Short-Chained Linear Scorpion Peptides: A Pool for Novel Antimicrobials
Abstract
:1. Introduction
2. The Emergence of Antibiotic Resistance
3. The Abundance of Antimicrobial Peptides in Nature
4. Characteristics and Antimicrobial Activity of Short Linear Peptides
| Systematic Name | Name | Amino Acid Sequence | Length | Gravy 1 | Hydrophobicity (kcal ∗ mol−1) 2 | pI 2 | Hemo Pi 3 | Net Charge 2 | References |
|---|---|---|---|---|---|---|---|---|---|
| NDBP-4.1 | IsCT | ILGKIWEGIKSLF | 13 | 0.78 | 10.23 | 9.74 | 0.73 | 2 | [49] |
| NDBP-4.2 | ISCT2 | IFGAIWNGIKSLF | 13 | 1.14 | 4.69 | 9.93 | 0.51 | 2 | [50] |
| NDBP-4.3 | BmKb1 | FLFSLIPSAISGLISAFK | 18 | 1.54 | 2.59 | 9.8 | 0.54 | 2 | [51] |
| NDBP-4.4 | BmKn2 | FIGAIANLLSKIF | 13 | 1.67 | 4.88 | 9.93 | 0.6 | 2 | [51] |
| NDBP-4.5 | mucroporin | LFGLIPSLIGGLVSAFK | 17 | 1.62 | 4.59 | 9.8 | 0.67 | 2 | [52] |
| NDBP-4.6 | meucin-13 | IFGAIAGLLKNIF | 13 | 1.7 | 5.57 | 9.93 | 0.52 | 2 | [53] |
| NDBP-4.7 | imcroporin | FFSLLPSLIGGLVSAIK | 17 | 1.59 | 3.9 | 9.8 | 0.64 | 2 | [54] |
| NDBP-4.8 | StCT1 | GFWGSLWEGVKSVV | 14 | 0.51 | 10.18 | 6.81 | 0.49 | 1 | [55] |
| NDBP-4.9 | HP1090 | IFKAIWSGIKSLF | 13 | 1.08 | 5.95 | 10.6 | 0.51 | 2 | [56] |
| NDBP-4.10 | ctriporin | FLWGLIPGAISAVTSLIKK | 19 | 1.16 | 6.74 | 10.6 | 0.74 | 3 | [57] |
| NDBP-4.11 | AamAP1 | FLFSLIPHAIGGLISAFK | 18 | 1.43 | 5.15 | 9.8 | 0.58 | 2 | [40] |
| NDBP-4.12 | AamAP2 | FPFSLIPHAIGGLISAIK | 18 | 1.23 | 7.13 | 9.8 | 0.48 | 2 | [40] |
| NDBP-4.13 | VmCT1 | FLGALWNVAKSVF | 13 | 1.21 | 5.23 | 9.93 | 0.51 | 2 | [58] |
| NDBP-4.14 | VmCT2 | FLSTLWNAAKSIF | 13 | 0.82 | 4.59 | 9.93 | 0.53 | 2 | [59] |
| NDBP-4.15 | StCT2 | GFWGKLWEGVKSAI | 14 | 0.14 | 12.82 | 9.94 | 0.52 | 2 | [60] |
| NDBP-4.16 | UyCT1 | GFWGKLWEGVKNAI | 14 | -0.05 | 13.21 | 9.94 | 0.49 | 2 | [39] |
| NDBP-4.17 | UyCT2 | FWGKLWEGVKNAI | 13 | -0.02 | 12.06 | 9.94 | 0.5 | 2 | [39] |
| NDBP-4.18 | UyCT3 | ILSAIWSGIKSLF | 13 | 1.39 | 4.07 | 9.93 | 0.6 | 2 | [39] |
| NDBP-4.19 | UyCT5 | IWSAIWSGIKGLL | 13 | 1.14 | 4.38 | 10.1 | 0.59 | 2 | [39] |
| NDBP-4.20 | pantinin-1 | GILGKLWEGFKSIV | 14 | 0.67 | 12.04 | 9.93 | 0.82 | 2 | [61] |
| NDBP-4.21 | pantinin-2 | IFGAIWKGISSLL | 13 | 1.42 | 4.76 | 10.1 | 0.83 | 2 | [61] |
| NDBP-4.22 | pantinin-3 | FLSTIWNGIKSLL | 13 | 0.94 | 4.08 | 10.1 | 0.83 | 2 | [61] |
| NDBP-4.23 | TsAP-1 | FLSLIPSLVGGSISAFK | 17 | 1.32 | 5.61 | 9.8 | 0.53 | 2 | [47] |
| NDBP-4.24 | TsAP-2 | FLGMIPGLIGGLISAFK | 17 | 1.55 | 5.2 | 9.8 | 0.52 | 2 | [47] |
| DRAMP18397 | Um4 | FFSALLSGIKSLF | 13 | 1.49 | 3.73 | 9.93 | 0.51 | 2 | [62] |
| DRAMP18398 | Um2 | ISQSDAILSAIWSGIKSLF | 19 | 0.83 | 8.78 | 6.55 | 0.51 | 1 | [62] |
| DRAMP18399 | Uy234 | FPFLLSLIPSAISAIKRL | 18 | 1.33 | 3.39 | 11.55 | 0.55 | 3 | [39] |
| DRAMP18400 | Uy192 | FLSTIWNGIKGLL | 13 | 0.97 | 4.77 | 10.14 | 0.65 | 2 | [39] |
| DRAMP18401 | Uy17 | ILSAIWSGIKGLL | 13 | 1.50 | 5.22 | 10.14 | 0.64 | 2 | [39] |
| DRAMP20775 | Ctry2459 | FLGFLKNLF | 9 | 1.33 | 3.82 | 9.93 | 0.49 | 2 | [56] |
| DRAMP21024 | stigmurin | FFSLIPSLVGGLISAFK | 17 | 1.53 | 3.44 | 9.80 | 0.56 | 2 | [63] |
| DRAMP21251 | AaeAP1 | FLFSLIPSVIAGLVSAIRN | 19 | 1.58 | 2.78 | 10.60 | 0.61 | 2 | [64] |
| DRAMP21252 | AaeAP2 | FLFSLIPSAIAGLVSAIRN | 19 | 1.46 | 3.74 | 10.60 | 0.66 | 2 | [64] |
| AP02518 | Cm38 | ARDGYIVDEKGCKFACFIN | 19 | −0.05 | 23.50 | 6.18 | 0.45 | 1 | [65] |
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- World Health Organization. WHO High Levels of Antibiotic Resistance Found Worldwide, New Data Shows. WHO, 23 January 2018; pp. 174–177. Available online: http://www.who.int/mediacentre/news/releases/2018/antibiotic-resistance-found/en/ (accessed on 1 March 2024).
- World Health Organization Europe. Resistant Bacteria Spreading in Hospitals across the European Region, According to a New WHO Report. World Health Organization, 2016. Available online: https://who-sandbox.squiz.cloud/en/media-centre/sections/press-releases/2016/11/resistant-bacteria-spreading-in-hospitals-across-the-european-region,-according-to-a-new-who-report (accessed on 19 November 2016).
- ECDC. Antimicrobial Resistance in the EU/EEA (EARS-Net)—Annual Epidemiological Report for 2022. 2023. Available online: https://atlas.ecdc.europa.eu/ (accessed on 1 March 2024).
- WHO. WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. WHO Media Centre. 2017. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 1 March 2024).
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. P T Peer-Rev. J. Formul. Manag. 2015, 40, 277–283. [Google Scholar]
- WHO. 2021 Antibacterial Agents in Clinical and Preclinical Development: An Overview and Analysis. 2022. Available online: https://www.who.int/publications/i/item/9789240047655 (accessed on 8 October 2022).
- European Medicines Agency. Committee for Medicinal Products for Human Use (CHMP). Guideline on the Evaluation of Medicinal Products Indicated for Treatment of Bacterial Infections. 2022. Available online: www.ema.europa.eu/contact (accessed on 1 March 2024).
- Frost, L.S. Conjugation, Bacterial. In Encyclopedia of Microbiology; Elsevier: Amsterdam, The Netherlands, 2014; pp. 729–742. [Google Scholar] [CrossRef]
- Sinha, S.; Mell, J.; Redfield, R.J. Bacterial Transformation. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Academic Press: Cambridge, MA, USA, 2013; pp. 271–273. [Google Scholar] [CrossRef]
- Bergman, J.M.; Fineran, P.C.; Petty, N.K.; Salmond, G.P.C. Transduction: The Transfer of Host DNA by Bacteriophages. In Encyclopedia of Microbiology; Academic Press: Cambridge, MA, USA, 2019; pp. 458–473. [Google Scholar] [CrossRef]
- Knobler, L.; Lemon, S.M.; Najafi, M. The Resistance Phenomenon in Microbes and Infectious Disease Vectors: Implications for Human Health and Strategies for Containment; Health (San Francisco); The National Academies Press: Washington, DC, USA, 1994. [Google Scholar] [CrossRef]
- Luyt, C.E.; Brechot, N.; Trouillet, J.L.; Chastre, J. Antibiotic stewardship in the intensive care unit. Crit. Care 2014, 18, 480. [Google Scholar] [CrossRef] [PubMed]
- Hargreaves, K.R.; Clokie, M.R.J. Clostridium difficile phages: Still difficult? Front. Microbiol. 2014, 5, 184. [Google Scholar] [CrossRef] [PubMed]
- Lakshmaiah Narayana, J.; Chen, J.-Y. Antimicrobial peptides: Possible anti-infective agents. Peptides 2015, 72, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Dutta, P.; Das, S. Mammalian Antimicrobial Peptides: Promising Therapeutic Targets against Infection and Chronic Inflammation. Curr. Top. Med. Chem. 2016, 16, 99–129. [Google Scholar] [CrossRef]
- Doiron, K.; Beaulieu, L.; Lemarchand, K. Reduction of bacterial biofilm formation using marine natural antimicrobial peptides. Colloids Surf. B Biointerfaces 2018, 167, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Almeida, J.R.; Mendes, B.; Lancellotti, M.; Marangoni, S.; Vale, N.; Passos, Ó.; Da Silva, S.L. A novel synthetic peptide inspired on Lys49 phospholipase A2 from Crotalus oreganus abyssus snake venom active against multidrug-resistant clinical isolates. Eur. J. Med. Chem. 2018, 149, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Domhan, C.; Uhl, P.; Meinhardt, A.; Zimmermann, S.; Kleist, C.; Lindner, T.; Wink, M. A novel tool against multiresistant bacterial pathogens—Lipopeptide modification of the natural antimicrobial peptide ranalexin for enhanced antimicrobial activity and improved pharmacokinetics. Int. J. Antimicrob. Agents 2018, 52, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Wang, G. Human antimicrobial peptides and proteins. Pharmaceuticals 2014, 7, 545–594. [Google Scholar] [CrossRef]
- Tonk, M.; Vilcinskas, A. The Medical Potential of Antimicrobial Peptides from Insects. Curr. Top. Med. Chem. 2017, 17, 554–575. [Google Scholar] [CrossRef]
- Fialho, T.L.; Carrijo, L.C.; Magalhães Júnior, M.J.; Baracat-Pereira, M.C.; Piccoli, R.H.; de Abreu, L.R. Extraction and identification of antimicrobial peptides from the Canastra artisanal minas cheese. Food Res. Int. 2018, 107, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Bo, J.; Yang, Y.; Zheng, R.; Fang, C.; Jiang, Y.; Liu, J.; Chen, M.; Hong, F.; Bailey, C.; Segner, H.; et al. Antimicrobial activity and mechanisms of multiple antimicrobial peptides isolated from rockfish Sebastiscus marmoratus. Fish Shellfish. Immunol. 2019, 93, 1007–1017. [Google Scholar] [CrossRef]
- Antimicrobial Peptides Market Size in 2023. Business Forecast up to 2029 with Strong Data Source & Driving Forces. Digital Journal. 2023. 114p. Available online: https://www.digitaljournal.com/pr/antimicrobial-peptides-market-size-in-2023-business-forecast-up-to-2029-with-strong-data-source-driving-forces-no-of-pages-114 (accessed on 12 January 2023).
- Chen, C.H.; Lu, T.K. Development and challenges of antimicrobial peptides for therapeutic applications. Antibiotics 2020, 9, 24. [Google Scholar] [CrossRef] [PubMed]
- Moretta, A.; Scieuzo, C.; Petrone, A.M.; Salvia, R.; Manniello, M.D.; Franco, A.; Falabella, P. Antimicrobial Peptides: A New Hope in Biomedical and Pharmaceutical Fields. Front. Cell. Infect. Microbiol. 2021, 11, 668632. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.B.; Seo, J. Antimicrobial peptides under clinical investigation. Pept. Sci. 2019, 111, e24122. [Google Scholar] [CrossRef]
- Askari, P.; Namaei, M.H.; Ghazvini, K.; Hosseini, M. In vitro and in vivo toxicity and antibacterial efficacy of melittin against clinical extensively drug-resistant bacteria. BMC Pharmacol. Toxicol. 2021, 22, 42. [Google Scholar] [CrossRef]
- Vetterli, S.U.; Zerbe, K.; Müller, M.; Urfer, M.; Mondal, M.; Wang, S.-Y.; Robinson, J.A. Thanatin targets the intermembrane protein complex required for lipopolysaccharide transport in Escherichia coli. Sci. Adv. 2018, 4, eaau2634. [Google Scholar] [CrossRef]
- Pletzer, D.; Hancock, R.E.W. Antibiofilm peptides: Potential as broadspectrum agents. J. Bacteriol. Am. Soc. Microbiol. 2016, 198, 2572–2578. [Google Scholar] [CrossRef]
- Guilhelmelli, F.; Vilela, N.; Albuquerque, P.; Derengowski, L.S. Antibiotic development challenges: The various mechanisms of action of antimicrobial peptides and of bacterial resistance. Front. Microbiol. 2013, 4, 353. [Google Scholar] [CrossRef]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial peptides: Diversity, mechanism of action and strategies to improve the activity and biocompatibility in vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef]
- Amorim-Carmo, B.; Parente, A.M.S.; Souza, E.S.; Silva-Junior, A.A.; Araújo, R.M.; Fernandes-Pedrosa, M.F. Antimicrobial Peptide Analogs from Scorpions: Modifications and Structure-Activity. Front. Mol. Biosci. 2022, 9, 887763. [Google Scholar] [CrossRef] [PubMed]
- Wimley, W.C. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem. Biol. 2010, 5, 905–917. [Google Scholar] [CrossRef] [PubMed]
- Gutsmann, T. Interaction between antimicrobial peptides and mycobacteria. Biochim. Et Biophys. Acta Biomembr. 2016, 1858, 1034–1043. [Google Scholar] [CrossRef] [PubMed]
- Fjell, C.D.; Hiss, J.A.; Hancock, R.E.W.; Schneider, G. Designing antimicrobial peptides: Form follows function. Nat. Rev. Drug Discov. 2012, 11, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.H.L.; Wang, C.H.; Liao, Y.T.; Chan, F.Y.; Kanaoka, Y.; Uchihashi, T.; Chen, R.P.Y. Visualizing the membrane disruption action of antimicrobial peptides by cryo-electron tomography. Nat. Commun. 2023, 14, 5464. [Google Scholar] [CrossRef] [PubMed]
- Swana, K.W.; Nagarajan, R.; Camesano, T.A. Atomic force microscopy to characterize antimicrobial peptide-induced defects in model supported lipid bilayers. Microorganisms 2021, 9, 1975. [Google Scholar] [CrossRef]
- Khamis, A.M.; Essack, M.; Gao, X.; Bajic, V.B. Structural bioinformatics Distinct profiling of antimicrobial peptide families. Bioinformatics 2015, 31, 849–856. [Google Scholar] [CrossRef] [PubMed]
- Luna-Ramírez, K.; Sani, M.A.; Silva-Sanchez, J.; Jiménez-Vargas, J.M.; Reyna-Flores, F.; Winkel, K.D.; Separovic, F. Membrane interactions and biological activity of antimicrobial peptides from Australian scorpion. Biochim. Biophys. Acta Biomembr. 2014, 1838, 2140–2148. [Google Scholar] [CrossRef] [PubMed]
- Almaaytah, A.; Zhou, M.; Wang, L.; Chen, T.; Walker, B.; Shaw, C. Antimicrobial/cytolytic peptides from the venom of the North African scorpion, Androctonus amoreuxi: Biochemical and functional characterization of natural peptides and a single site-substituted analog. Peptides 2012, 35, 291–299. [Google Scholar] [CrossRef]
- Mullen, G.R.; Sissom, W.D. Scorpions (scorpiones). In Medical and Veterinary Entomology; Elsevier: Amsterdam, The Netherlands, 2018; pp. 489–504. [Google Scholar] [CrossRef]
- White, S.H.; Wimley, W.C. Hydrophobic interactions of peptides with membrane interfaces. Biochim. Biophys. Acta BBA Rev. Biomembr. 1998, 1376, 339–352. [Google Scholar] [CrossRef]
- Parente, A.M.S.; Daniele-Silva, A.; Furtado, A.A.; Melo, M.A.; Lacerda, A.F.; Queiroz, M.; Fernandes-Pedrosa, M.D.F. Analogs of the scorpion venom peptide Stigmurin: Structural assessment, toxicity, and increased antimicrobial activity. Toxins 2018, 10, 161. [Google Scholar] [CrossRef]
- Uggerhøj, L.E.; Poulsen, T.J.; Munk, J.K.; Fredborg, M.; Sondergaard, T.E.; Frimodt-Moller, N.; Wimmer, R. Rational design of alpha-helical antimicrobial peptides: Do’s and don’ts. ChemBioChem 2015, 16, 242–253. [Google Scholar] [CrossRef]
- Xie, J.; Zhao, Q.; Li, S.; Yan, Z.; Li, J.; Li, Y.; Wang, R. Novel antimicrobial peptide CPF-C1 analogs with superior stabilities and activities against multidrug-resistant bacteria. Chem. Biol. Drug Des. 2017, 90, 690–702. [Google Scholar] [CrossRef]
- De La Salud Bea, R.; Petraglia, A.F.; De Johnson, L.E.L. Synthesis, antimicrobial activity and toxicity of analogs of the scorpion venom BmKn peptides. Toxicon 2015, 101, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Ma, C.; Du, Q.; Wei, R.; Wang, L.; Zhou, M.; Shaw, C. Two peptides, TsAP-1 and TsAP-2, from the venom of the Brazilian yellow scorpion, Tityus serrulatus: Evaluation of their antimicrobial and anticancer activities. Biochimie 2013, 95, 1784–1794. [Google Scholar] [CrossRef]
- Chaudhary, K.; Kumar, R.; Singh, S.; Tuknait, A.; Gautam, A.; Mathur, D.; Raghava, G.P.S. A web server and mobile app for computing hemolytic potency of peptides. Sci. Rep. 2016, 6, 22843. [Google Scholar] [CrossRef]
- Lee, K.; Shin, S.Y.; Kim, K.; Lim, S.S.; Hahm, K.S.; Kim, Y. Antibiotic activity and structural analysis of the scorpion-derived antimicrobial peptide IsCT and its analogs. Biochem. Biophys. Res. Commun. 2004, 323, 712–719. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Corzo, G.; Naoki, H.; Andriantsiferana, M.; Nakajima, T. Purification, structure-function analysis, and molecular characterization of novel linear peptides from scorpion Opisthacanthus madagascariensis. Biochem. Biophys. Res. Commun. 2002, 293, 1514–1522. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.C.; Wang, S.X.; Zhu, Y.; Zhu, S.Y.; Li, W.X. Identification and functional characterization of novel scorpion venom peptides with no disulfide bridge from Buthus martensii Karsch. Peptides 2004, 25, 143–150. [Google Scholar] [CrossRef]
- Dai, C.; Ma, Y.; Zhao, Z.; Zhao, R.; Wang, Q.; Wu, Y.; Li, W. Mucroporin, the first cationic host defense peptide from the venom of Lychas mucronatus. Antimicrob. Agents Chemother. 2008, 52, 3967–3972. [Google Scholar] [CrossRef]
- Gao, B.; Sherman, P.; Luo, L.; Bowie, J.; Zhu, S. Structural and functional characterization of two genetically related meucin peptides highlights evolutionary divergence and convergence in antimicrobial peptides. FASEB J. 2009, 23, 1230–1245. [Google Scholar] [CrossRef]
- Zhao, Z.; Ma, Y.; Dai, C.; Zhao, R.; Li, S.R.; Wu, Y.; Li, W. Imcroporin, a new cationic antimicrobial peptide from the venom of the scorpion Isometrus maculates. Antimicrob. Agents Chemother. 2009, 53, 3472–3477. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Cao, L.; Ma, Y.; Mao, P.; Wang, W.; Zhao, R.; Li, W. Cloning and functional characterization of a new antimicrobial peptide gene StCT1 from the venom of the scorpion Scorpiops tibetanus. Peptides 2010, 31, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Zhao, Z.; He, Y.; Wu, L.; Cai, D.; Hong, W.; Li, W. A new natural α-helical peptide from the venom of the scorpion Heterometrus petersii kills HCV. Peptides 2011, 32, 11–19. [Google Scholar] [CrossRef]
- Fan, Z.; Cao, L.; He, Y.; Hu, J.; Di, Z.; Wu, Y.; Cao, Z. Ctriporin, a new anti-methicillin-resistant Staphylococcus aureus peptide from the venom of the scorpion Chaerilus tricostatus. Antimicrob. Agents Chemother. 2011, 55, 5220–5229. [Google Scholar] [CrossRef]
- Pedron, C.N.; Torres, M.D.T.; Lima, J.A.d.S.; Silva, P.I.; Silva, F.D.; Oliveira, V.X. Novel designed VmCT1 analogs with increased antimicrobial activity. Eur. J. Med. Chem. 2017, 126, 456–463. [Google Scholar] [CrossRef]
- Ramírez-Carreto, S.; Quintero-Hernández, V.; Jiménez-Vargas, J.M.; Corzo, G.; Possani, L.D.; Becerril, B.; Ortiz, E. Gene cloning and functional characterization of four novel antimicrobial-like peptides from scorpions of the family Vaejovidae. Peptides 2012, 34, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Li, Z.; Zhang, R.; Wu, Y.; Li, W.; Cao, Z. StCT2, a new antibacterial peptide characterized from the venom of the scorpion Scorpiops tibetanus. Peptides 2012, 36, 213–220. [Google Scholar] [CrossRef]
- Zeng, X.C.; Zhou, L.; Shi, W.; Luo, X.; Zhang, L.; Nie, Y.; Cao, H. Three new antimicrobial peptides from the scorpion Pandinus imperator. Peptides 2013, 45, 28–34. [Google Scholar] [CrossRef]
- Luna-Ramirez, K.; Tonk, M.; Rahnamaeian, M.; Vilcinskas, A. Bioactivity of natural and engineered antimicrobial peptides from venom of the scorpions urodacus yaschenkoi and U. Manicatus. Toxins 2017, 9, 22. [Google Scholar] [CrossRef]
- De Melo, E.T.; Estrela, A.B.; Santos, E.C.G.; Machado, P.R.L.; Farias, K.J.S.; Torres, T.M.; Fernandes-Pedrosa, M.D.F. Structural characterization of a novel peptide with antimicrobial activity from the venom gland of the scorpion Tityus stigmurus: Stigmurin. Peptides 2015, 68, 3–10. [Google Scholar] [CrossRef]
- Du, Q.; Hou, X.; Wang, L.; Zhang, Y.; Xi, X.; Wang, H.; Shaw, C. AaeAP1 and AaeAP2: Novel antimicrobial peptides from the venom of the scorpion, Androctonus aeneas: Structural characterisation, molecular cloning of biosynthetic precursor-encoding cDNAS and engineering of analogues with enhanced antimicrobial and anticancer activities. Toxins 2015, 7, 219–237. [Google Scholar] [CrossRef]
- Duenas-Cuellar, R.A.; Kushmerick, C.; Naves, L.A.; Batista, I.F.C.; Guerrero-Vargas, J.A.; Pires, O.R., Jr.; Fontes, W.; Castro, M.S. Cm38: A New Antimicrobial Peptide Active Against Klebsiella pneumoniae is Homologous to Cn11. Protein Pept. Lett. 2015, 22, 164–172. [Google Scholar] [CrossRef]
- Lamiable, A.; Thevenet, P.; Rey, J.; Vavrusa, M.; Derreumaux, P.; Tuffery, P. PEP-FOLD3: Faster denovo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016, 44, W449–W454. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, J.K.; Kathuria, M.; Kumar, A.; Mitra, K.; Ghosh, J.K. An Unprecedented alteration in mode of action of IsCT resulting its translocation into bacterial cytoplasm and inhibition of macromolecular syntheses. Sci. Rep. 2015, 5, 9127. [Google Scholar] [CrossRef]
- Zhao, Z.; Hong, W.; Zeng, Z.; Wu, Y.; Hu, K.; Tian, X.; Cao, Z. Mucroporin-M1 inhibits hepatitis B virus replication by activating the mitogen-activated protein kinase (MAPK) pathway and down-regulating HNF4α in vitro and in vivo. J. Biol. Chem. 2012, 287, 30181–30190. [Google Scholar] [CrossRef] [PubMed]
- Rostaminejad, M.; Savardashtaki, A.; Mortazavi, M.; Khajeh, S. Animal Gene Identification and characterization of new putative antimicrobial peptides from scorpion Chaerilus tricostatus revealed by in silico analysis and structure modeling. Anim. Gene 2022, 26, 200137. [Google Scholar] [CrossRef]
- Mahnam, K.; Lotfi, M.; Shapoorabadi, F.A. Examining the interactions scorpion venom peptides (HP1090, Meucin-13, and Meucin-18) with the receptor binding domain of the coronavirus spike protein to design a mutated therapeutic peptide. J. Mol. Graph. Model. 2021, 107, 107952. [Google Scholar] [CrossRef] [PubMed]
- Hong, W.; Zhang, R.; Di, Z.; He, Y.; Zhao, Z.; Hu, J.; Cao, Z. Design of histidine-rich peptides with enhanced bioavailability and inhibitory activity against hepatitis C virus. Biomaterials 2013, 34, 3511–3522. [Google Scholar] [CrossRef]
- Alcolea-Medina, A.; Snell, L.B.; Alder, C.; Charalampous, T.; Williams, T.G.S.; Athitha, V.; Edgeworth, J.D. The ongoing Streptococcus pyogenes (Group A Streptococcus) outbreak in London, United Kingdom, in December 2022: A molecular epidemiology study. Clin. Microbiol. Infect. 2023, 29, 887–890. [Google Scholar] [CrossRef]
- Williamson, D.A.; Chow, E.P.F.; Gorrie, C.L.; Seemann, T.; Ingle, D.J.; Higgins, N.; Howden, B.P. Bridging of Neisseria gonorrhoeae lineages across sexual networks in the HIV pre-exposure prophylaxis era. Nat. Commun. 2019, 10, 3988. [Google Scholar] [CrossRef] [PubMed]
- Lassoued, Y.; Assad, Z.; Ouldali, N.; Caseris, M.; Mariani, P.; Birgy, A.; Faye, A. Unexpected Increase in Invasive Group A Streptococcal Infections in Children After Respiratory Viruses Outbreak in France: A 15-Year Time-Series Analysis. Open Forum Infect. Dis. 2023, 10, ofad188. [Google Scholar] [CrossRef] [PubMed]
- Whittles, L.K.; White, P.J.; Paul, J.; Didelot, X. Epidemiological trends of antibiotic resistant Gonorrhoea in the United Kingdom. Antibiotics 2018, 7, 60. [Google Scholar] [CrossRef] [PubMed]

| Priority 1: CRITICAL | |
| i. | Acinetobacter baumannii, carbapenem-resistant |
| ii. | Pseudomonas aeruginosa, carbapenem-resistant |
| iii. | Enterobacteriaceae, carbapenem-resistant, ESBL-producing |
| Priority 2: HIGH | |
| iv. | Enterococcus faecium, vancomycin-resistant |
| v. | Staphylococcus aureus, methicillin-resistant, vancomycin-intermediate and resistant |
| vi. | Helicobacter pylori, clarithromycin-resistant |
| vii. | Campylobacter spp., fluoroquinolone-resistant |
| viii | Salmonellae, fluoroquinolone-resistant |
| ix. | Neisseria gonorrhoeae, cephalosporin-resistant, fluoroquinolone-resistant |
| Priority 3: MEDIUM | |
| x. | Streptococcus pneumoniae, penicillin-non-susceptible |
| xi. | Haemophilus influenzae, ampicillin-resistant |
| xii. | Shigella spp., fluoroquinolone-resistant |
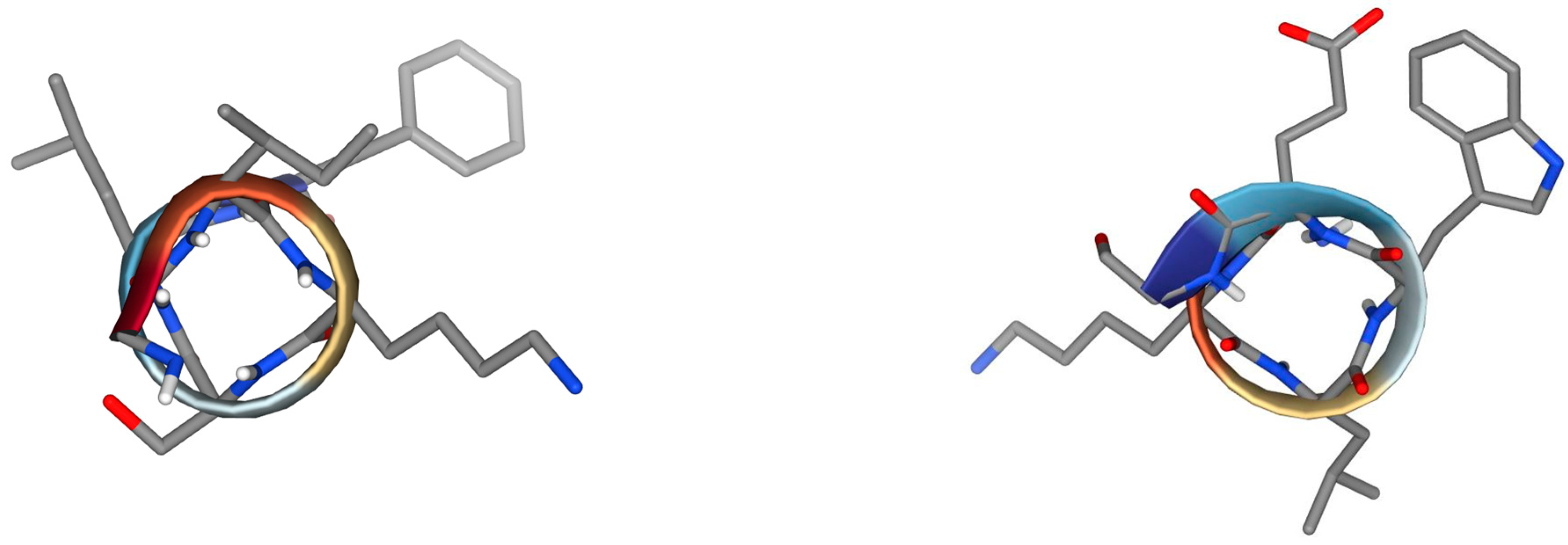
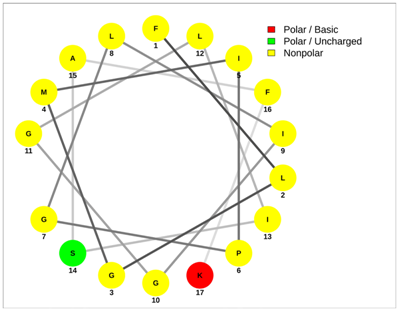
| ISCT2 | BmKn2 |
 |  |
| IFGAIWNGIKSLF | FIGAIANLLSKIF |
| AamAP1 | TSAP-2 |
 |  |
| FLFSLIPHAIGGLISAFK | FLGMIPGLIGGLISAFK |
| UyCT2 | Pantinin-2 |
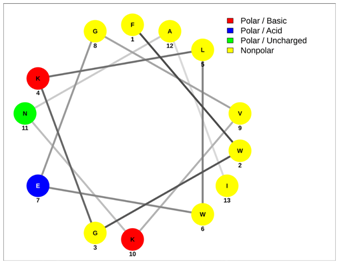 | 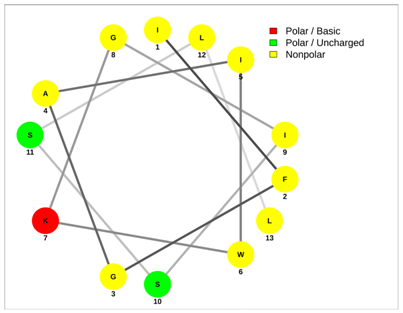 |
| FWGKLWEGVKNAI | IFGAIWKGISSLL |
| Mucroporin | Ctriporin |
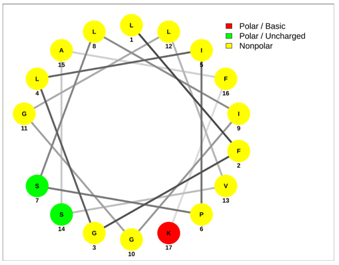 |  |
| LFGLIPSLIGGLVSAFK | FLWGLIPGAISAVTSLIKK |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panayi, T.; Diavoli, S.; Nicolaidou, V.; Papaneophytou, C.; Petrou, C.; Sarigiannis, Y. Short-Chained Linear Scorpion Peptides: A Pool for Novel Antimicrobials. Antibiotics 2024, 13, 422. https://doi.org/10.3390/antibiotics13050422
Panayi T, Diavoli S, Nicolaidou V, Papaneophytou C, Petrou C, Sarigiannis Y. Short-Chained Linear Scorpion Peptides: A Pool for Novel Antimicrobials. Antibiotics. 2024; 13(5):422. https://doi.org/10.3390/antibiotics13050422
Chicago/Turabian StylePanayi, Tolis, Spiridoula Diavoli, Vicky Nicolaidou, Christos Papaneophytou, Christos Petrou, and Yiannis Sarigiannis. 2024. "Short-Chained Linear Scorpion Peptides: A Pool for Novel Antimicrobials" Antibiotics 13, no. 5: 422. https://doi.org/10.3390/antibiotics13050422
APA StylePanayi, T., Diavoli, S., Nicolaidou, V., Papaneophytou, C., Petrou, C., & Sarigiannis, Y. (2024). Short-Chained Linear Scorpion Peptides: A Pool for Novel Antimicrobials. Antibiotics, 13(5), 422. https://doi.org/10.3390/antibiotics13050422











