Exploring Structure–Activity Relationships of Niclosamide-Based Colistin Potentiators in Colistin-Resistant Gram-Negative Bacteria
Abstract
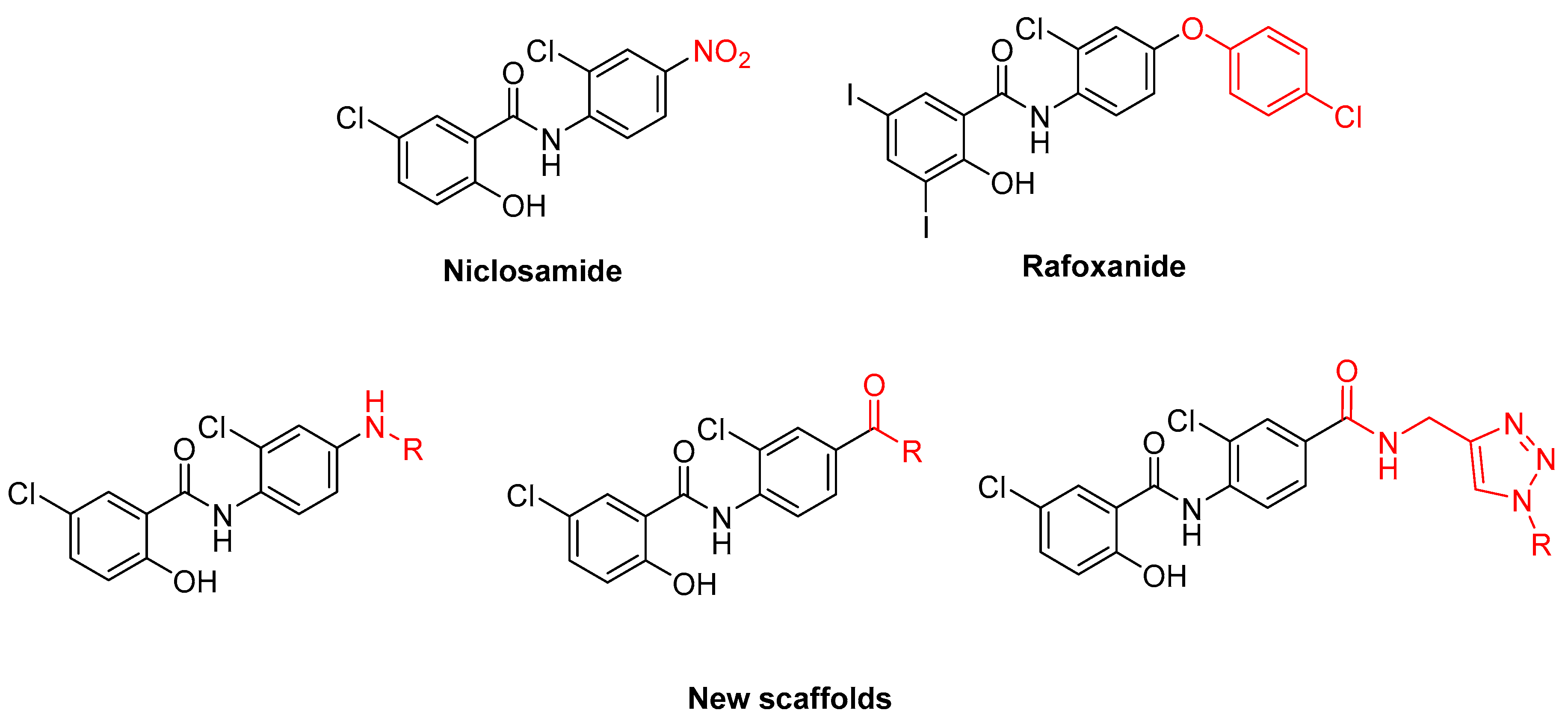
:1. Introduction
2. Results and Discussion
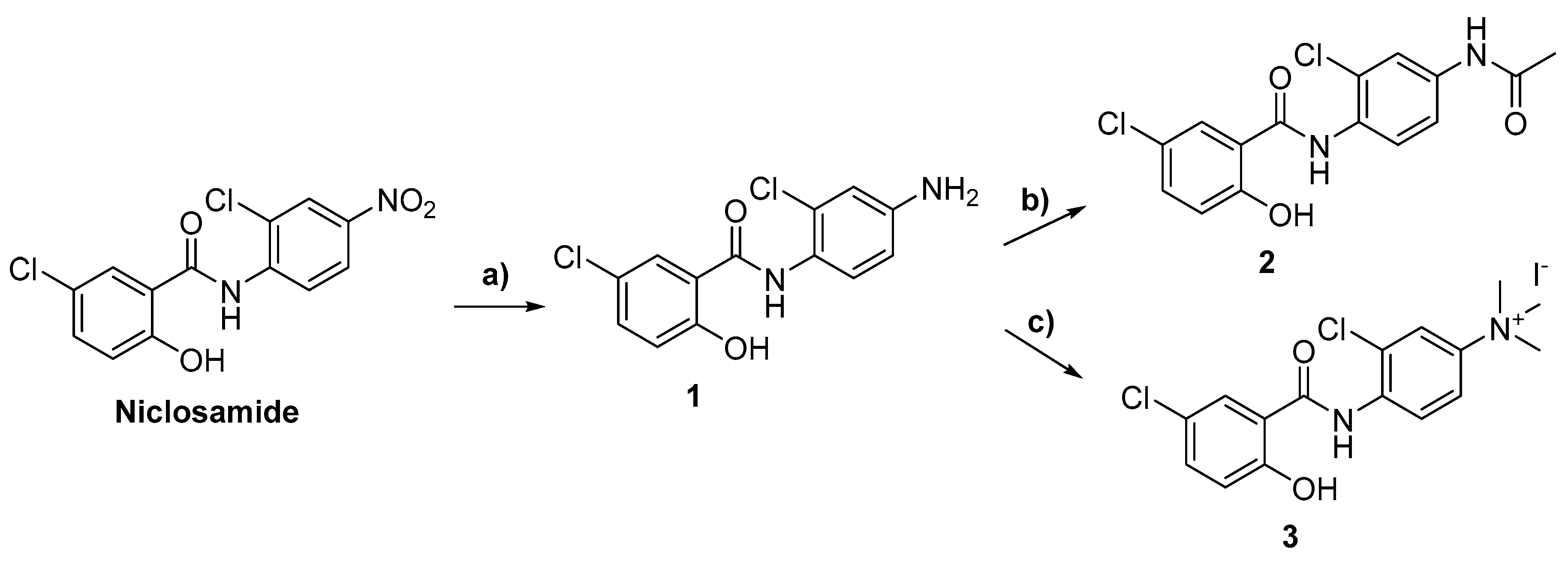
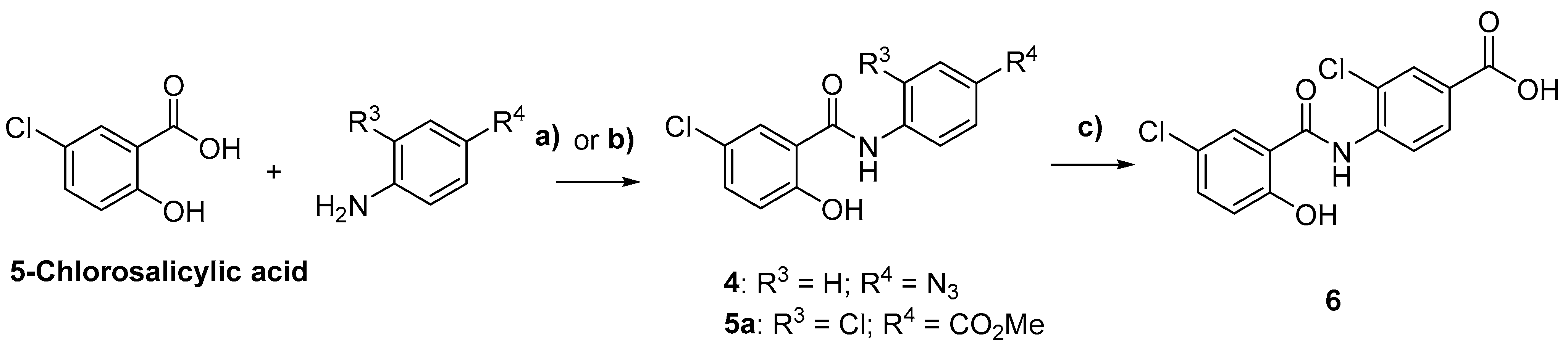
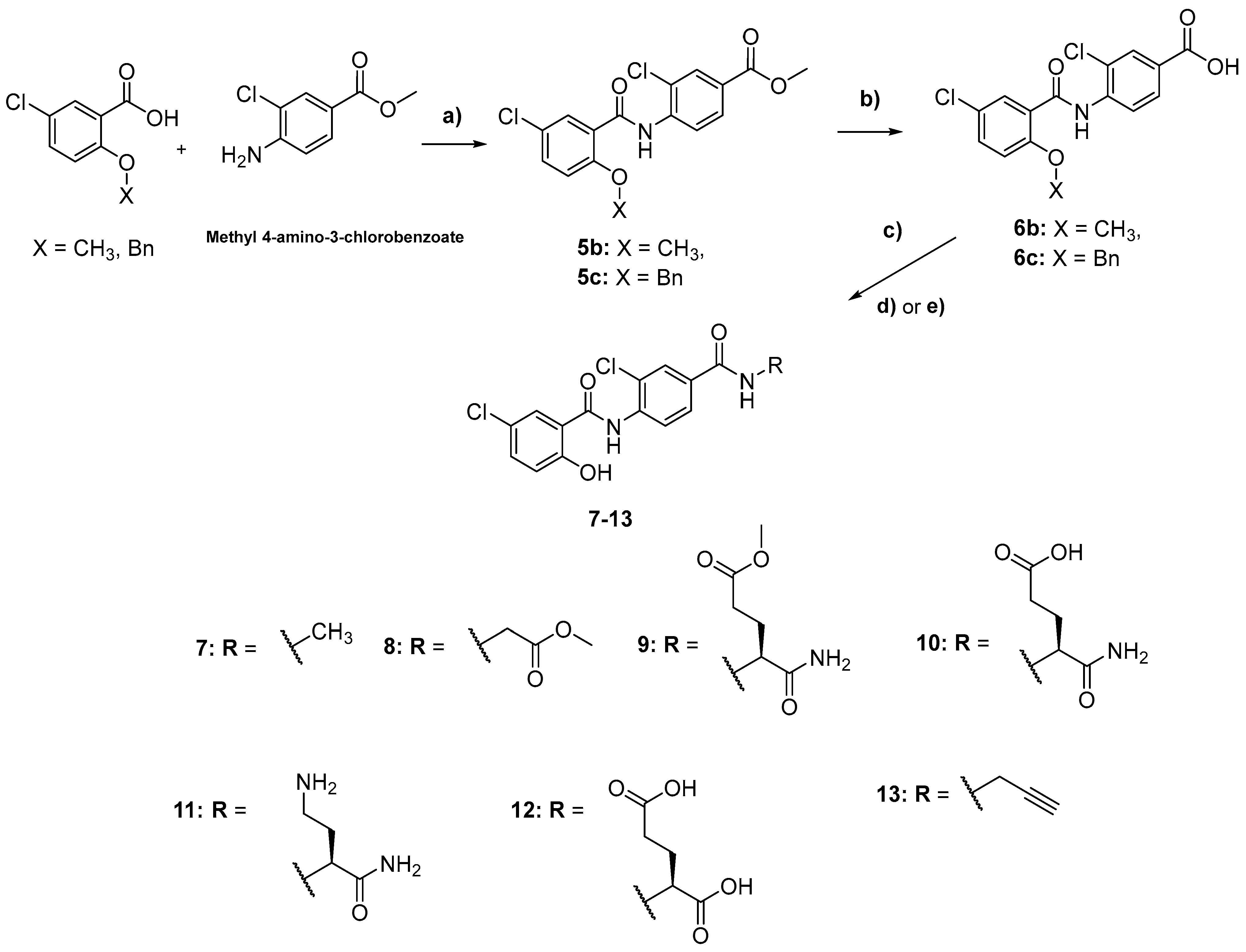
2.1. Chemistry
2.2. Biological Evaluations
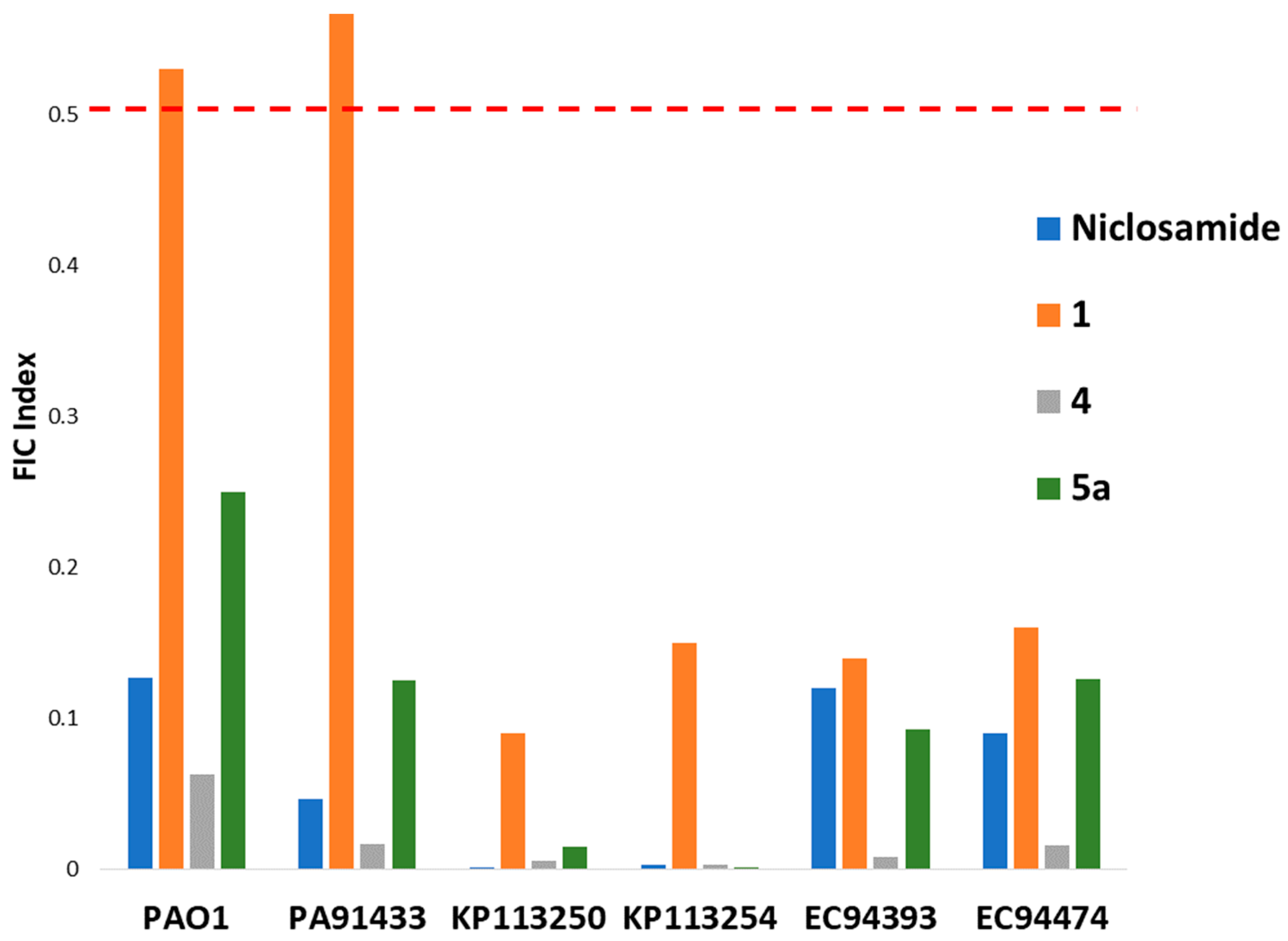
2.2.1. Initial Screen for Synergy with Colistin and SAR
2.2.2. Synergy between 5a and Colistin against P. aeruginosa and A. baumannii
2.2.3. Cytotoxicity of 1 and 5a against Eukaryotic Cells
3. Materials and Methods
3.1. Materials
3.2. Synthetic Methods
General Synthetic Procedures
3.3. Antibacterial Activity
3.3.1. Antimicrobial Susceptibility Assay
3.3.2. Checkerboard Assay
3.4. Cell Proliferation Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Theuretzbacher, U. Global Antimicrobial Resistance in Gram-Negative Pathogens and Clinical Need. Curr. Opin. Microbiol. 2017, 39, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.D.; Wright, G.D. Antibacterial Drug Discovery in the Resistance Era. Nature 2016, 529, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Todd, R.; Kiehlbauch, J.; Chen, L.; Walters, M.; Kallen, A. Notes from the Field: Pan-Resistant New Delhi Metallo-Beta-Lactamase-Producing Klebsiella Pneumoniae—Washoe County, Nevada, 2016. MMWR Morb. Mortal Wkly. Rep. 2017, 66, 33. [Google Scholar] [CrossRef]
- Prasad, V.; Mailankody, S. Research and Development Spending to Bring a Single Cancer Drug to Market and Revenues after Approval. JAMA Intern. Med. 2017, 177, 1569. [Google Scholar] [CrossRef]
- Fernandes, P.; Martens, E. Antibiotics in Late Clinical Development. Biochem. Pharmacol. 2017, 133, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Ejim, L.; Farha, M.A.; Falconer, S.B.; Wildenhain, J.; Coombes, B.K.; Tyers, M.; Brown, E.D.; Wright, G.D. Combinations of Antibiotics and Nonantibiotic Drugs Enhance Antimicrobial Efficacy. Nat. Chem. Biol. 2011, 7, 348–350. [Google Scholar] [CrossRef] [PubMed]
- Rajamuthiah, R.; Fuchs, B.B.; Conery, A.L.; Kim, W.; Jayamani, E.; Kwon, B.; Ausubel, F.M.; Mylonakis, E. Repurposing Salicylanilide Anthelmintic Drugs to Combat Drug Resistant Staphylococcus Aureus. PLoS ONE 2015, 10, e0124595. [Google Scholar] [CrossRef]
- Imperi, F.; Massai, F.; Pillai, C.R.; Longo, F.; Zennaro, E.; Rampioni, G.; Visc, P.; Leoni, L. New Life for an Old Drug: The Anthelmintic Drug Niclosamide Inhibits Pseudomonas Aeruginosa Quorum Sensing. Antimicrob. Agents Chemother. 2013, 57, 996–1005. [Google Scholar] [CrossRef]
- Domalaon, R.; Malaka De Silva, P.; Kumar, A.; Zhanel, G.G.; Schweizer, F. The Anthelmintic Drug Niclosamide Synergizes with Colistin and Reverses Colistin Resistance in Gram-Negative Bacilli. Antimicrob. Agents Chemother. 2019, 63, e02574-18. [Google Scholar] [CrossRef]
- Jara, M.O.; Warnken, Z.N.; Sahakijpijarn, S.; Thakkar, R.; Kulkarni, V.R.; Christensen, D.J.; Koleng, J.J.; Williams, R.O. Oral Delivery of Niclosamide as an Amorphous Solid Dispersion That Generates Amorphous Nanoparticles during Dissolution. Pharmaceutics 2022, 14, 2568. [Google Scholar] [CrossRef]
- Chen, W.; Mook, R.A.; Premont, R.T.; Wang, J. Niclosamide: Beyond an Antihelminthic Drug. Cell Signal. 2018, 41, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Mattingly, A.E.; Jania, L.A.; Smith, R.; Melander, R.J.; Ernst, R.K.; Koller, B.H.; Melander, C. Benzimidazole Isosteres of Salicylanilides Are Highly Active Colistin Adjuvants. ACS Infect. Dis. 2021, 7, 3303–3313. [Google Scholar] [CrossRef] [PubMed]
- Copp, J.N.; Pletzer, D.; Brown, A.S.; Van der Heijden, J.; Miton, C.M.; Edgar, R.J.; Rich, M.H.; Little, R.F.; Williams, E.M.; Hancock, R.E.W.; et al. Mechanistic Understanding Enables the Rational Design of Salicylanilide Combination Therapies for Gram-Negative Infections. mBio 2020, 11, e02068-20. [Google Scholar] [CrossRef] [PubMed]
- Ngai, T.W.; Elfar, G.A.; Yeo, P.; Phua, N.; Hor, J.H.; Chen, S.; Ho, Y.S.; Cheok, C.F. Nitro-Deficient Niclosamide Confers Reduced Genotoxicity and Retains Mitochondrial Uncoupling Activity for Cancer Therapy. Int. J. Mol. Sci. 2021, 22, 10420. [Google Scholar] [CrossRef] [PubMed]
- Domalaon, R.; Okunnu, O.; Zhanel, G.G.; Schweizer, F. Synergistic Combinations of Anthelmintic Salicylanilides Oxyclozanide, Rafoxanide, and Closantel with Colistin Eradicates Multidrug-Resistant Colistin-Resistant Gram-Negative Bacilli. J. Antibiot. 2019, 72, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Huisgen, R. 1,3-Dipolar Cycloadditions. Past and Future. Angew. Chem. Int. Ed. Engl. 1963, 2, 565–598. [Google Scholar] [CrossRef]
- Mook, R.A.; Ren, X.-R.; Wang, J.; Piao, H.; Barak, L.S.; Lyerly, H.K.; Chen, W. Benzimidazole Inhibitors from the Niclosamide Chemotype Inhibit Wnt/β-Catenin Signaling with Selectivity over Effects on ATP Homeostasis. Bioorganic Med. Chem. 2017, 25, 1804–1816. [Google Scholar] [CrossRef]
- Walkty, A.; Karlowsky, J.A.; Adam, H.J.; Lagace-Wiens, P.; Baxter, M.; Mulvey, M.R.; McCracken, M.; Poutanen, S.M.; Roscoe, D.; Zhanel, G.G. Frequency of MCR-1-Mediated Colistin Resistance among Escherichia Coli Clinical Isolates Obtained from Patients in Canadian Hospitals (CANWARD 2008-2015). CMAJ Open 2016, 4, E641–E645. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: A Microbiological and Molecular Biological Study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Shangguan, F.; Liu, Y.; Ma, L.; Qu, G.; Lv, Q.; An, J.; Yang, S.; Lu, B.; Cao, Q. Niclosamide Inhibits Ovarian Carcinoma Growth by Interrupting Cellular Bioenergetics. J. Cancer 2020, 11, 3454–3466. [Google Scholar] [CrossRef]
- Bittman, R.; Li, Z.; Samadder, P.; Arthur, G. Anticancer Activity of a Ceramide Analog Containing a Disulfide Linkage. Cancer Lett. 2007, 251, 53–58. [Google Scholar] [CrossRef]
- Zhanel, G.G.; DeCorby, M.; Adam, H.; Mulvey, M.R.; McCracken, M.; Lagacé-Wiens, P.; Nichol, K.A.; Wierzbowski, A.; Baudry, P.J.; Tailor, F.; et al. Prevalence of Antimicrobial-Resistant Pathogens in Canadian Hospitals: Results of the Canadian Ward Surveillance Study (CANWARD 2008). Antimicrob. Agents Chemother. 2010, 54, 4684–4693. [Google Scholar] [CrossRef]
- Hoban, D.J.; Zhanel, G.G. Introduction to the CANWARD Study (2007–11). J. Antimicrob. Chemother. 2013, 68 (Suppl. S1), i3–i5. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018; ISBN 1562389238. [Google Scholar]
- Berry, L.; Brizuela, M.; Jackson, G.; Schweizer, F. A Niclosamide-Tobramycin Hybrid Adjuvant Potentiates Cefiderocol AgainstP. Aeruginosa. RSC Med. Chem. 2021, 12, 1565–1573. [Google Scholar] [CrossRef]
- Wang, D.; Zou, L.; Jin, Q.; Hou, J.; Ge, G.; Yang, L. Human Carboxylesterases: A Comprehensive Review. Acta Pharm. Sin. B 2018, 8, 699–712. [Google Scholar] [CrossRef]



 | |||||
|---|---|---|---|---|---|
| Colistin MIC with 4 μM Compound (μg/mL) | |||||
| Compound | R | KP113250 | KP113254 | EC94393 | EC94474 |
| Niclosamide | NO2 | 0.25 | 0.5 | 0.25 | 0.5 |
| 1 | NH2 | 16 | 32 | 0.5 | 2 |
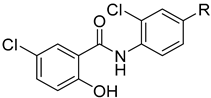
| 2 |  | 256 | 256 | 8 | 16 |
| 3 | 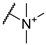 | 256 | 256 | 8 | 16 |
| Colistin MIC alone | 256 | 256 | 8 | 16 | |
 | ||||||
|---|---|---|---|---|---|---|
| Colistin MIC with 4 μM Compound (μg/mL) | ||||||
| Compound | R1 | R2 | KP113250 | KP113254 | EC94393 | EC94474 |
| Niclosamide | Cl | NO2 | 0.25 | 0.5 | 0.25 | 0.5 |
| 4 | H | N3 | 0.25 | 0.5 | 0.016 | 0.25 |
| 5a | Cl |  | 1 | 1 | 0.25 | 1 |
| 6 | Cl |  | 256 | 256 | 8 | 16 |
| 7 | Cl | 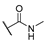 | 256 | 256 | 8 | 16 |
| 8 | Cl |  | 256 | 256 | 8 | 16 |
| 9 | Cl | 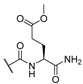 | 256 | 256 | 8 | 16 |
| 10 | Cl | 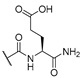 | 256 | 256 | 8 | 16 |
| 11 | Cl | 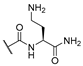 | 256 | 256 | 8 | 16 |
| 12 | Cl |  | 256 | 256 | 8 | 16 |
| 14 | Cl |  | 256 | 256 | 8 | 16 |
| 15 | Cl | 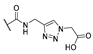 | 256 | 256 | 8 | 16 |
| 16 | Cl | 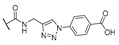 | 256 | 256 | 8 | 16 |
| 17 | Cl |  | 256 | 256 | 8 | 16 |
| Colistin MIC alone | 256 | 256 | 8 | 16 | ||
 | |||||||
|---|---|---|---|---|---|---|---|
| Colistin MIC with 4 μM Compound (μg/mL) | |||||||
| Compound | R1 | R2 | R3 | KP113250 | KP113254 | EC94393 | EC94474 |
| Niclosamide | Cl | H | NO2 | 0.25 | 0.5 | 0.25 | 0.5 |
| 18 | H | OH | NO2 | 256 | 256 | 8 | 16 |
| 19 | OH | OH | NO2 | 256 | 256 | 8 | 16 |
| 20 | Cl | H | 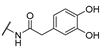 | 256 | 256 | 8 | 16 |
| 21 | Cl | H | 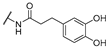 | 256 | 256 | 8 | 16 |
| Colistin MIC alone | 256 | 256 | 8 | 16 | |||
| Strain | MICCol | MICCombi | MIC5a | MICCombi | FICI | Interpretation |
|---|---|---|---|---|---|---|
| PAO1 | 1 | 0.25 | >256 | 0.25 | 0.250 < x < 0.251 | Synergy |
| PA095 | 0.25 | 0.016 | >256 | 16 | 0.063 < x < 0.125 | Synergy |
| PA259 | 0.25 | 0.25 | >256 | 1 | 1.000 < x < 1.004 | Additive |
| PA260 | 0.25 | 0.125 | >256 | 1 | 0.500 < x < 0.504 | Additive |
| PA264 | 1 | 0.25 | >256 | 0.5 | 0.250 < x < 0.252 | Synergy |
| PA262 | 4 | 0.25 | >256 | 1 | 0.063 < x < 0.066 | Synergy |
| PA100036 | 2 | 0.25 | >256 | 1 | 0.125 < x < 0.129 | Synergy |
| PA101885 | 4 | 0.5 | >256 | 1 | 0.125 < x < 0.129 | Synergy |
| PA91433 | 4 | 0.5 | >256 | 0.25 | 0.125 < x < 0.126 | Synergy |
| PA101243 | 1024 | 0.5 | >256 | 1 | 0.0005 < x < 0.0045 | Synergy |
| Strain | MICCol | MICCombi | MIC5a | MICCombi | FICI | Interpretation |
|---|---|---|---|---|---|---|
| A. baumannii ATCC 17978 | 1 | 0.25 | >256 | 0.25 | 0.250 < x < 0.251 | Synergy |
| A baumannii 110193 | 4 | 0.5 | >256 | 0.25 | 0.125 < x < 0.126 | Synergy |
| AB027 | 1024 | 0.5 | >256 | 1 | 0.0005 < x < 0.004 | Synergy |
| AB031 | 0.25 | 0.015625 | >256 | 16 | 0.063 < x < 0.125 | Synergy |
| LAC-4 | 0.25 | 0.25 | >256 | 1 | 1.000 < x < 1.004 | Additive |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berry, L.; Neale, Q.; Arora, R.; Ramirez, D.; Brizuela, M.; Domalaon, R.; Arthur, G.; Schweizer, F. Exploring Structure–Activity Relationships of Niclosamide-Based Colistin Potentiators in Colistin-Resistant Gram-Negative Bacteria. Antibiotics 2024, 13, 43. https://doi.org/10.3390/antibiotics13010043
Berry L, Neale Q, Arora R, Ramirez D, Brizuela M, Domalaon R, Arthur G, Schweizer F. Exploring Structure–Activity Relationships of Niclosamide-Based Colistin Potentiators in Colistin-Resistant Gram-Negative Bacteria. Antibiotics. 2024; 13(1):43. https://doi.org/10.3390/antibiotics13010043
Chicago/Turabian StyleBerry, Liam, Quinn Neale, Rajat Arora, Danyel Ramirez, Marc Brizuela, Ronald Domalaon, Gilbert Arthur, and Frank Schweizer. 2024. "Exploring Structure–Activity Relationships of Niclosamide-Based Colistin Potentiators in Colistin-Resistant Gram-Negative Bacteria" Antibiotics 13, no. 1: 43. https://doi.org/10.3390/antibiotics13010043
APA StyleBerry, L., Neale, Q., Arora, R., Ramirez, D., Brizuela, M., Domalaon, R., Arthur, G., & Schweizer, F. (2024). Exploring Structure–Activity Relationships of Niclosamide-Based Colistin Potentiators in Colistin-Resistant Gram-Negative Bacteria. Antibiotics, 13(1), 43. https://doi.org/10.3390/antibiotics13010043








