Decline in ESBL Production and Carbapenem Resistance in Urinary Tract Infections among Key Bacterial Species during the COVID-19 Pandemic
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design, Setting, and Duration
2.2. Inclusion and Exclusion Criteria
2.3. Methodology for the Cultivation and Analysis of Urine Samples: Adhering to WHO Guidelines and Utilizing Semi-Quantitative Techniques
2.4. Application of the BD Phoenix System in Bacterial Isolate Identification
2.5. Antimicrobial Susceptibility Testing of E. coli, Pseudomonas, and Klebsiella Strains Using the Kirby–Bauer Method for Carbapenem Resistance Identification
2.6. Application of the Double-Disc Synergy Test (DDST) for ESBL Detection
2.7. Statistical Analysis
3. Results
3.1. ESBL Production
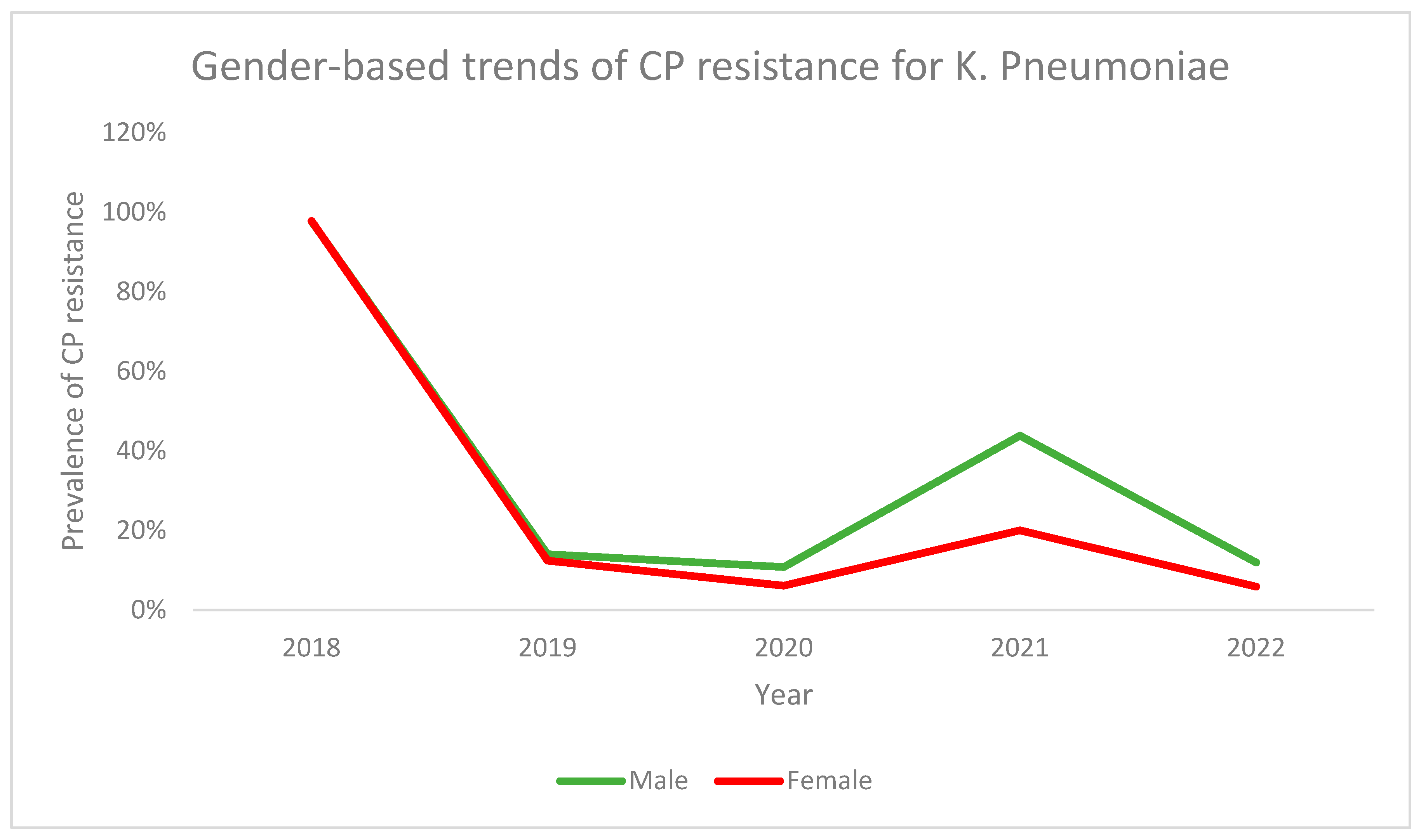
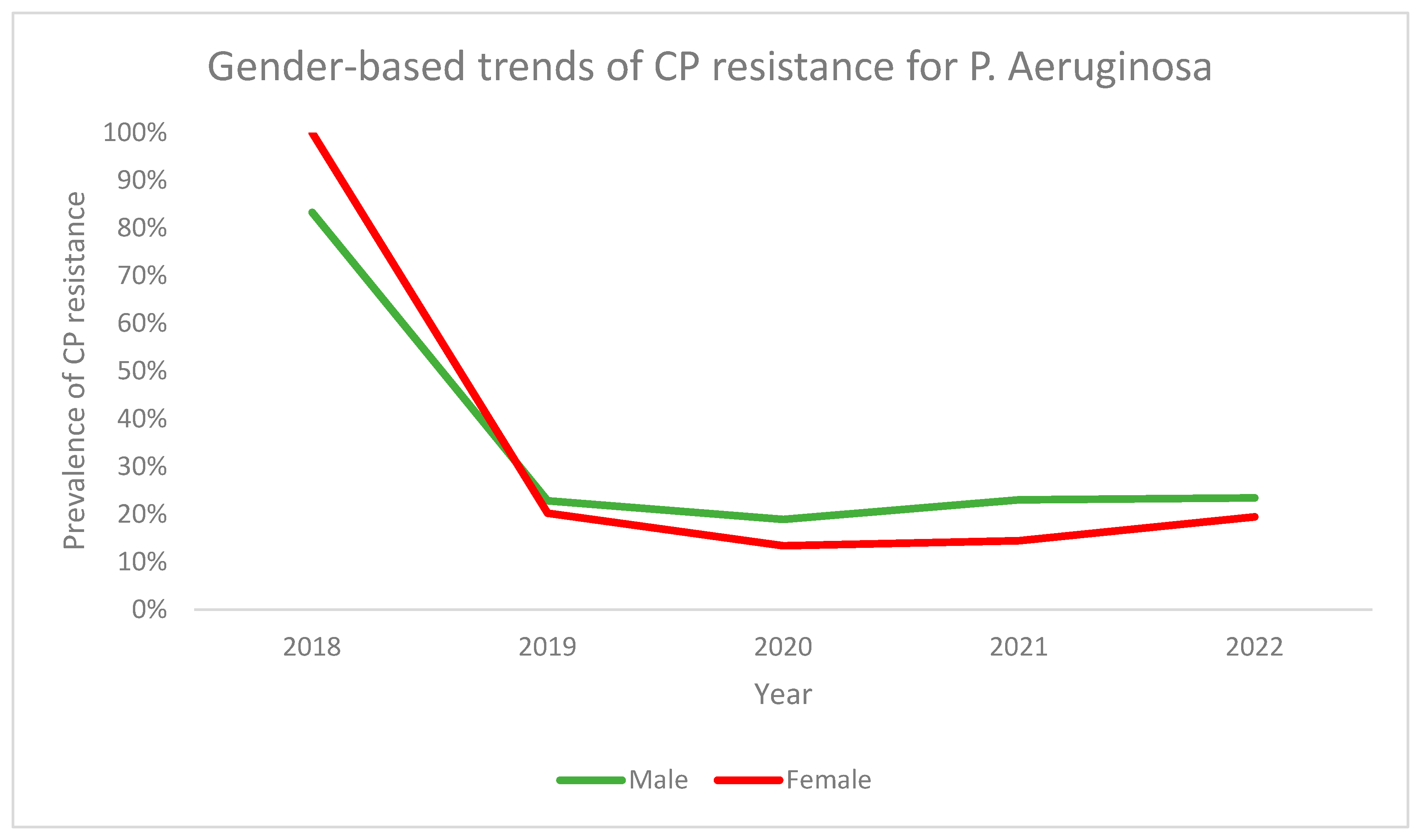
3.2. Carbapenem (CP) Resistance
4. Discussion
Strengths and Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Stamm, W.E.; Norrby, S.R. Urinary Tract Infections: Disease Panorama and Challenges. J. Infect. Dis. 2001, 183, S1–S4. [Google Scholar] [CrossRef] [PubMed]
- Flores-Mireles, A.; Hreha, T.N.; Hunstad, D.A. Pathophysiology, Treatment, and Prevention of Catheter-Associated Urinary Tract Infection. Top. Spinal Cord Inj. Rehabil. 2019, 25, 228–240. [Google Scholar] [CrossRef] [PubMed]
- Ku, J.H.; Bruxvoort, K.J.; Salas, S.B.; Varley, C.D.; Casey, J.A.; Raphael, E.; Robinson, S.C.; Nachman, K.E.; Lewin, B.J.; Contreras, R.; et al. Multidrug Resistance of Escherichia coli from Outpatient Uncomplicated Urinary Tract Infections in a Large United States Integrated Healthcare Organization. Open Forum Infect. Dis. 2023, 10, ofad287. [Google Scholar] [CrossRef] [PubMed]
- Sabih, A.; Leslie, S.W. Complicated Urinary Tract Infections; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Dai, B.; Liu, Y.; Jia, J.; Mei, C. Long-Term Antibiotics for the Prevention of Recurrent Urinary Tract Infection in Children: A Systematic Review and Meta-Analysis. Arch. Dis. Child. 2010, 95, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary Tract Infections: Epidemiology, Mechanisms of Infection and Treatment Options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Agegnehu, A.; Worku, M.; Nigussie, D.; Lulu, B.; Tadesse, B.T. Pediatric Febrile Urinary Tract Infection Caused by ESBL Producing Enterobacteriaceae Species. BioMed Res. Int. 2020, 2020, 6679029. [Google Scholar] [CrossRef]
- Goyal, D.; Dean, N.; Neill, S.; Jones, P.; Dascomb, K. Risk Factors for Community-Acquired Extended-Spectrum Beta-Lactamase–Producing Enterobacteriaceae Infections—A Retrospective Study of Symptomatic Urinary Tract Infections. Open Forum Infect. Dis. 2019, 6, ofy357. [Google Scholar] [CrossRef]
- Tabasi, M.; Karam, M.R.A.; Habibi, M.; Mostafavi, E.; Bouzari, S. Genotypic Characterization of Virulence Factors in Escherichia coli Isolated from Patients with Acute Cystitis, Pyelonephritis and Asymptomatic Bacteriuria. J. Clin. Diagn. Res. 2016, 10, DC01. [Google Scholar] [CrossRef]
- Pana, Z.D.; Zaoutis, T. Treatment of Extended-Spectrum β-Lactamase-Producing (ESBLs) Infections: What Have We Learned until Now? F1000Research 2018, 7, 1347. [Google Scholar] [CrossRef]
- Kawamura, K.; Nagano, N.; Suzuki, M.; Wachino, J.; Kimura, K.; Arakawa, Y. ESBL-Producing Escherichia coli and Its Rapid Rise among Healthy People. Food Saf. 2017, 5, 122–150. [Google Scholar] [CrossRef]
- Vachvanichsanong, P.; McNeil, E.B.; Dissaneewate, P. Extended-Spectrum Beta-Lactamase Escherichia coli and Klebsiella pneumoniae Urinary Tract Infections. Epidemiol. Infect. 2021, 149, e12. [Google Scholar] [CrossRef] [PubMed]
- Abalkhail, A.; AlYami, A.S.; Alrashedi, S.F.; Almushayqih, K.M.; Alslamah, T.; Alsalamah, Y.A.; Elbehiry, A. The Prevalence of Multidrug-Resistant Escherichia coli Producing ESBL among Male and Female Patients with Urinary Tract Infections in Riyadh Region, Saudi Arabia. Healthcare 2022, 10, 1778. [Google Scholar] [CrossRef] [PubMed]
- De Kraker, M.E.A.; Stewardson, A.J.; Harbarth, S. Will 10 Million People Die a Year Due to Antimicrobial Resistance by 2050? PLoS Med. 2016, 13, e1002184. [Google Scholar] [CrossRef]
- Ventola, C.L. The Antibiotic Resistance Crisis: Part 1: Causes and Threats. Pharm. Ther. 2015, 40, 277. [Google Scholar]
- Knight, G.M.; Glover, R.E.; McQuaid, C.F.; Olaru, I.D.; Gallandat, K.; Leclerc, Q.J.; Fuller, N.M.; Willcocks, S.J.; Hasan, R.; van Kleef, E.; et al. Antimicrobial Resistance and COVID-19: Intersections and Implications. eLife 2021, 10, e64139. [Google Scholar] [CrossRef]
- Altamimi, I.; Almazyed, A.; Alshammary, S.; Altamimi, A.; Alhumimidi, A.; Alnutaifi, R.; Malhis, M.; Altamimi, A. Bacterial Pathogens and Antimicrobial Susceptibility Patterns of Urinary Tract Infections in Children during COVID-19 2019–2020: A Large Tertiary Care Center in Saudi Arabia. Children 2023, 10, 971. [Google Scholar] [CrossRef]
- Mansouri, F.; Sheibani, H.; Javedani Masroor, M.; Afsharian, M. Extended-Spectrum Beta-Lactamase (ESBL)-Producing Enterobacteriaceae and Urinary Tract Infections in Pregnant/Postpartum Women: A Systematic Review and Meta-Analysis. Int. J. Clin. Pract. 2019, 73, e13422. [Google Scholar] [CrossRef]
- Langford, B.J.; So, M.; Simeonova, M.; Leung, V.; Lo, J.; Kan, T.; Raybardhan, S.; Sapin, M.E.; Mponponsuo, K.; Farrell, A.; et al. Antimicrobial Resistance in Patients with COVID-19: A Systematic Review and Meta-Analysis. Lancet Microbe 2023, 4, e179–e191. [Google Scholar] [CrossRef]
- Lai, C.C.; Chen, S.Y.; Ko, W.C.; Hsueh, P.R. Increased Antimicrobial Resistance during the COVID-19 Pandemic. Int. J. Antimicrob. Agents 2021, 57, 106324. [Google Scholar] [CrossRef]
- Hassounah, M.; Raheel, H.; Alhefzi, M. Digital Response during the COVID-19 Pandemic in Saudi Arabia. J. Med. Internet Res. 2020, 22, e19338. [Google Scholar] [CrossRef]
- Bono, M.J.; Leslie, S.W.; Reygaert, W.C.; Doerr, C. Uncomplicated Urinary Tract Infections (Nursing); StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Giesen, L.G.; Cousins, G.; Dimitrov, B.D.; Van De Laar, F.A.; Fahey, T. Predicting Acute Uncomplicated Urinary Tract Infection in Women: A Systematic Review of the Diagnostic Accuracy of Symptoms and Signs. BMC Fam. Pract. 2010, 11, 78. [Google Scholar] [CrossRef]
- Hay, A.D.; Birnie, K.; Busby, J.; Delaney, B.; Downing, H.; Dudley, J.; Durbaba, S.; Fletcher, M.; Harman, K.; Hollingworth, W.; et al. Microbiological Diagnosis of Urinary Tract Infection by NHS and Research Laboratories; NIHR Journals Library: Southampton, UK, 2016. [Google Scholar]
- Cornaglia, G.; Courcol, R.; Herrmann, J.L.; Kahlmeter, G.; Peigue-Lafeuille, H.; Jordi, V. European Manual of Clinical Microbiology; European Society for Clinical Microbiology and Infectious Diseases: Basel, Switzerland, 2012. [Google Scholar]
- Vandepitte, J. Basic Laboratory Procedures in Clinical Bacteriology; World Health Organization: Geneva, Switzerland, 2003; Google Books; Available online: https://books.google.com.pk/books?hl=en&lr=&id=UHY0DgAAQBAJ&oi=fnd&pg=PR5&dq=Basic+laboratory+procedures+in+clinical+bacteriology%3B+World+Health+Organization-+2003.&ots=dD7eQvo9vA&sig=Vj9FHU8_2J9plrL3FtHt2kzl8dQ&redir_esc=y#v=onepage&q=Basic%20laboratory%20procedures%20in%20clinical%20bacteriology%3B%20World%20Health%20Organization-%202003.&f=false (accessed on 12 February 2024).
- Public Health England. UK Standards for Microbiology Investigations. Investigation of urine. B 41 Issue 8.7. Available online: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/770688/B_41i8.7.pdf (accessed on 21 August 2020).
- BioMérieux USA. API® Reference Guide. Available online: https://www.biomerieux-usa.com/sites/subsidiary_us/files/18_api-ref-guide_v7.pdf (accessed on 12 February 2024).
- Carroll, K.C.; Glanz, B.D.; Borek, A.P.; Burger, C.; Bhally, H.S.; Henciak, S.; Flayhart, D. Evaluation of the BD Phoenix Automated Microbiology System for Identification and Antimicrobial Susceptibility Testing of Enterobacteriaceae. J. Clin. Microbiol. 2006, 44, 3506–3509. [Google Scholar] [CrossRef]
- BD PhoenixTM Panels—BD. Available online: https://www.bd.com/en-eu/offerings/capabilities/microbiology-solutions/clinical-microbiology/identification-and-susceptibility-testing/bd-phoenix-automated-identification-and-susceptibility-testing-system/bd-phoenix-panels (accessed on 12 February 2024).
- Kosikowska, U.; Andrzejczuk, S.; Grywalska, E.; Chwiejczak, E.; Winiarczyk, S.; Pietras-Ożga, D.; Stępień-Pyśniak, D. Prevalence of Susceptibility Patterns of Opportunistic Bacteria in Line with CLSI or EUCAST among Haemophilus parainfluenzae Isolated from Respiratory Microbiota. Sci. Rep. 2020, 10, 11512. [Google Scholar] [CrossRef]
- Giske, C.G. EUCAST Subcommitee for Detection of Resistance Mechanisms; EUCAST: Stockholm, Sweden, 2012. [Google Scholar]
- Bradford, P.A. Extended-Spectrum β-Lactamases in the 21st Century: Characterization, Epidemiology, and Detection of This Important Resistance Threat. Clin. Microbiol. Rev. 2001, 14, 933–951. [Google Scholar] [CrossRef] [PubMed]
- WHO. Antimicrobial Resistance; Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014; pp. 12–28. [Google Scholar]
- Karanika, S.; Karantanos, T.; Arvanitis, M.; Grigoras, C.; Mylonakis, E. Fecal Colonization with Extended-Spectrum Beta-Lactamase–Producing Enterobacteriaceae and Risk Factors Among Healthy Individuals: A Systematic Review and Metaanalysis. Clin. Infect. Dis. 2016, 63, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Lemenand, O.; Coeffic, T.; Thibaut, S.; Colomb Cotinat, M.; Caillon, J.; Birgand, G. Decreasing Proportion of Extended-Spectrum Beta-Lactamase among E. coli Infections during the COVID-19 Pandemic in France. J. Infect. 2021, 83, 664–670. [Google Scholar] [CrossRef]
- Wardoyo, E.H.; Suardana, I.W.; Yasa, I.W.P.S.; Sukrama, I.D.M. Antibiotics Susceptibility of Escherichia coli Isolates from Clinical Specimens before and during COVID-19 Pandemic. Iran. J. Microbiol. 2021, 13, 156. [Google Scholar] [CrossRef]
- Nordmann, P.; Dortet, L.; Poirel, L. Carbapenem Resistance in Enterobacteriaceae: Here Is the Storm! Trends Mol. Med. 2012, 18, 263–272. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Adegoke, A.A.; Ikott, W.E.; Okoh, A.I. Carbapenem Resistance Associated with Coliuria among Outpatient and Hospitalised Urology Patients. New Microbes New Infect. 2022, 48, 101019. [Google Scholar] [CrossRef]
- Mareș, C.; Petca, R.C.; Petca, A.; Popescu, R.I.; Jinga, V. Does the COVID-19 Pandemic Modify the Antibiotic Resistance of Uropathogens in Female Patients? A New Storm? Antibiotics 2022, 11, 376. [Google Scholar] [CrossRef] [PubMed]
- Taha, R.; Mowallad, A.; Mufti, A.; Althaqafi, A.; Fatani, A.A.; El-Hossary, D.; Ossenkopp, J.; AlhajHussein, B.; Kaaki, M.; Jawi, N.; et al. Prevalence of Carbapenem-Resistant Enterobacteriaceae in Western Saudi Arabia and Increasing Trends in the Antimicrobial Resistance of Enterobacteriaceae. Cureus 2023, 15, e35050. [Google Scholar] [CrossRef] [PubMed]
- Mareș, C.; Petca, R.C.; Popescu, R.I.; Petca, A.; Geavlete, B.F.; Jinga, V. Uropathogens’ Antibiotic Resistance Evolution in a Female Population: A Sequential Multi-Year Comparative Analysis. Antibiotics 2023, 12, 948. [Google Scholar] [CrossRef] [PubMed]
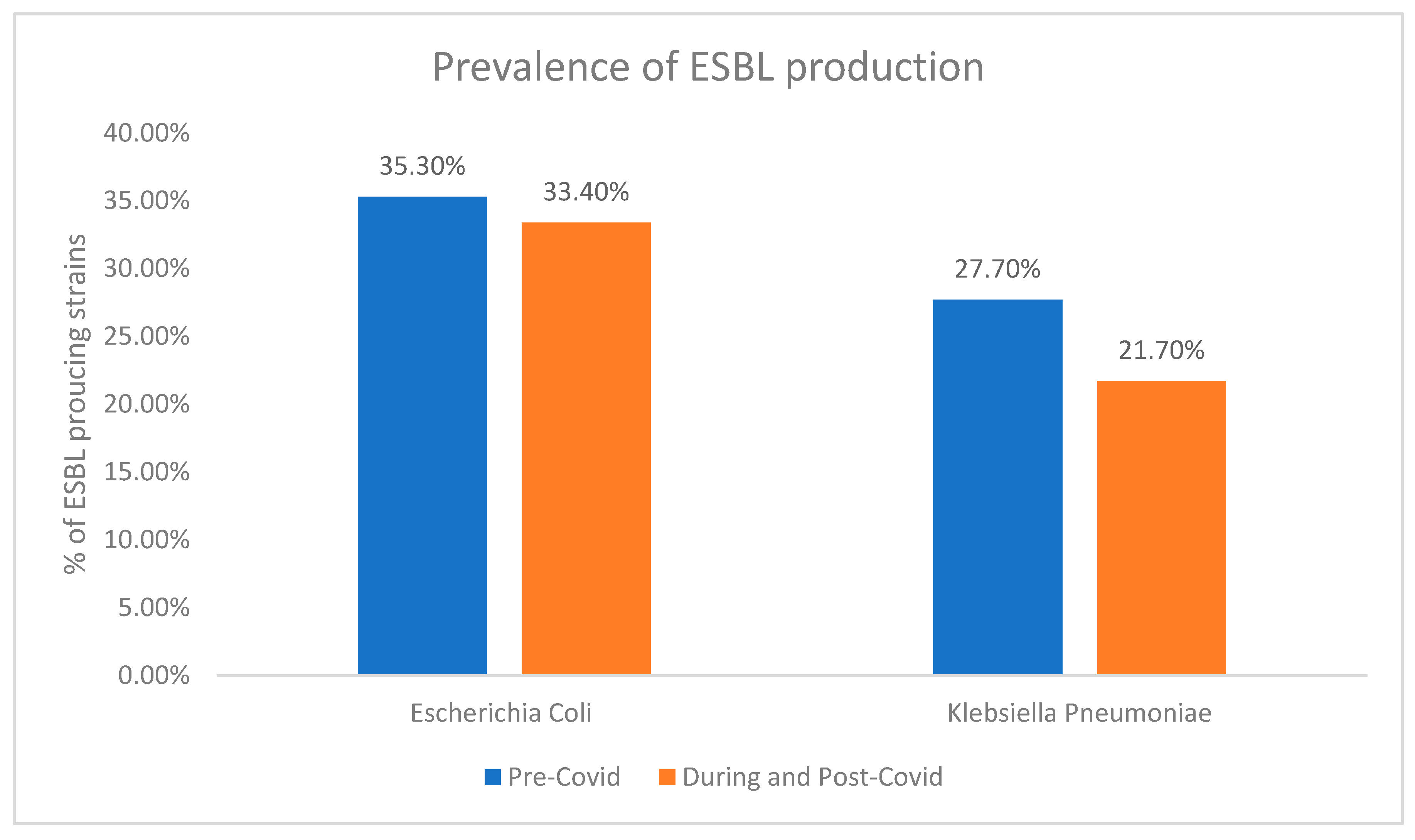

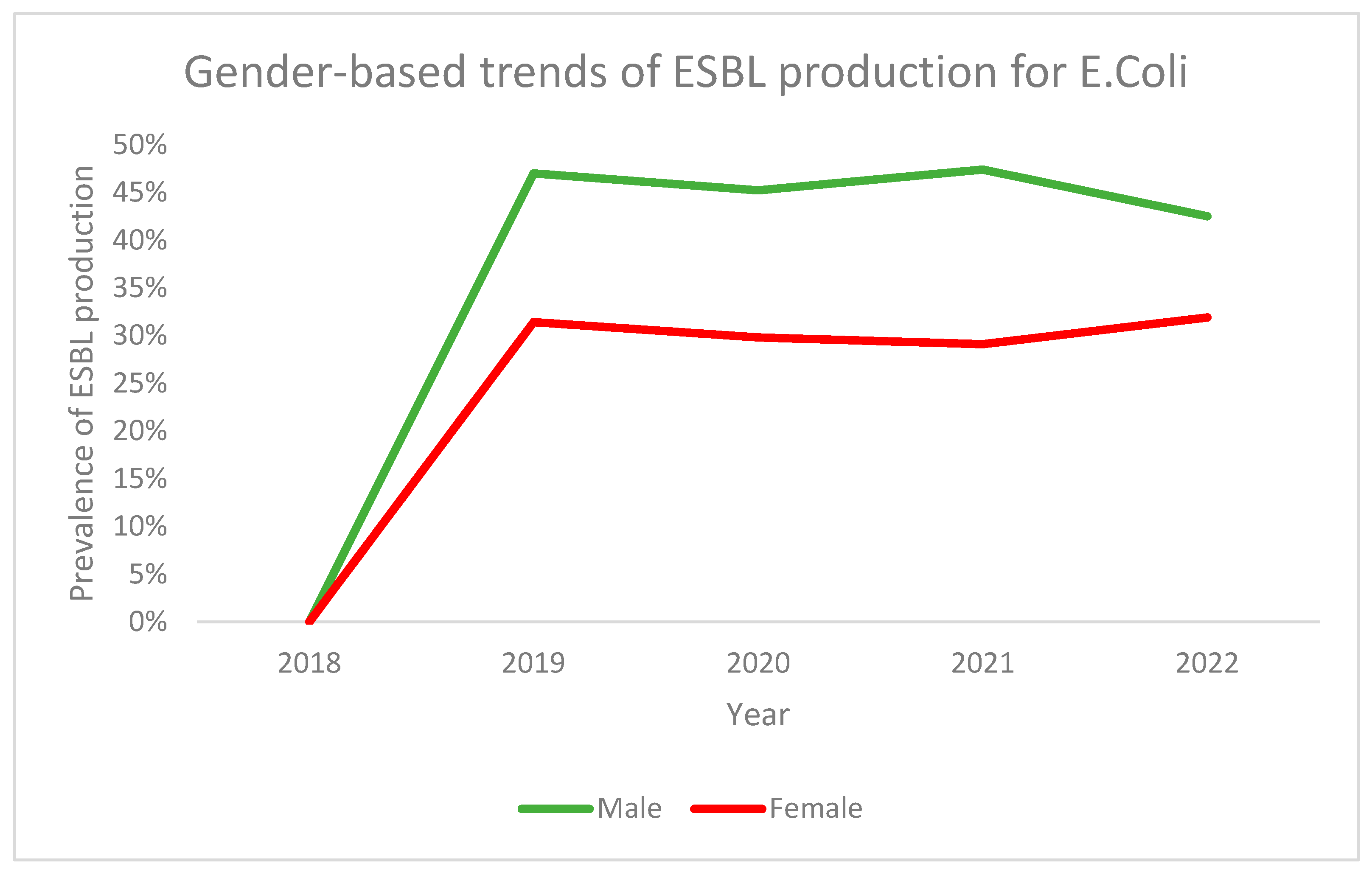
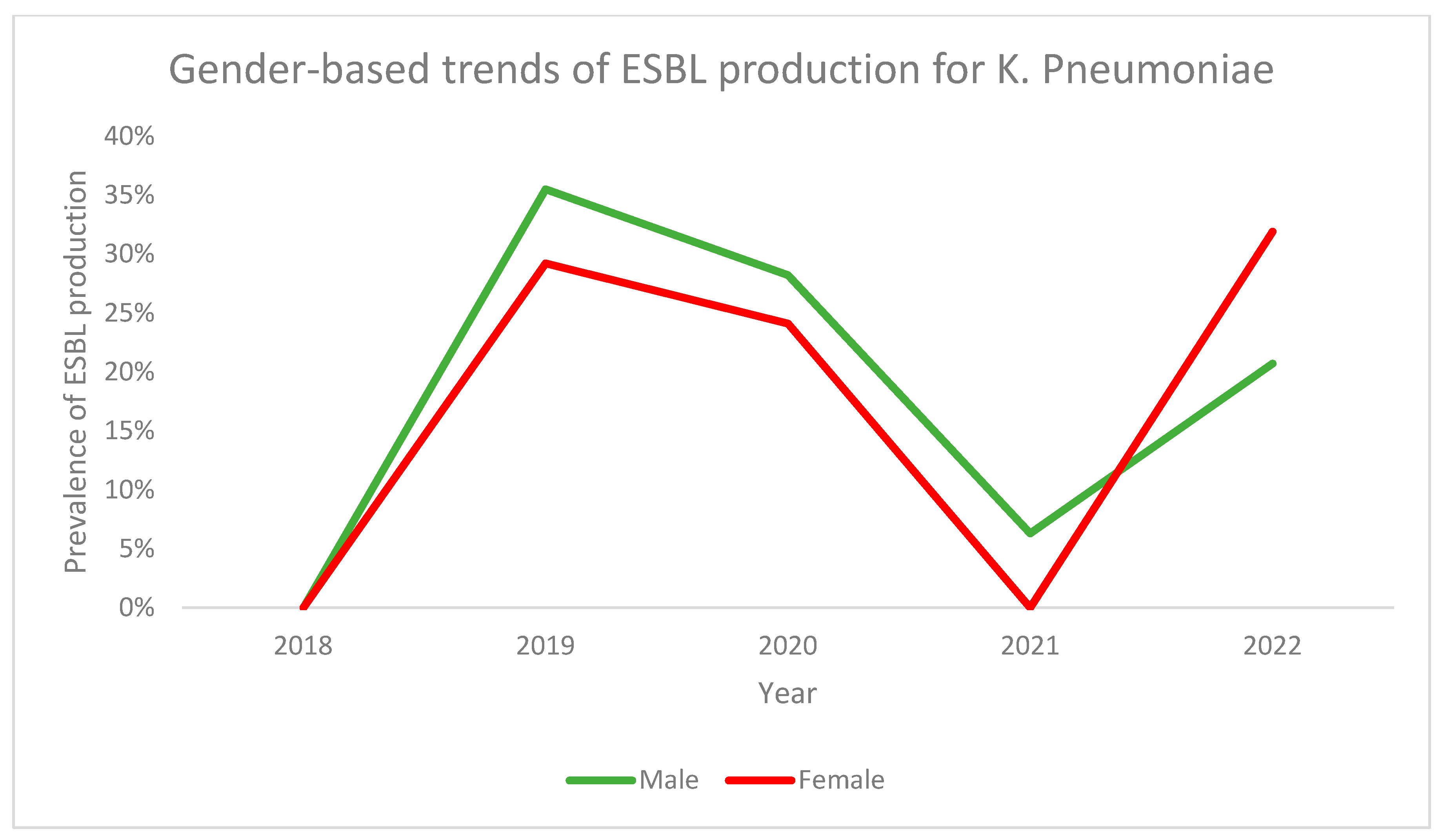
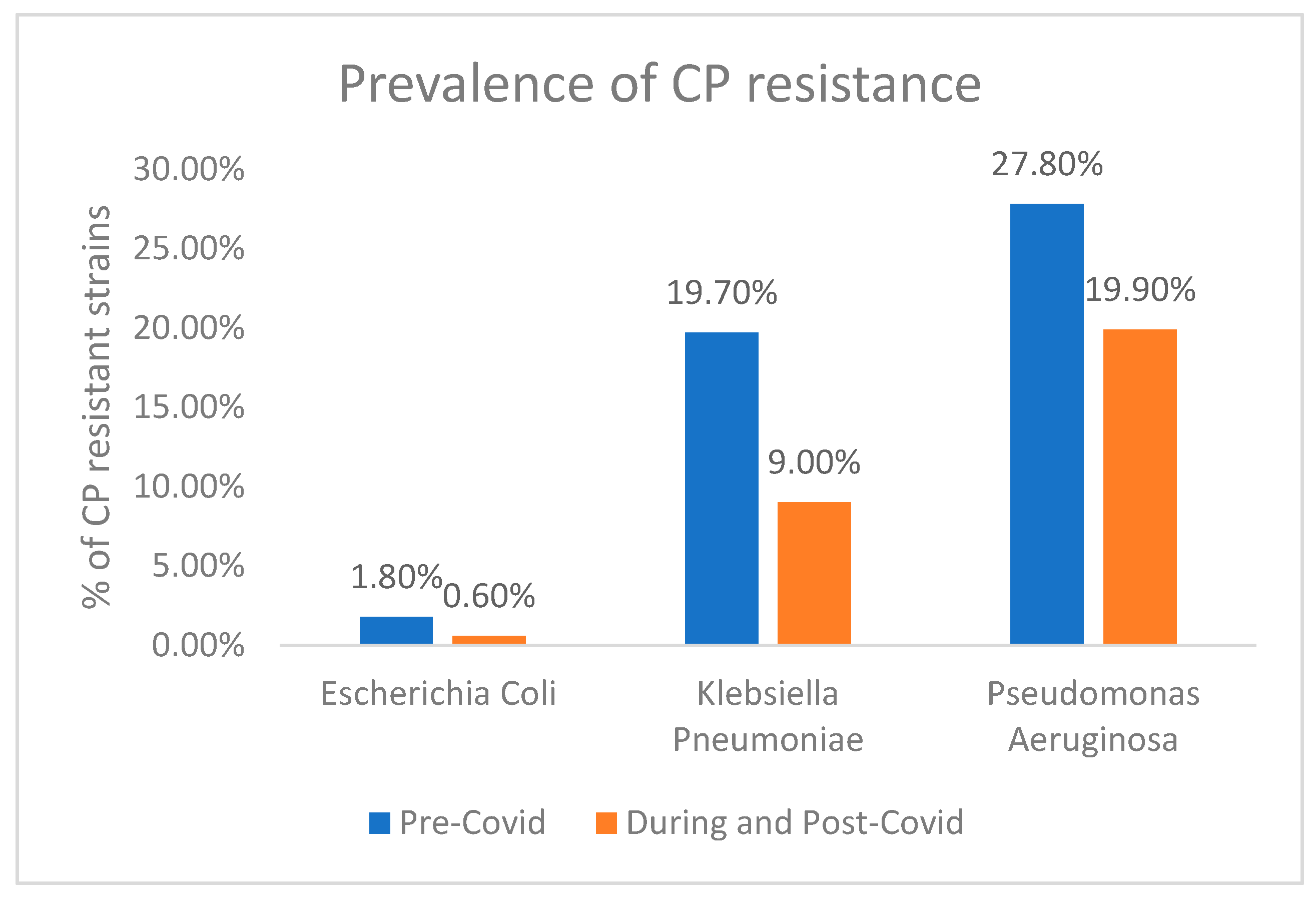


| Variable | ESBL Producing Strains (n = 801) | Non-ESBL Producing Strains (n = 1466) | p-Value |
|---|---|---|---|
Gender
| (n, %) 262 (47.1) 539 (31.5) | (n, %) 294 (52.9) 1172 (68.5) | 0.000 |
| Age in years (Mean ± S.D) | 44.3 ± 26.5 | 40.3 ± 24.3 | 0.000 |
Patient type
| (n, %) 766 (34.8) 35 (55.6) | (n, %) 1438 (65.2) 28 (44.4) | 0.001 |
| Variable | ESBL Producing Strains (n = 1349) | Non-ESBL Producing Strains (n = 2680) | p-Value |
|---|---|---|---|
Gender
| (n, %) 418 (44.1) 931 (30.2) | (n, %) 529 (55.9) 2151 (69.8) | 0.000 |
| Age in years (Mean ± S.D) | 47.8 ± 26.0 | 42.2 ± 25.0 | 0.006 |
Patient type
| (n, %) 1306 (33.2) 43 (43.9) | (n, %) 2629 (66.8) 55 (56.1) | 0.027 |
| Variable | ESBL Producing Strains (n = 279) | Non-ESBL Producing Strains (n = 727) | p-Value |
|---|---|---|---|
Gender
| (n, %) 110 (30.2) 169 (26.3) | (n, %) 254 (69.8) 473 (73.7) | 0.185 |
| Age in years (Mean ± S.D) | 41.6 ± 28.3 | 45.9 ± 27.9 | 0.028 |
Patient type
| (n, %) 273 (28.1) 6 (17.6) | (n, %) 699 (71.9) 28 (82.4) | 0.181 |
| Variable | ESBL Producing Strains (n = 266) | Non-ESBL Producing Strains (n = 962) | p-Value |
|---|---|---|---|
Gender
| (n, %) 97 (22.8) 169 (21.0) | (n, %) 328 (77.2) 634 (79.0) | 0.472 |
| Age in years (Mean ± S.D) | 45.9 ± 28.5 | 48.5 ± 25.2 | 0.149 |
Patient type
| (n, %) 256 (21.7) 10 (21.7) | (n, %) 926 (78.3) 36 (78.3) | 0.990 |
| Variable | OR (95% CI) | p-Value |
|---|---|---|
| E. coli | 1.60 (1.44–1.79) | 0.000 |
| Male | 1.51 (1.37–1.67) | 0.000 |
| Age | 1.00 (1.00–1.04) | 0.000 |
| Inpatients | 0.74 (0.57–0.96) | 0.024 |
| During COVID | 0.91 (0.83–0.99) | 0.040 |
| Variable | CP Resistant Strains (n = 41) | CP Sensitive Strains (n = 2226) | p-Value |
|---|---|---|---|
Gender
| (n, %) 14 (2.6) 27 (1.6) | (n, %) 534 (97.4) 1682 (98.4) | 0.177 |
| Age in years (Mean ± S.D) | 47.2 ± 26.9 | 41.7 ± 25.5 | 0.179 |
Patient type
| (n, %) 41 (1.9) 0 | (n, %) 2173 (98.1) 53 (100) | 0.327 |
| Variable | CP Resistant Strains (n = 24) | CP Sensitive Strains (n = 4009) | p-Value |
|---|---|---|---|
Gender
| (n, %) 6 (0.6) 18 (0.6) | (n, %) 942 (99.4) 3067 (99.4) | 0.953 |
| Age in years (Mean ± S.D) | 41.3 ± 23.1 | 44.1 ± 25.5 | 0.588 |
Patient type
| (n, %) 24 (0.6) 0 | (n, %) 3911 (99.4) 98 (100) | 0.429 |
| Variable | CP Resistant Strains (n = 198) | CP Sensitive Strains (n = 808) | p-Value |
|---|---|---|---|
Gender
| (n, %) 84 (23.5) 114 (17.6) | (n, %) 273 (76.5) 535 (82.4) | 0.027 |
| Age in years (Mean ± S.D) | 53.3 ± 27.0 | 42.7 ± 27.9 | 0.000 |
Patient type
| (n, %) 187 (19.3) 11 (30.6) | (n, %) 782 (80.7) 26 (69.4) | 0.096 |
| Variable | CP Resistant Strains (n = 110) | CP Sensitive Strains (n = 1117) | p-Value |
|---|---|---|---|
Gender
| (n, %) 57 (13.4) 53 (6.6) | (n, %) 367 (86.6) 750 (93.4) | 0.000 |
| Age in years (Mean ± S.D) | 54.7 ± 23.7 | 47.4 ± 26.1 | 0.005 |
Patient type
| (n, %) 106 (8.9) 5 (10.6) | (n, %) 1075 (91.1) 42 (89.4) | 0.643 |
| Variable | CP Resistant Strains (n = 115) | CP Sensitive Strains (n = 298) | p-Value |
|---|---|---|---|
Gender
| (n, %) 69 (27.6) 46 (28.2) | (n, %) 181(72.4) 117 (71.8) | 0.891 |
| Age in years (Mean ± S.D) | 46.9 ± 25.9 | 49.3 ± 28.2 | 0.430 |
Patient type
| (n, %) 111 (28.2) 4 (20.0) | (n, %) 282 (71.8) 16 (80.0) | 0.422 |
| Variable | CP Resistant Strains (n = 128) | CP Sensitive Strains (n = 515) | p-Value |
|---|---|---|---|
Gender
| (n, %) 79 (22.3) 49 (17.0) | (n, %) 276 (77.7) 239 (83.0) | 0.098 |
| Age in years (Mean ± S.D) | 55.0 ± 24.2 | 53.2 ± 27.0 | 0.491 |
Patient type
| (n, %) 123 (19.6) 5 (27.8) | (n, %) 503 (80.4) 13 (72.2) | 0.394 |
| Variable | OR (95% CI) | p-Value |
|---|---|---|
| E. coli - with reference to K. pneumoniae - with reference to P. aeruginosa | 0.06 (0.05–0.08) 0.03 (0.02–0.05) | 0.000 0.000 |
| Klebsiella pneumoniae - with reference to E. coli - with reference to P. aeruginosa | 15.31 (11.62–20.14) 0.52 (0.50–0.63) | 0.000 0.000 |
| Pseudomonas aeruginosa - with reference to E. coli - with reference to K. pneumoniae | 28.84 (21.62–38.20) 1.93 (1.66–2.34) | 0.000 0.000 |
| Variable | OR (95% CI) | p-Value |
|---|---|---|
| Male | 2.50 (2.12–2.90) | 0.000 |
| Age | 1.10 (1.04–1.12) | 0.000 |
| Inpatients | 0.69 (0.46–1.05) | 0.084 |
| During COVID | 0.43 (0.37–0.51) | 0.000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Altamimi, I.; Binkhamis, K.; Alhumimidi, A.; Alabdulkarim, I.M.; Almugren, A.; Alhemsi, H.; Altamimi, A.; Almazyed, A.; Elbih, S.; Alghunaim, R.; et al. Decline in ESBL Production and Carbapenem Resistance in Urinary Tract Infections among Key Bacterial Species during the COVID-19 Pandemic. Antibiotics 2024, 13, 216. https://doi.org/10.3390/antibiotics13030216
Altamimi I, Binkhamis K, Alhumimidi A, Alabdulkarim IM, Almugren A, Alhemsi H, Altamimi A, Almazyed A, Elbih S, Alghunaim R, et al. Decline in ESBL Production and Carbapenem Resistance in Urinary Tract Infections among Key Bacterial Species during the COVID-19 Pandemic. Antibiotics. 2024; 13(3):216. https://doi.org/10.3390/antibiotics13030216
Chicago/Turabian StyleAltamimi, Ibraheem, Khalifa Binkhamis, Abdullah Alhumimidi, Ibrahim M. Alabdulkarim, Abdulrahman Almugren, Hadi Alhemsi, Abdulaziz Altamimi, Abeer Almazyed, Seham Elbih, Razan Alghunaim, and et al. 2024. "Decline in ESBL Production and Carbapenem Resistance in Urinary Tract Infections among Key Bacterial Species during the COVID-19 Pandemic" Antibiotics 13, no. 3: 216. https://doi.org/10.3390/antibiotics13030216
APA StyleAltamimi, I., Binkhamis, K., Alhumimidi, A., Alabdulkarim, I. M., Almugren, A., Alhemsi, H., Altamimi, A., Almazyed, A., Elbih, S., Alghunaim, R., & Altamimi, A. (2024). Decline in ESBL Production and Carbapenem Resistance in Urinary Tract Infections among Key Bacterial Species during the COVID-19 Pandemic. Antibiotics, 13(3), 216. https://doi.org/10.3390/antibiotics13030216






