Change in Diagnosis of Helicobacter pylori Infection in the Treatment-Failure Era
Abstract
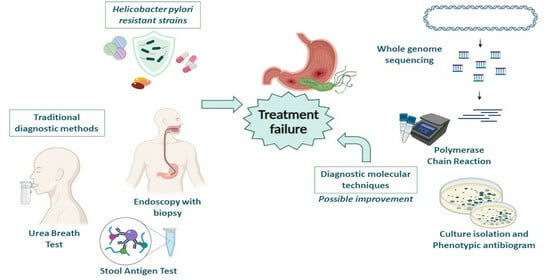
:1. Introduction
2. Molecular Mechanisms of Antimicrobial Resistance
2.1. Single Drug Resistance
2.2. Multidrug Resistance
2.3. Hetero-Resistance
3. Conventional Diagnostic Approaches
3.1. Non-Invasive Methods
3.1.1. Urea Breath Test
3.1.2. Stool Antigen Test
3.1.3. Serological Test
3.2. Invasive Test
Endoscopy with Biopsy
4. New Perspectives in Diagnostics and Applicability in Real Life
4.1. Conventional Microbiological Approaches
4.2. Molecular Diagnostic Approach
4.3. Whole Genome Sequencing
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Wen, J.; Lau, H.C.; Peppelenbosch, M.; Yu, J. Gastric Microbiota beyond H. pylori: An Emerging Critical Character in Gastric Carcinogenesis. Biomedicines 2021, 9, 1680. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Choi, H.; Leung, K.; Jiang, F.; Graham, D.Y.; Leung, W.K. Global prevalence of Helicobacter pylori infection between 1980 and 2022: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2023, 8, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Zamani, M.; Ebrahimtabar, F.; Zamani, V.; Miller, W.H.; Alizadeh-Navaei, R.; Shokri-Shirvani, J.; Derakhshan, M.H. Systematic review with meta-analysis: The worldwide prevalence of Helicobacter pylori infection. Aliment. Pharmacol. Ther. 2018, 47, 868. [Google Scholar] [CrossRef] [PubMed]
- FitzGerald, R.; Smith, S.M. An Overview of Helicobacter pylori Infection. Methods Mol. Biol. 2021, 2283, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Chmiela, M.; Kupcinskas, J. Review: Pathogenesis of Helicobacter pylori infection. Helicobacter 2019, 24, e12638. [Google Scholar] [CrossRef] [PubMed]
- Larussa, T.; Leone, I.; Suraci, E.; Imeneo, M.; Luzza, F. Helicobacter pylori and T Helper Cells: Mechanisms of Immune Escape and Tolerance. J. Immunol. Res. 2015, 2015, 981328. [Google Scholar] [CrossRef] [PubMed]
- Alipour, M. Molecular Mechanism of Helicobacter pylori-Induced Gastric Cancer. J. Gastrointest. Cancer 2021, 52, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Cao, X.S.; Guo, G.Y.; Zhou, M.G.; Yu, B. Effect of Helicobacter Pylori Eradication on Human Gastric Microbiota: A Systematic Review and Meta-Analysis. Front. Cell. Infect. Microbiol. 2022, 12, 899248. [Google Scholar] [CrossRef] [PubMed]
- Abenavoli, L.; Procopio, A.C.; Paravati, M.R.; Costa, G.; Milić, N.; Alcaro, S.; Luzza, F. Mediterranean Diet: The Beneficial Effects of Lycopene in Non-Alcoholic Fatty Liver Disease. J. Clin. Med. 2022, 11, 3477. [Google Scholar] [CrossRef]
- Abenavoli, L.; Giubilei, L.; Procopio, A.C.; Spagnuolo, R.; Luzza, F.; Boccuto, L.; Scarpellini, E. Gut Microbiota in Non-Alcoholic Fatty Liver Disease Patients with Inflammatory Bowel Diseases: A Complex Interplay. Nutrients 2022, 14, 5323. [Google Scholar] [CrossRef]
- Abo-Amer, Y.E.; Sabal, A.; Ahmed, R.; Hasan, N.F.E.; Refaie, R.; Mostafa, S.M.; Mohamed, A.A.; Khalil, M.; Elagawy, W.; Abd-Elsalam, S. Relationship between Helicobacter pylori Infection and Nonalcoholic Fatty Liver Disease (NAFLD) in a Developing Country: A Cross-Sectional Study. Diabetes Metab. Syndr. Obes. 2020, 13, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Dore, M.P.; Pes, G.M. What Is New in Helicobacter pylori Diagnosis. An Overview. J. Clin. Med. 2021, 10, 2091. [Google Scholar] [CrossRef] [PubMed]
- Malfertheiner, P.; Megraud, F.; Rokkas, T.; Gisbert, J.P.; Liou, J.M.; Schulz, C.; Gasbarrini, A.; Hunt, R.H.; Leja, M.; O’Morain, C.; et al. Management of Helicobacter pylori infection: The Maastricht VI/Florence consensus report. Gut 2022, 71, 1724–1762. [Google Scholar] [CrossRef] [PubMed]
- Savoldi, A.; Carrara, E.; Graham, D.Y.; Conti, M.; Tacconelli, E. Prevalence of Antibiotic Resistance in Helicobacter pylori: A Systematic Review and Meta-analysis in World Health Organization Regions. Gastroenterology 2018, 155, 1372–1382.e17. [Google Scholar] [CrossRef]
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Srisuphanunt, M.; Wilairatana, P.; Kooltheat, N.; Duangchan, T.; Katzenmeier, G.; Rose, J.B. Molecular Mechanisms of Antibiotic Resistance and Novel Treatment Strategies for Helicobacter pylori Infections. Trop. Med. Infect. Dis. 2023, 8, 163. [Google Scholar] [CrossRef]
- Jearth, V.; Rath, M.M.; Chatterjee, A.; Kale, A.; Panigrahi, M.K. Drug-Resistant Helicobacter pylori: Diagnosis and Evidence-Based Approach. Diagnostics 2023, 13, 2944. [Google Scholar] [CrossRef]
- Quirino, A.; Cicino, C.; Scarlata, G.G.M.; Marascio, N.; Di Gennaro, G.; Matera, G.; Licata, F.; Bianco, A. Prevalence of Colonization with Multidrug-Resistant Bacteria: Results of a 5-Year Active Surveillance in Patients Attending a Teaching Hospital. Antibiotics 2023, 12, 1525. [Google Scholar] [CrossRef] [PubMed]
- Romano, M.; Gravina, A.G.; Eusebi, L.H.; Pellegrino, R.; Palladino, G.; Frazzoni, L.; Dajti, E.; Gasbarrini, A.; Di Mario, F.; Zagari, R.M.; et al. Management of Helicobacter pylori infection: Guidelines of the Italian Society of Gastroenterology (SIGE) and the Italian Society of Digestive Endoscopy (SIED). Dig. Liver Dis. 2022, 54, 1153–1161. [Google Scholar] [CrossRef]
- Boyanova, L.; Hadzhiyski, P.; Gergova, R.; Markovska, R. Evolution of Helicobacter pylori Resistance to Antibiotics: A Topic of Increasing Concern. Antibiotics 2023, 12, 332. [Google Scholar] [CrossRef]
- Tshibangu-Kabamba, E.; Yamaoka, Y. Helicobacter pylori infection and antibiotic resistance—From biology to clinical implications. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 613–629. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.C.; Iyer, P.G.; Moss, S.F. AGA Clinical Practice Update on the Management of Refractory Helicobacter pylori Infection: Expert Review. Gastroenterology 2021, 160, 1831–1841. [Google Scholar] [CrossRef] [PubMed]
- Ailloud, F.; Didelot, X.; Woltemate, S.; Pfaffinger, G.; Overmann, J.; Bader, R.C.; Schulz, C.; Malfertheiner, P.; Suerbaum, S. Within-host evolution of Helicobacter pylori shaped by niche-specific adaptation, intragastric migrations and selective sweeps. Nat. Commun. 2019, 10, 2273. [Google Scholar] [CrossRef]
- de Marco, B.A.; Natori, J.S.H.; Fanelli, S.; Tótoli, E.G.; Salgado, H.R.N. Characteristics, Properties and Analytical Methods of Amoxicillin: A Review with Green Approach. Crit. Rev. Anal. Chem. 2017, 47, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.J.; Ke, J.N.; Kuo, T.; Lin, C.Y.; Hsieh, S.Y.; Chiu, Y.F.; Wu, H.Y.; Huang, M.Z.; Bui, N.N.; Chiu, C.H.; et al. Multiple amino acid substitutions in penicillin-binding protein-1A confer amoxicillin resistance in refractory Helicobacter pylori infection. J. Microbiol. Immunol. Infect. 2023, 56, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Windham, I.H.; Merrell, D.S. Interplay between Amoxicillin Resistance and Osmotic Stress in Helicobacter pylori. J. Bacteriol. 2022, 204, e0004522. [Google Scholar] [CrossRef] [PubMed]
- Losurdo, G.; Giorgio, F.; Pricci, M.; Girardi, B.; Russo, F.; Riezzo, G.; Martulli, M.; Piazzolla, M.; Cocomazzi, F.; Abbruzzi, F.; et al. Helicobacter pylori Primary and Secondary Genotypic Resistance to Clarithromycin and Levofloxacin Detection in Stools: A 4-Year Scenario in Southern Italy. Antibiotics 2020, 9, 723. [Google Scholar] [CrossRef] [PubMed]
- Anderson, V.R.; Perry, C.M. Levofloxacin: A review of its use as a high-dose, short-course treatment for bacterial infection. Drugs 2008, 68, 535–565. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wen, Y.; Xiao, Q.; Zheng, W.; Long, G.; Chen, B.; Shu, X.; Jiang, M. Mutations in the Antibiotic Target Genes Related to Clarithromycin, Metronidazole and Levofloxacin Resistance in Helicobacter pylori Strains from Children in China. Infect. Drug Resist. 2020, 13, 311–322. [Google Scholar] [CrossRef]
- Marques, A.T.; Vítor, J.M.B.; Santos, A.; Oleastro, M.; Vale, F.F. Trends in Helicobacter pylori resistance to clarithromycin: From phenotypic to genomic approaches. Microb. Genom. 2020, 6, e000344. [Google Scholar] [CrossRef]
- Kocsmár, É.; Buzás, G.M.; Szirtes, I.; Kocsmár, I.; Kramer, Z.; Szijártó, A.; Fadgyas-Freyler, P.; Szénás, K.; Rugge, M.; Fassan, M.; et al. Primary and secondary clarithromycin resistance in Helicobacter pylori and mathematical modeling of the role of macrolides. Nat. Commun. 2021, 12, 2255. [Google Scholar] [CrossRef] [PubMed]
- Serapide, F.; Quirino, A.; Scaglione, V.; Morrone, H.L.; Longhini, F.; Bruni, A.; Garofalo, E.; Matera, G.; Marascio, N.; Scarlata, G.G.M.; et al. Is the Pendulum of Antimicrobial Drug Resistance Swinging Back after COVID-19? Microorganisms 2022, 10, 957. [Google Scholar] [CrossRef]
- Huang, Z.; Zhu, Y.; Li, X.; Yao, Z.; Ge, R. The mechanisms of metronidazole resistance of Helicobacter pylori: A transcriptomic and biochemical study. Microb. Pathog. 2023, 183, 106303. [Google Scholar] [CrossRef] [PubMed]
- Contreras, M.; Benejat, L.; Mujica, H.; Peña, J.; García-Amado, M.A.; Michelangeli, F.; Lehours, P. Real-time PCR detection of a 16S rRNA single mutation of Helicobacter pylori isolates associated with reduced susceptibility and resistance to tetracycline in the gastroesophageal mucosa of individual hosts. J. Med. Microbiol. 2019, 68, 1287–1291. [Google Scholar] [CrossRef] [PubMed]
- Gerrits, M.M.; de Zoete, M.R.; Arents, N.L.; Kuipers, E.J.; Kusters, J.G. 16S rRNA mutation-mediated tetracycline resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 2002, 46, 2996–3000. [Google Scholar] [CrossRef] [PubMed]
- Gisbert, J.P. Rifabutin for the Treatment of Helicobacter pylori Infection: A Review. Pathogens 2020, 10, 15. [Google Scholar] [CrossRef] [PubMed]
- Heep, M.; Beck, D.; Bayerdörffer, E.; Lehn, N. Rifampin and rifabutin resistance mechanism in Helicobacter pylori. Antimicrob. Agents Chemother. 1999, 43, 1497–1499. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Park, S.Y.; Jeong, S.J.; Jung, D.H.; Kim, J.H.; Jeong, S.H.; Kang, I.M.; Song, Y.G. Can Aminoglycosides Be Used as a New Treatment for Helicobacter pylori? In vitro Activity of Recently Isolated Helicobacter pylori. Infect. Chemother. 2019, 51, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhang, M.; Lu, B.; Dai, J. Helicobacter pylori and Antibiotic Resistance, A Continuing and Intractable Problem. Helicobacter 2016, 21, 349–363. [Google Scholar] [CrossRef]
- Dascălu, R.I.; Bolocan, A.; Păduaru, D.N.; Constantinescu, A.; Mitache, M.M.; Stoica, A.D.; Andronic, O. Multidrug resistance in Helicobacter pylori infection. Front. Microbiol. 2023, 14, 1128497. [Google Scholar] [CrossRef]
- Hathroubi, S.; Servetas, S.L.; Windham, I.; Merrell, D.S.; Ottemann, K.M. Helicobacter pylori Biofilm Formation and Its Potential Role in Pathogenesis. Microbiol. Mol. Biol. Rev. 2018, 82, e00001-18. [Google Scholar] [CrossRef] [PubMed]
- Kadkhodaei, S.; Siavoshi, F.; Akbari Noghabi, K. Mucoid and coccoid Helicobacter pylori with fast growth and antibiotic resistance. Helicobacter 2020, 25, e12678. [Google Scholar] [CrossRef] [PubMed]
- Keikha, M.; Karbalaei, M. Prevalence of antibiotic heteroresistance associated with Helicobacter pylori infection: A systematic review and meta-analysis. Microb. Pathog. 2022, 170, 105720. [Google Scholar] [CrossRef] [PubMed]
- Castelle, C.J.; Banfield, J.F. Major New Microbial Groups Expand Diversity and Alter our Understanding of the Tree of Life. Cell 2018, 172, 1181–1197. [Google Scholar] [CrossRef] [PubMed]
- Cardos, A.I.; Maghiar, A.; Zaha, D.C.; Pop, O.; Fritea, L.; Miere Groza, F.; Cavalu, S. Evolution of Diagnostic Methods for Helicobacter pylori Infections: From Traditional Tests to High Technology, Advanced Sensitivity and Discrimination Tools. Diagnostics 2022, 12, 508. [Google Scholar] [CrossRef] [PubMed]
- Bordin, D.S.; Voynovan, I.N.; Andreev, D.N.; Maev, I.V. Current Helicobacter pylori Diagnostics. Diagnostics 2021, 11, 1458. [Google Scholar] [CrossRef] [PubMed]
- Kayali, S.; Aloe, R.; Bonaguri, C.; Gaiani, F.; Manfredi, M.; Leandro, G.; Fornaroli, F.; Di Mario, F.; De’ Angelis, G.L. Non-invasive tests for the diagnosis of Helicobacter pylori: State of the art. Acta Biomed. 2018, 89, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Ferwana, M.; Abdulmajeed, I.; Alhajiahmed, A.; Madani, W.; Firwana, B.; Hasan, R.; Altayar, O.; Limburg, P.J.; Murad, M.H.; Knawy, B. Accuracy of urea breath test in Helicobacter pylori infection: Meta-analysis. World J. Gastroenterol. 2015, 21, 1305–1314. [Google Scholar] [CrossRef] [PubMed]
- Lemos, F.F.B.; Castro, C.T.; Silva Luz, M.; Rocha, G.R.; Correa Santos, G.L.; de Oliveira Silva, L.G.; Calmon, M.S.; Souza, C.L.; Zarpelon-Schutz, A.C.; Teixeira, K.N.; et al. Urea breath test for Helicobacter pylori infection in adult dyspeptic patients: A meta-analysis of diagnostic test accuracy. World J. Gastroenterol. 2024, 30, 579–598. [Google Scholar] [CrossRef]
- Han, Y.H.; Zhang, W.; Wang, Y.T.; Xiong, Z.J.; Du, Q.; Xie, Y.; Lu, H. Performance evaluation of a novel 14C-urea breath test (solid scintillation) for the diagnosis of Helicobacter pylori infection. Medicine 2023, 102, e33107. [Google Scholar] [CrossRef]
- Alzoubi, H.; Al-Mnayyis, A.; Al Rfoa, I.; Aqel, A.; Abu-Lubad, M.; Hamdan, O.; Jaber, K. The Use of 13C-Urea Breath Test for Non-Invasive Diagnosis of Helicobacter pylori Infection in Comparison to Endoscopy and Stool Antigen Test. Diagnostics 2020, 10, 448. [Google Scholar] [CrossRef]
- Alradhawi, M.; Zakeri, N.; Negus, R.; Mack, D.; Morgan, M.Y. PTH-79 Post-eradication Retesting for Helicobacter Pylori Infection in Patients Undergoing Upper Gastro-intestinal Endoscopy: Compliance with Guidelines. Gut 2021, 70, A139. [Google Scholar] [CrossRef]
- Shimoyama, T. Stool antigen tests for the management of Helicobacter pylori infection. World J. Gastroenterol. 2013, 19, 8188–8191. [Google Scholar] [CrossRef]
- Gisbert, J.P.; Pajares, J.M. 13C-urea breath test in the diagnosis of Helicobacter pylori infection—A critical review. Aliment. Pharmacol. Ther. 2004, 20, 1001–1017. [Google Scholar] [CrossRef] [PubMed]
- Chatrangsun, B.; Vilaichone, R.K. Endoscopic Diagnosis for H. pylori Infection: White Light Imaging (WLI) vs. Image-Enhanced Endoscopy (IEE). Asian Pac. J. Cancer Prev. 2021, 22, 3031–3038. [Google Scholar] [CrossRef]
- East, J.E.; Vleugels, J.L.; Roelandt, P.; Bhandari, P.; Bisschops, R.; Dekker, E.; Hassan, C.; Horgan, G.; Kiesslich, R.; Longcroft-Wheaton, G.; et al. Advanced endoscopic imaging: European Society of Gastrointestinal Endoscopy (ESGE) Technology Review. Endoscopy 2016, 48, 1029–1045. [Google Scholar] [CrossRef] [PubMed]
- Dohi, O.; Yagi, N.; Onozawa, Y.; Kimura-Tsuchiya, R.; Majima, A.; Kitaichi, T.; Horii, Y.; Suzuki, K.; Tomie, A.; Okayama, T.; et al. Linked color imaging improves endoscopic diagnosis of active Helicobacter pylori infection. Endosc. Int. Open. 2016, 4, E800–E805. [Google Scholar] [CrossRef]
- Destura, R.V.; Labio, E.D.; Barrett, L.J.; Alcantara, C.S.; Gloria, V.I.; Daez, M.L.; Guerrant, R.L. Laboratory diagnosis and susceptibility profile of Helicobacter pylori infection in the Philippines. Ann. Clin. Microbiol. Antimicrob. 2004, 3, 25. [Google Scholar] [CrossRef] [PubMed]
- De Francesco, V.; Zullo, A.; Manta, R.; Satriano, A.; Fiorini, G.; Pavoni, M.; Saracino, I.M.; Giostra, F.; Monti, G.; Vaira, D. Culture-based antibiotic susceptibility testing for Helicobacter pylori infection: A systematic review. Ann. Gastroenterol. 2022, 35, 127–134. [Google Scholar] [CrossRef]
- Thomas, J.E.; Gibson, G.R.; Darboe, M.K.; Dale, A.; Weaver, L.T. Isolation of Helicobacter pylori from human faeces. Lancet 1992, 340, 1194–1195. [Google Scholar] [CrossRef]
- Kelly, S.M.; Pitcher, M.C.; Farmery, S.M.; Gibson, G.R. Isolation of Helicobacter pylori from feces of patients with dyspepsia in the United Kingdom. Gastroenterology 1994, 107, 1671–1674. [Google Scholar] [CrossRef] [PubMed]
- Guaman, J.F.; Bayas-Morejon, I.F.; Arcos, V.; Tigre-Leon, A.; Lucio-Quintana, A.; Salazar, S.; Gaibor-Chavez, J.; Curay, R.R. Detection of Helicobacter pylori from human biological samples (feces) by antigenic screening and culture. Jundishapur J. Microbiol. 2018, 11, e66721. [Google Scholar] [CrossRef]
- Bujanda, L.; Nyssen, O.P.; Vaira, D.; Saracino, I.M.; Fiorini, G.; Lerang, F.; Georgopoulos, S.; Tepes, B.; Heluwaert, F.; Gasbarrini, A.; et al. Antibiotic Resistance Prevalence and Trends in Patients Infected with Helicobacter pylori in the Period 2013–2020: Results of the European Registry on H. pylori Management (Hp-EuReg). Antibiotics 2021, 10, 1058. [Google Scholar] [CrossRef] [PubMed]
- Testerman, T.L.; McGee, D.J.; Mobley, H.L. Helicobacter pylori growth and urease detection in the chemically defined medium Ham’s F-12 nutrient mixture. J. Clin. Microbiol. 2001, 39, 3842–3850. [Google Scholar] [CrossRef] [PubMed]
- Posteraro, B.; Posteraro, P.; Sanguinetti, M. Helicobacter pylori. In Molecular Medical Microbiology, 2nd ed.; Tang, Y.E., Sussman, M., Liu, D., Poxton, I., Schwartzman, J., Eds.; Academic Press: New York, NY, USA, 2015; Chapter 68; pp. 1237–1258. [Google Scholar]
- Zullo, A.; Francesco, V.; Gatta, L. Helicobacter pylori culture: From bench to bedside. Ann. Gastroenterol. 2022, 35, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Garibyan, L.; Avashia, N. Polymerase chain reaction. J. Investig. Dermatol. 2013, 133, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Medakina, I.; Tsapkova, L.; Polyakova, V.; Nikolaev, S.; Yanova, T.; Dekhnich, N.; Khatkov, I.; Bordin, D.; Bodunova, N. Helicobacter pylori Antibiotic Resistance: Molecular Basis and Diagnostic Methods. Int. J. Mol. Sci. 2023, 24, 9433. [Google Scholar] [CrossRef] [PubMed]
- Gong, R.J.; Xu, C.X.; Li, H.; Liu, X.M. Polymerase chain reaction-based tests for detecting Helicobacter pylori clarithromycin resistance in stool samples: A meta-analysis. World J. Clin. Cases 2021, 9, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Falsey, A.R.; Branche, A.R.; Croft, D.P.; Formica, M.A.; Peasley, M.R.; Walsh, E.E. Real-life Assessment of BioFire FilmArray Pneumonia Panel in Adults Hospitalized With Respiratory Illness. J. Infect Dis. 2024, 229, 214–222. [Google Scholar] [CrossRef]
- Leonardi, M.; La Marca, G.; Pajola, B.; Perandin, F.; Ligozzi, M.; Pomari, E. Assessment of real-time PCR for Helicobacter pylori DNA detection in stool with co-infection of intestinal parasites: A comparative study of DNA extraction methods. BMC Microbiol. 2020, 20, 131. [Google Scholar] [CrossRef]
- Wang, Y.H.; Li, Z.; Wang, L.; Zhu-Ge, L.Y.; Zhao, R.L.; Wu, S.; Wang, Y.; An, Y.; Xie, Y. A systematic review and meta-analysis of genotypic methods for detecting antibiotic resistance in Helicobacter pylori. Helicobacter 2018, 23, e12467. [Google Scholar] [CrossRef] [PubMed]
- Binmaeil, H.; Hanafiah, A.; Mohamed Rose, I.; Raja Ali, R.A. Development and Validation of Multiplex Quantitative PCR Assay for Detection of Helicobacter pylori and Mutations Conferring Resistance to Clarithromycin and Levofloxacin in Gastric Biopsy. Infect. Drug Resist. 2021, 14, 4129–4145. [Google Scholar] [CrossRef] [PubMed]
- Shan, H.; Zhu, G.; Zhang, Y.; Ke, L.; Yang, X.; Qiao, A.; Wei, B.; Wang, Y.; Fan, Y.; Du, M. Multiplex PCR-ASE functionalized microfluidic diagnostic platform for the detection of clarithromycin resistance mutations in Helicobacter pylori. Sens. Actuators B Chem. 2023, 387, 133808. [Google Scholar] [CrossRef]
- Gisbert, J.P. Empirical or susceptibility-guided treatment for Helicobacter pylori infection? A comprehensive review. Ther. Adv. Gastroenterol. 2020, 13, 1756284820968736. [Google Scholar] [CrossRef] [PubMed]
- Quirino, A.; Marascio, N.; Scarlata, G.G.M.; Cicino, C.; Pavia, G.; Pantanella, M.; Carlisi, G.; Mercurio, M.; Familiari, F.; Rotundo, S.; et al. Orthopedic Device-Related Infections Due to Emerging Pathogens Diagnosed by a Combination of Microbiological Approaches: Case Series and Literature Review. Diagnostics 2022, 12, 3224. [Google Scholar] [CrossRef] [PubMed]
- Costache, C.; Colosi, H.A.; Grad, S.; Paștiu, A.I.; Militaru, M.; Hădărean, A.P.; Țoc, D.A.; Neculicioiu, V.S.; Baciu, A.M.; Opris, R.V.; et al. Antibiotic Resistance in Helicobacter pylori Isolates from Northwestern and Central Romania Detected by Culture-Based and PCR-Based Methods. Antibiotics 2023, 12, 1672. [Google Scholar] [CrossRef] [PubMed]
- Bongiorno, D.; Bivona, D.A.; Cicino, C.; Trecarichi, E.M.; Russo, A.; Marascio, N.; Mezzatesta, M.L.; Musso, N.; Privitera, G.F.; Quirino, A.; et al. Omic insights into various ceftazidime-avibactam-resistant Klebsiella pneumoniae isolates from two southern Italian regions. Front. Cell. Infect. Microbiol. 2023, 12, 1010979. [Google Scholar] [CrossRef] [PubMed]
- Domanovich-Asor, T.; Motro, Y.; Khalfin, B.; Craddock, H.A.; Peretz, A.; Moran-Gilad, J. Genomic Analysis of Antimicrobial Resistance Genotype-to-Phenotype Agreement in Helicobacter pylori. Microorganisms 2020, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Domanovich-Asor, T.; Craddock, H.A.; Motro, Y.; Khalfin, B.; Peretz, A.; Moran-Gilad, J. Unraveling antimicrobial resistance in Helicobacter pylori: Global resistome meets global phylogeny. Helicobacter 2021, 26, e12782. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, S.; Yang, F.; Chi, W.; Ding, L.; Liu, T.; Zhu, F.; Ji, D.; Zhou, J.; Fang, Y.; et al. Antimicrobial resistance patterns and genetic elements associated with the antibiotic resistance of Helicobacter pylori strains from Shanghai. Gut Pathog. 2022, 14, 14. [Google Scholar] [CrossRef]
- Berberich, A.J.; Ho, R.; Hegele, R.A. Whole genome sequencing in the clinic: Empowerment or too much information? CMAJ. 2018, 190, E124–E125. [Google Scholar] [CrossRef] [PubMed]
- Boccabella, L.; Palma, E.G.; Abenavoli, L.; Scarlata, G.G.M.; Boni, M.; Ianiro, G.; Santori, P.; Tack, J.F.; Scarpellini, E. Post-Coronavirus Disease 2019 Pandemic Antimicrobial Resistance. Antibiotics 2024, 13, 233. [Google Scholar] [CrossRef] [PubMed]
- Franklin, A.M.; Brinkman, N.E.; Jahne, M.A.; Keely, S.P. Twenty-first century molecular methods for analyzing antimicrobial resistance in surface waters to support One Health assessments. J. Microbiol. Methods 2021, 184, 106174. [Google Scholar] [CrossRef] [PubMed]
- Baquero, F.; Martínez, J.L.F.; Lanza, V.; Rodríguez-Beltrán, J.; Galán, J.C.; San Millán, A.; Cantón, R.; Coque, T.M. Evolutionary Pathways and Trajectories in Antibiotic Resistance. Clin. Microbiol. Rev. 2021, 34, e0005019. [Google Scholar] [CrossRef] [PubMed]
- Sijbom, M.; Büchner, F.L.; Saadah, N.H.; Numans, M.E.; De Boer, M.G.J. Trends in antibiotic selection pressure generated in primary care and their association with sentinel antimicrobial resistance patterns in Europe. J. Antimicrob. Chemother. 2023, 78, 1245–1252. [Google Scholar] [CrossRef] [PubMed]
- Quirino, A.; Cicino, C.; Scaglione, V.; Marascio, N.; Serapide, F.; Scarlata, G.G.M.; Lionello, R.; Divenuto, F.; La Gamba, V.; Pavia, G.; et al. In vitro Activity of Cefiderocol Against Carbapenem-Resistant Acinetobacter baumannii Clinical Isolates: A Single Center Experience. Mediterr. J. Hematol. Infect. Dis. 2023, 15, e2023043. [Google Scholar] [CrossRef] [PubMed]
- Patel, T.S.; Pogue, J.M.; Mills, J.P.; Kaye, K.S. Meropenem-vaborbactam: A new weapon in the war against infections due to resistant Gram-negative bacteria. Future Microbiol. 2018, 13, 971–983. [Google Scholar] [CrossRef]
- Wang, Y.K.; Kuo, F.C.; Liu, C.J.; Wu, M.C.; Shih, H.Y.; Wang, S.S.; Wu, J.Y.; Kuo, C.H.; Huang, Y.K.; Wu, D.C. Diagnosis of Helicobacter pylori infection: Current options and developments. World J. Gastroenterol. 2015, 21, 11221–11235. [Google Scholar] [CrossRef]


| Non-Invasive Methods | Sensibility | Specificity |
|---|---|---|
| 13C-UBT | 96.60% | 96.93% |
| 14C-UBT | 96.15% | 89.84% |
| SAT | 95.5% | 97.6% |
| Serological test | 80–95% | 80–95% |
| Endoscopic Techniques | Sensibility | Specificity | Accuracy |
|---|---|---|---|
| WLI | 90.00% | 70.00% | 78.00% |
| NBI | 85.00% | 80.00% | 82.00% |
| LCI | 95.00% | 76.70% | 84.00% |
| BLI | 95.00% | 80.00% | 86.00% |
| Gene Target | Applicability |
|---|---|
| cagA, vacA, ureA, ureC | Identification |
| A2143G, A2142G, A2142C | Detection of clarithromycin resistance |
| gyrA, gyrB | Detection of levofloxacin resistance |
| pbp1A, pbp2, pbp3, hefC, hopC, hofH | Detection of amoxicillin re-sistance |
| TET-1 | Detection of tetracycline re-sistance |
| Microbiological Approach | Advantages | Disadvantages |
|---|---|---|
| Culture isolation and phenotypic antibiogram | Diagnostic gold standard Defines a MIC Falls within the Maastricht IV/Florence Consensus Report | Difficult to perform Higher TAT (>7 days) Low sensitivity |
| PCR and genotypic antibiogram | High sensitivity and specificity Performed directly on biological sample Significant reduction in TAT (<1 day) Promotes the differential diagnosis with other gastro-intestinal tract infection Falls within the Maastricht IV/Florence Consensus Report | Needs of a confirmation through conventional microbiological approach Limited resistance gene detection Does not define a MIC |
| WGS | Simultaneous detection of more genes with an elevated depth of sequencing Useful for the identification of new variants and epidemiological surveillance | Higher costs Requirement of highly trained staff Need to evaluate a large quantity of data Necessity to continuously update the database to avoid a possible underestimation of the data Higher TAT (~7 days) Not included within the Maastricht IV/Florence Consensus Report |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spagnuolo, R.; Scarlata, G.G.M.; Paravati, M.R.; Abenavoli, L.; Luzza, F. Change in Diagnosis of Helicobacter pylori Infection in the Treatment-Failure Era. Antibiotics 2024, 13, 357. https://doi.org/10.3390/antibiotics13040357
Spagnuolo R, Scarlata GGM, Paravati MR, Abenavoli L, Luzza F. Change in Diagnosis of Helicobacter pylori Infection in the Treatment-Failure Era. Antibiotics. 2024; 13(4):357. https://doi.org/10.3390/antibiotics13040357
Chicago/Turabian StyleSpagnuolo, Rocco, Giuseppe Guido Maria Scarlata, Maria Rosaria Paravati, Ludovico Abenavoli, and Francesco Luzza. 2024. "Change in Diagnosis of Helicobacter pylori Infection in the Treatment-Failure Era" Antibiotics 13, no. 4: 357. https://doi.org/10.3390/antibiotics13040357