b-2 | 2 | 16 | 64 | 4 | 16 | 64 | 8 | 16 | 16 | 32 | 4 | >64 |
| 3b-3 | 16 | 64 | >64 | 16 | 32 | 64 | 64 | 16 | 16 | 64 | 4 | >64 |
| 3b-6 | 16 | >64 | >64 | 32 | 64 | 64 | >32 | 16 | 16 | >64 | 16 | >64 |
| Azithromycin | ≤0.125 | 16 | >64 | ≤0.125 | 16 | >64 | 1 | >64 | >64 | 8 | ≤0.125 | >64 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Habinovec, I.; Mikulandra, I.; Pranjić, P.; Kazazić, S.; Paljetak, H.Č.; Barišić, A.; Bertoša, B.; Bukvić, M.; Novak, P. Screening of Novel Antimicrobial Diastereomers of Azithromycin–Thiosemicarbazone Conjugates: A Combined LC-SPE/Cryo NMR, MS/MS and Molecular Modeling Approach. Antibiotics 2022, 11, 1738. https://doi.org/10.3390/antibiotics11121738
Habinovec I, Mikulandra I, Pranjić P, Kazazić S, Paljetak HČ, Barišić A, Bertoša B, Bukvić M, Novak P. Screening of Novel Antimicrobial Diastereomers of Azithromycin–Thiosemicarbazone Conjugates: A Combined LC-SPE/Cryo NMR, MS/MS and Molecular Modeling Approach. Antibiotics. 2022; 11(12):1738. https://doi.org/10.3390/antibiotics11121738
Chicago/Turabian StyleHabinovec, Iva, Ivana Mikulandra, Paula Pranjić, Saša Kazazić, Hana Čipčić Paljetak, Antun Barišić, Branimir Bertoša, Mirjana Bukvić, and Predrag Novak. 2022. "Screening of Novel Antimicrobial Diastereomers of Azithromycin–Thiosemicarbazone Conjugates: A Combined LC-SPE/Cryo NMR, MS/MS and Molecular Modeling Approach" Antibiotics 11, no. 12: 1738. https://doi.org/10.3390/antibiotics11121738
APA StyleHabinovec, I., Mikulandra, I., Pranjić, P., Kazazić, S., Paljetak, H. Č., Barišić, A., Bertoša, B., Bukvić, M., & Novak, P. (2022). Screening of Novel Antimicrobial Diastereomers of Azithromycin–Thiosemicarbazone Conjugates: A Combined LC-SPE/Cryo NMR, MS/MS and Molecular Modeling Approach. Antibiotics, 11(12), 1738. https://doi.org/10.3390/antibiotics11121738







