Antimicrobial Resistance Surveillance of Pigs and Chickens in the Lao People’s Democratic Republic, 2018–2021
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling Design
2.2. Sample Collection
2.3. Sample Processing and Bacterial Recovery
2.4. Antimicrobial Susceptibility Testing (AST)
2.5. Data Entry and Storage
2.6. Data Analysis and Interpretation
2.6.1. Validation and Data Preparation
2.6.2. Descriptive Statistics of MIC and Binary Resistance Information
2.6.3. Comparison between Chickens and Pigs, and Subanalysis of Chicken Types and Sampling Rounds in Pigs
3. Results
3.1. Bacterial Isolation
3.2. Antimicrobial Resistance in Escherichia coli Isolates
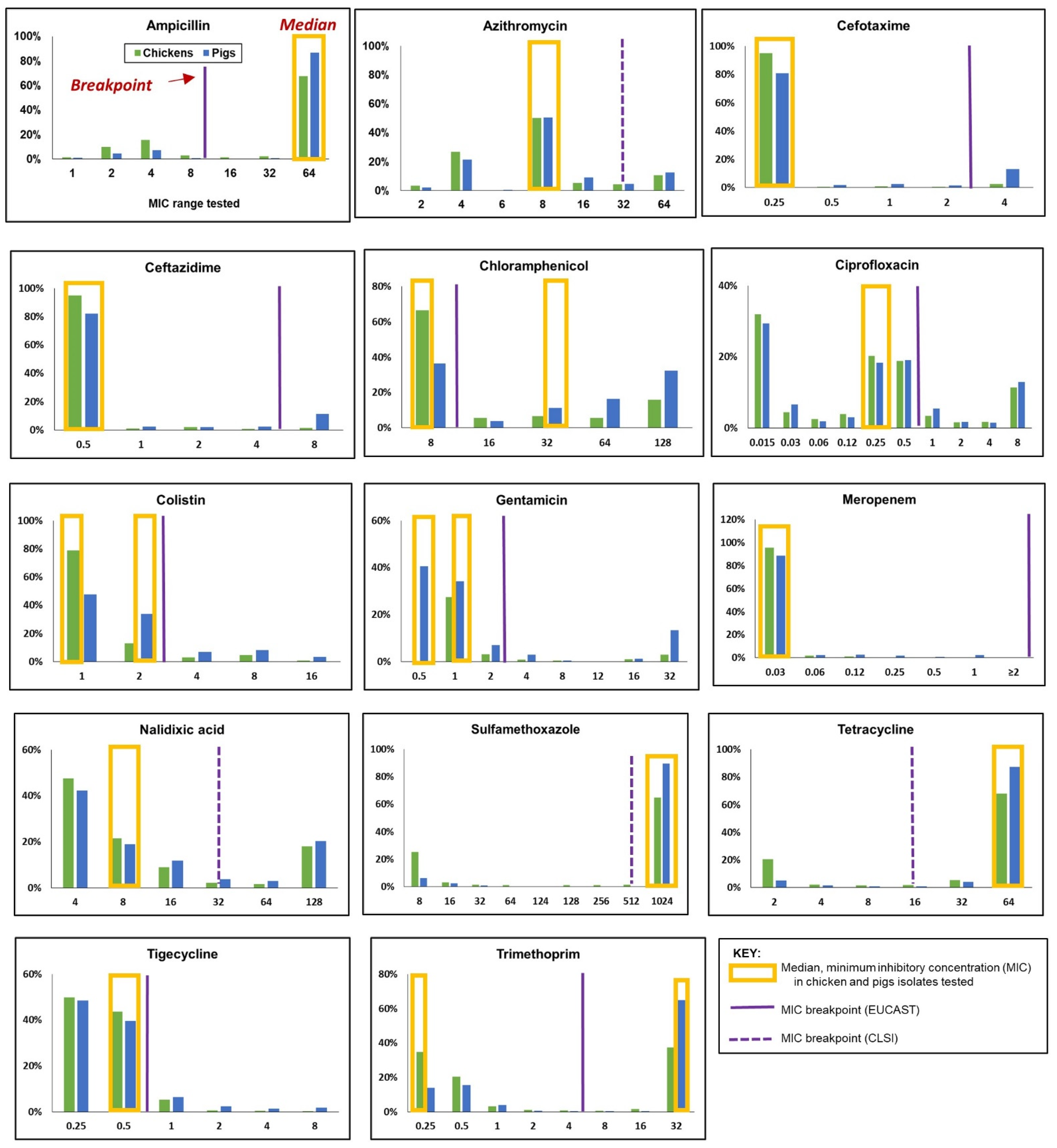
3.2.1. Minimum Inhibitory Concentration (MIC) Distribution
3.2.2. Resistance to Single (Homologous) Antimicrobials
Comparison between Different Chicken Production Types
Comparison between Two Time Periods/Geographical Regions of the Lao PDR in Pig Isolates
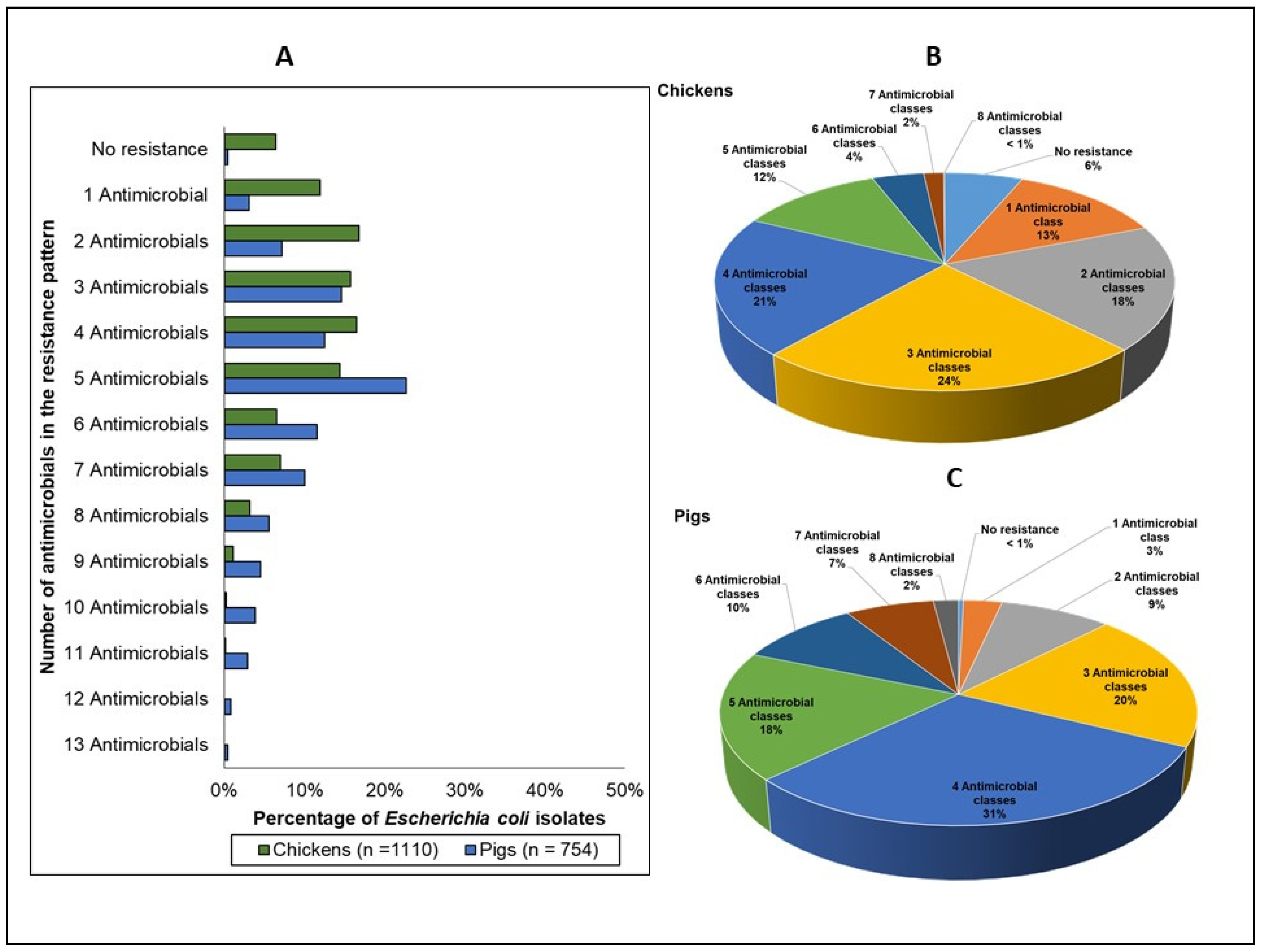
3.2.3. Multidrug and Multiclass Resistance
3.3. Antimicrobial Resistance in Salmonella Isolates
3.3.1. MIC Distribution
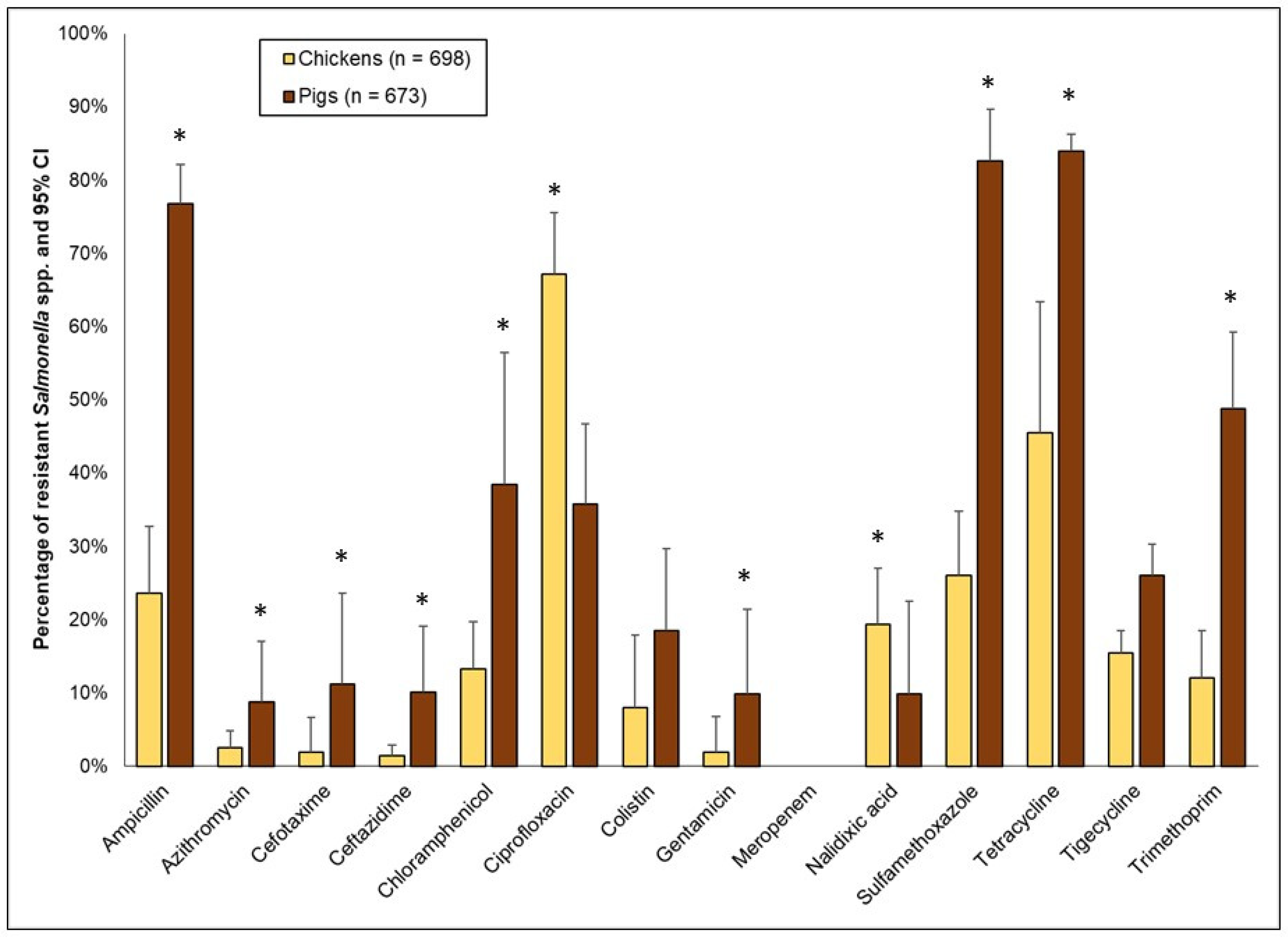
3.3.2. Resistance to Single (Homologous) Antimicrobials
Comparison between Different Chicken Production Types
Comparison between Two Time Periods/Geographical Regions of the Lao PDR in Pig Isolates
3.3.3. Multidrug and Multiclass Resistance
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nhung, N.T.; Cuong, N.V.; Thwaites, G.; Carrique-Mas, J. Antimicrobial Usage and Antimicrobial Resistance in Animal Production in Southeast Asia: A Review. Antibiotics 2016, 5, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coyne, L.; Arief, R.; Benigno, C.; Giang, V.N.; Huong, L.Q.; Jeamsripong, S.; Kalpravidh, W.; McGrane, J.; Padungtod, P.; Patrick, I.; et al. Characterizing Antimicrobial Use in the Livestock Sector in Three South East Asian Countries (Indonesia, Thailand, and Vietnam). Antibiotics 2019, 8, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strom, G.; Boqvist, S.; Albihn, A.; Fernstrom, L.L.; Andersson Djurfeldt, A.; Sokerya, S.; Sothyra, T.; Magnusson, U. Antimicrobials in small-scale urban pig farming in a lower middle-income country—Arbitrary use and high resistance levels. Antimicrob. Resist. Infect. Control 2018, 7, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuat, C.V.; Hue, P.T.; Loan, N.T.P.; Thuy, N.T.; Hue, L.T.; Giang, V.N.; Erickson, V.I.; Padungtod, P. Antimicrobial Resistance Pilot Surveillance of Pigs and Chickens in Vietnam, 2017–2019. Front. Vet. Sci. 2021, 8, 618497. [Google Scholar] [CrossRef]
- Holmes, A.H.; Moore, L.S.; Sundsfjord, A.; Steinbakk, M.; Regmi, S.; Karkey, A.; Guerin, P.J.; Piddock, L.J. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 2016, 387, 176–187. [Google Scholar] [CrossRef]
- Graham, D.W.; Bergeron, G.; Bourassa, M.W.; Dickson, J.; Gomes, F.; Howe, A.; Kahn, L.H.; Morley, P.S.; Scott, H.M.; Simjee, S.; et al. Complexities in understanding antimicrobial resistance across domesticated animal, human, and environmental systems. Ann. N. Y. Acad. Sci. 2019, 1441, 17–30. [Google Scholar] [CrossRef]
- Pungpian, C.; Sinwat, N.; Angkititrakul, S.; Prathan, R.; Chuanchuen, R. Presence and Transfer of Antimicrobial Resistance Determinants in Escherichia coli in Pigs, Pork, and Humans in Thailand and Lao PDR Border Provinces. Microb. Drug Resist. 2021, 27, 571–584. [Google Scholar] [CrossRef]
- Morel, C.M.; Alm, R.A.; Ardal, C.; Bandera, A.; Bruno, G.M.; Carrara, E.; Colombo, G.L.; de Kraker, M.E.A.; Essack, S.; Frost, I.; et al. A one health framework to estimate the cost of antimicrobial resistance. Antimicrob. Resist. Infect. Control 2020, 9, 187. [Google Scholar] [CrossRef]
- Nguyen, N.T.; Nguyen, H.M.; Nguyen, C.V.; Nguyen, T.V.; Nguyen, M.T.; Thai, H.Q.; Ho, M.H.; Thwaites, G.; Ngo, H.T.; Baker, S.; et al. Use of Colistin and Other Critical Antimicrobials on Pig and Chicken Farms in Southern Vietnam and Its Association with Resistance in Commensal Escherichia coli Bacteria. Appl. Environ. Microbiol. 2016, 82, 3727–3735. [Google Scholar] [CrossRef] [Green Version]
- Trongjit, S.; Angkititrakul, S.; Tuttle, R.E.; Poungseree, J.; Padungtod, P.; Chuanchuen, R. Prevalence and antimicrobial resistance in Salmonella enterica isolated from broiler chickens, pigs and meat products in Thailand-Cambodia border provinces. Microbiol. Immunol. 2017, 61, 23–33. [Google Scholar] [CrossRef] [Green Version]
- Moe, A.Z.; Paulsen, P.; Pichpol, D.; Fries, R.; Irsigler, H.; Baumann, M.P.O.; Oo, K.N. Prevalence and Antimicrobial Resistance of Salmonella Isolates from Chicken Carcasses in Retail Markets in Yangon, Myanmar. J. Food Prot. 2017, 80, 947–951. [Google Scholar] [CrossRef] [PubMed]
- Gibson, J.S.; Wai, H.; Oo, S.; Hmwe, E.M.M.; Wai, S.S.; Htun, L.L.; Lim, H.P.; Latt, Z.M.; Henning, J. Antimicrobials use and resistance on integrated poultry-fish farming systems in the Ayeyarwady Delta of Myanmar. Sci. Rep. 2020, 10, 16149. [Google Scholar] [CrossRef]
- Lay, K.K.; Jeamsripong, S.; Sunn, K.P.; Angkititrakul, S.; Prathan, R.; Srisanga, S.; Chuanchuen, R. Colistin Resistance and ESBL Production in Salmonella and Escherichia coli from Pigs and Pork in the Thailand, Cambodia, Lao PDR, and Myanmar Border Area. Antibiotics 2021, 10, 657. [Google Scholar] [CrossRef] [PubMed]
- Munkholm, L.; Rubin, O.; Baekkeskov, E.; Humboldt-Dachroeden, S. Attention to the Tripartite’s one health measures in national action plans on antimicrobial resistance. J. Public Health Policy 2021, 42, 236–248. [Google Scholar] [CrossRef]
- Ministry of Health and Ministry of Agriculture and Forestry. National Strategic Plan on Antimicrobial Resistance in Lao PDR 2019–2023. Government of Lao People’s Democratic Republic. Available online: https://www.who.int/publications/m/item/lao-pdr-national-strategic-plan-on-antimicrobial-resistance-in-lao-pdr-2019-2023 (accessed on 31 October 2021).
- Munkholm, L.; Rubin, O. The global governance of antimicrobial resistance: A cross-country study of alignment between the global action plan and national action plans. Glob. Health 2020, 16, 109. [Google Scholar] [CrossRef] [PubMed]
- FAO. The FAO Action Plan on Antimicrobial Resistance 2021–2025. Available online: https://www.fao.org/documents/card/en/c/cb5545en/ (accessed on 26 December 2021).
- FAO. The FAO Action Plan on Antimicrobial Resistance 2016–2020. Available online: http://www.fao.org/3/a-i5996e.pdf (accessed on 12 October 2021).
- OIE. The OIE Strategy on Antimicrobial Resistance and the Prudent Use of Antimicrobials. Available online: https://www.oie.int/fileadmin/Home/eng/Media_Center/docs/pdf/PortailAMR/EN_OIE-AMRstrategy.pdf (accessed on 15 December 2021).
- Houattongkham, S.; Yamamoto, E.; Sithivong, N.; Inthaphatha, S.; Kariya, T.; Saw, Y.M.; Vongduangchanh, A.; Keosavanh, O.; Hamajima, N. Etiologic agents of acute diarrhea in sentinel surveillance sites in Vientiane Capital, Lao People’s Democratic Republic, 2012–2015. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1115–1122. [Google Scholar] [CrossRef]
- EFSA. Technical specifications on harmonised monitoring of antimicrobial resistance in zoonotic and indicator bacteria from food-producing animals and food. EFSA J. 2019, 17, e05709. [Google Scholar] [CrossRef] [Green Version]
- Tu, L.T.; Hoang, N.V.; Cuong, N.V.; Campbell, J.; Bryant, J.E.; Hoa, N.T.; Kiet, B.T.; Thompson, C.; Duy, D.T.; Phat, V.V.; et al. High levels of contamination and antimicrobial-resistant non-typhoidal Salmonella serovars on pig and poultry farms in the Mekong Delta of Vietnam. Epidemiol. Infect. 2015, 143, 3074–3086. [Google Scholar] [CrossRef] [Green Version]
- FAO. FAO Assessment Tool for Laboratories and AMR Surveillance Systems (FAO-ATLASS). Available online: https://www.fao.org/antimicrobial-resistance/resources/tools/fao-atlass/en/ (accessed on 31 October 2021).
- FAO. Monitoring and Surveillance of Antimicrobial Resistance in Bacteria from Healthy Food Animals Intended for Consumption. Available online: http://www.fao.org/3/ca6897en/CA6897EN.pdf (accessed on 18 June 2018).
- Thermo Fisher Scientific. Sensititre Antimicrobial Susceptibility Testing System. Available online: https://www.thermofisher.com/la/en/home/clinical/clinical-microbiology/antimicrobial-susceptibility-testing/sensititre-antimicrobial-susceptibility-testing-system.html (accessed on 31 October 2021).
- CLSI (Ed.) Performance Standards for Antimicrobial Susceptibility Testing, CLSI supplement M100, 31st ed.; Clinical Laboratory Standard Institute: Wayne, PA, USA, 2021. [Google Scholar]
- Government of Canada. Canadian Integrated Program for Antimicrobial Resistance Surveillance 2018: Design And Methods. Available online: https://www.canada.ca/en/public-health/services/surveillance/canadian-integrated-program-antimicrobial-resistance-surveillance-cipars/cipars-reports/2018-annual-report-design-methods.html (accessed on 24 December 2021).
- United States Center for Disease Control and Prevention; National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS). Antibiotics Tested by NARMS. Available online: https://www.cdc.gov/narms/antibiotics-tested.html (accessed on 24 December 2021).
- European Centre for Disease Prevention and Control. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2018/2019. EFSA J. 2021, 19, e06490. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [Green Version]
- WHO. Critically Important Antimicrobials for Human Medicine 6th Revision. Available online: https://www.who.int/publications/i/item/9789241515528 (accessed on 31 October 2021).
- Chansamouth, V.; Mayxay, M.; Dance, D.A.; Roberts, T.; Phetsouvanh, R.; Vannachone, B.; Vongsouvath, M.; Davong, V.; Inthavong, P.; Khounsy, S.; et al. Antimicrobial use and resistance data in human and animal sectors in the Lao PDR: Evidence to inform policy. BMJ Glob. Health 2021, 6, e007009. [Google Scholar] [CrossRef] [PubMed]
- Poupaud, M.; Putthana, V.; Patriarchi, A.; Caro, D.; Agunos, A.; Tansakul, N.; Goutard, F.L. Understanding the veterinary antibiotics supply chain to address antimicrobial resistance in Lao PDR: Roles and interactions of involved stakeholders. Acta Trop. 2021, 220, 105943. [Google Scholar] [CrossRef] [PubMed]
- EFSA. Antimicrobial Resistance in Europe (Interactive Data Platform). Available online: https://multimedia.efsa.europa.eu/dataviz-2018/index.htm (accessed on 24 December 2021).
- United States Food and Drugs Administration. NARMS Interactive Data Displays. Available online: https://www.fda.gov/animal-veterinary/national-antimicrobial-resistance-monitoring-system/narms-now-integrated-data (accessed on 26 December 2021).
- National Steering Committee on Antimicrobial Resistance of Thailand. Thailand’s First One Health Report on Antimicrobial Consumption and Antimicrobial Resistance in 2017. Available online: http://ihppthaigov.net/DB/publication/attachresearch/433/chapter1.pdf (accessed on 24 December 2021).
- National Steering Committee on Antimicrobial Resistance of Thailand. Thailand’s One Health Report on Antimicrobial Consumption and Antimicrobial Resistance in 2018. Available online: http://ihppthaigov.net/DB/publication/attachresearch/432/chapter1.pdf (accessed on 24 December 2021).
- FAO. FAO Urges Countries to Discontinue the Use of Chloramphenicol in Animal Production. Available online: https://www.fao.org/asiapacific/news/detail-events/en/c/47419/ (accessed on 2 November 2021).
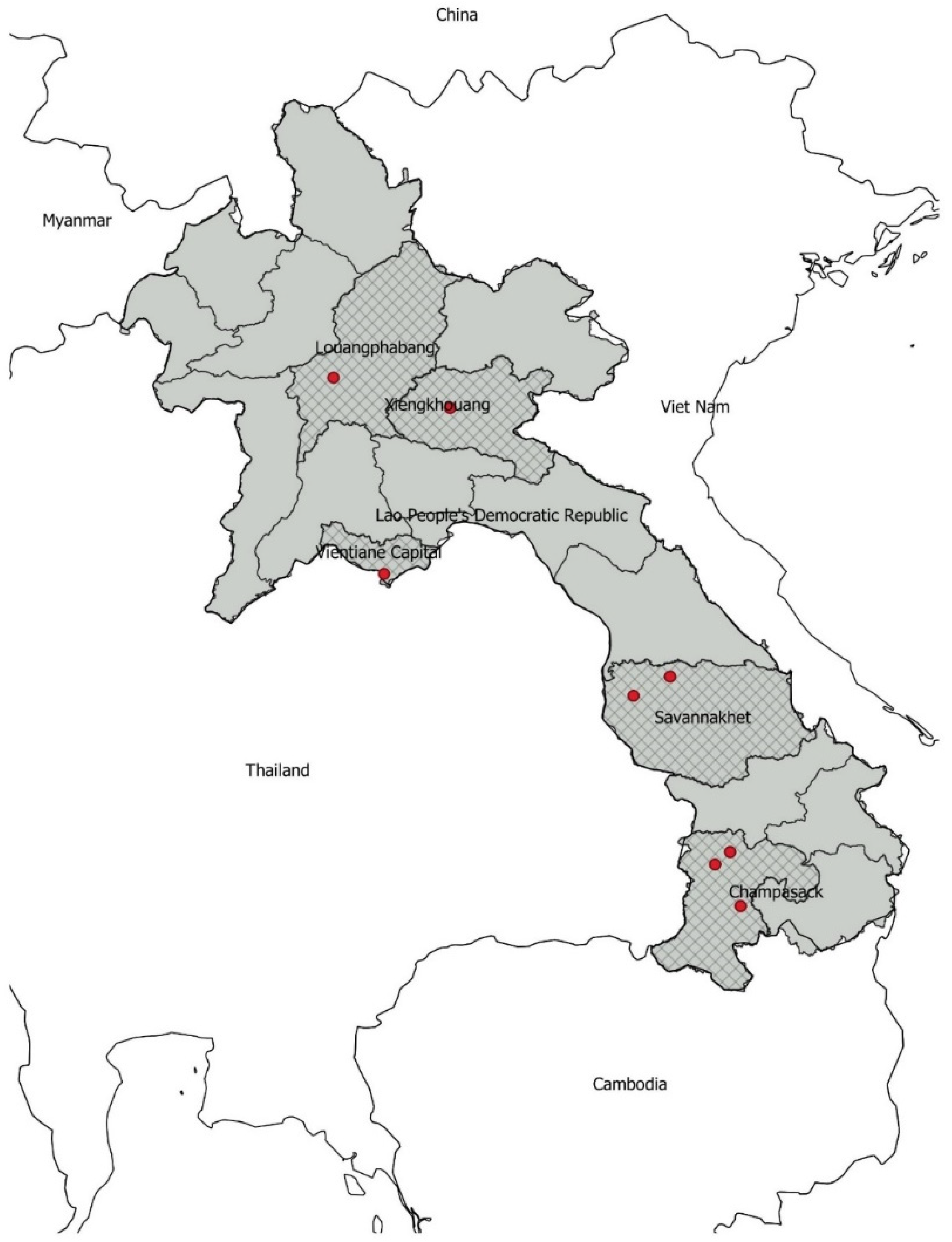


| Province | Pig (Round 1, 2018–2019) | Pig (Round 2, 2020–2021) | Poultry (Round 2, 2020–2021) | Total (by Province) | ||
|---|---|---|---|---|---|---|
| Native | Broiler | Layer | ||||
| Louangprabang | 0 | 262 | 164 | 0 | 91 | 517 |
| Xiengkhouang | 0 | 125 | 75 | 0 | 0 | 200 |
| Vientiane Capital | 160 | 0 | 0 | 255 | 510 | 925 |
| Savannakhet | 350 | 0 | 224 | 0 | 0 | 574 |
| Champasak | 262 | 0 | 362 | 574 | 224 | 1422 |
| Total (by species and breed): | 772 | 387 | 825 | 829 | 825 | 3638 |
| Resistance Phenotypes | Chickens % Isolates | Pigs % Isolates | Pigs Compared to Chickens (Odds Ratios, 95% CIs, and p-Values) |
|---|---|---|---|
| No resistance 1 | 6% | <1% | OR 0.06 (0.01–0.38), p = 0.003 |
| 1 Antimicrobial class | 13% | 3% | OR: 0.220 (0.09–0.52), p = 0.001 |
| 2 Multiclass resistance | 19% | 9% | OR: 0.43 (0.22–0.83), p = 0.011 |
| 3 Multiclass resistance | 24% | 20% | OR: 0.79(065–0.98), p = 0.036 |
| 4 Multiclass resistance | 21% | 31% | OR: 1.68 (1.49–1.90), p < 0.0001 |
| 5 Multiclass resistance | 12% | 18% | OR 1.68 (1.18–2.42), p = 0.004 |
| 6 Multiclass resistance | 4% | 10% | OR 2.4 2 (1.62–3.63), p < 0.0001 |
| 7 Multiclass resistance | 2% | 7% | OR 4.56 (2.54–8.29), p < 0.0001 |
| 8 Multiclass resistance | 0.1% | 2% | OR 22.51 (2.70–187.45), p = 0.004 |
| ≥3 Multiclass resistance 2 | 62% | 88% | OR 4.29 (1.82–10.12), p = 0.001 |
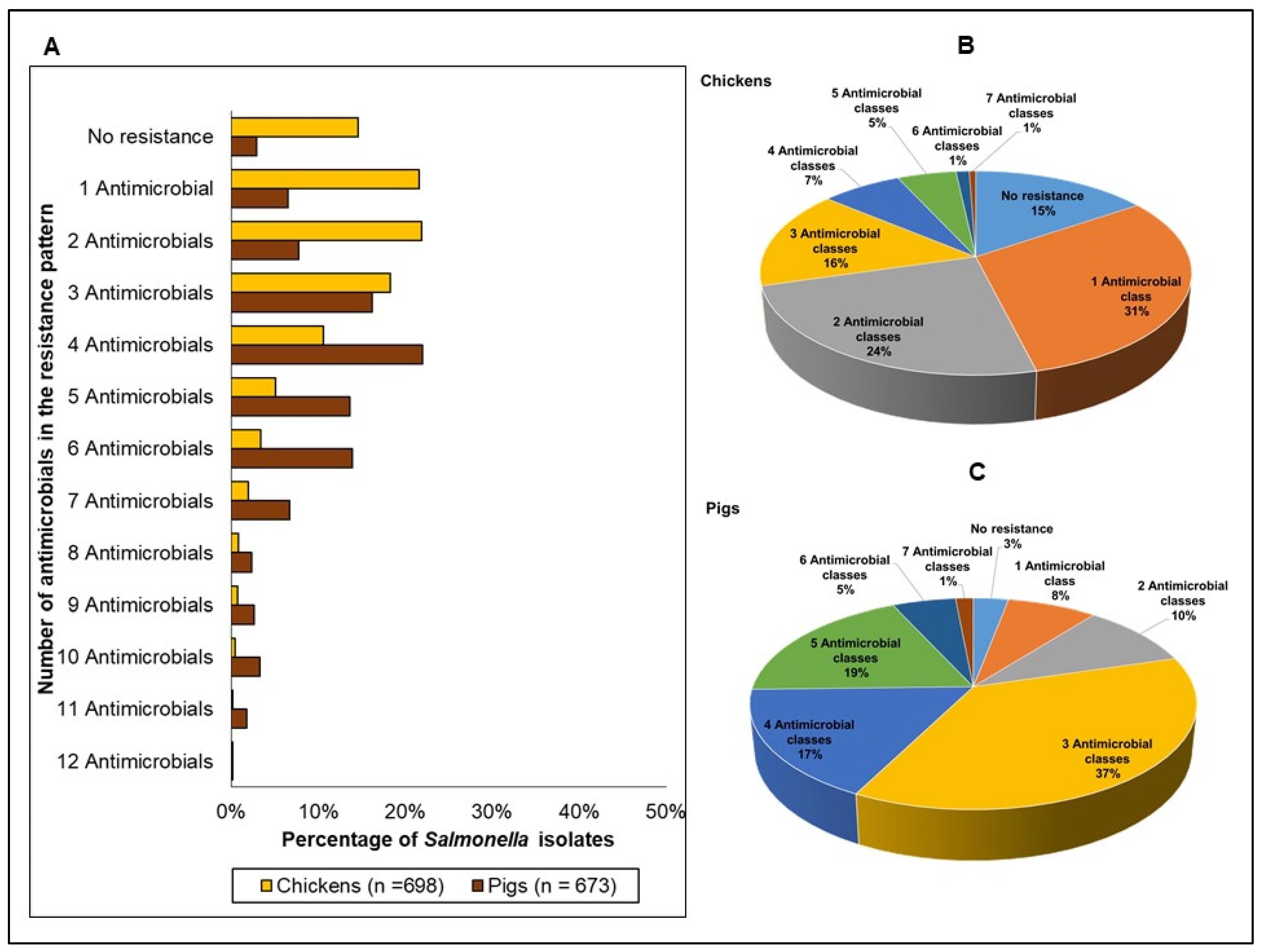
| Resistance Phenotypes | Chickens | Pigs | Pigs Compared to Chickens (Odds Ratios, 95% CIs, and p-Values) |
|---|---|---|---|
| No resistance 1 | 15% | 3% | OR 0.18 (0.07–0.46), p < 0.0001 |
| 1 Antimicrobial class | 25% | 7% | OR: 0.20 (0.10–0.46), p < 0.0001 |
| 2 Multiclass resistance | 27% | 10% | OR: 0.29 (0.16–0.51), p < 0.0001 |
| 3 Multiclass resistance | 17% | 36% | OR: 2.70 (1.86–3.93), p < 0.0001 |
| 4 Multiclass resistance | 7% | 16% | OR: 2.38 (1.72–3.29), p < 0.0001 |
| 5 Multiclass resistance | 5% | 18% | OR 3.78 (1.52–12.35), p = 0.028 |
| 6 Multiclass resistance | 1% | 6% | OR 1.58 (2.73–9.34), p < 0.0001 |
| 7 Multiclass resistance | 1% | 4% | OR 5.3 (1.10–25.44), p = 0.037 |
| 8 Multiclass resistance | <1% | <1% | OR 1.03 (0.12–8.26), p = 0.979 |
| ≥3 Multiclass resistance 2 | 33% | 81% | OR 8.53 (4.1–17.5), p < 0.0001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Inthavong, P.; Chanthavong, S.; Nammanininh, P.; Phommachanh, P.; Theppangna, W.; Agunos, A.; Wagenaar, J.A.; Douangngeun, B.; Loth, L. Antimicrobial Resistance Surveillance of Pigs and Chickens in the Lao People’s Democratic Republic, 2018–2021. Antibiotics 2022, 11, 177. https://doi.org/10.3390/antibiotics11020177
Inthavong P, Chanthavong S, Nammanininh P, Phommachanh P, Theppangna W, Agunos A, Wagenaar JA, Douangngeun B, Loth L. Antimicrobial Resistance Surveillance of Pigs and Chickens in the Lao People’s Democratic Republic, 2018–2021. Antibiotics. 2022; 11(2):177. https://doi.org/10.3390/antibiotics11020177
Chicago/Turabian StyleInthavong, Phouth, Somphaivanh Chanthavong, Phounsavanh Nammanininh, Phouvong Phommachanh, Watthana Theppangna, Agnes Agunos, Jaap A. Wagenaar, Bounlom Douangngeun, and Leo Loth. 2022. "Antimicrobial Resistance Surveillance of Pigs and Chickens in the Lao People’s Democratic Republic, 2018–2021" Antibiotics 11, no. 2: 177. https://doi.org/10.3390/antibiotics11020177
APA StyleInthavong, P., Chanthavong, S., Nammanininh, P., Phommachanh, P., Theppangna, W., Agunos, A., Wagenaar, J. A., Douangngeun, B., & Loth, L. (2022). Antimicrobial Resistance Surveillance of Pigs and Chickens in the Lao People’s Democratic Republic, 2018–2021. Antibiotics, 11(2), 177. https://doi.org/10.3390/antibiotics11020177






