Abstract
The rise of multi-drug-resistant (MDR) pathogenic bacteria presents a grave challenge to global public health, with antimicrobial resistance ranking as the third leading cause of mortality worldwide. Understanding the mechanisms underlying antibiotic resistance is crucial for developing effective treatments. Efflux pumps, particularly those of the resistance-nodulation-cell division (RND) superfamily, play a significant role in expelling molecules from bacterial cells, contributing to the emergence of multi-drug resistance. These are transmembrane transporters naturally produced by Gram-negative bacteria. This review provides comprehensive insights into the modulation of RND efflux pump expression in bacterial pathogens by numerous and common molecules (bile, biocides, pharmaceuticals, additives, plant extracts, etc.). The interplay between these molecules and efflux pump regulators underscores the complexity of antibiotic resistance mechanisms. The clinical implications of efflux pump induction by non-antibiotic compounds highlight the challenges posed to public health and the urgent need for further investigation. By addressing antibiotic resistance from multiple angles, we can mitigate its impact and preserve the efficacy of antimicrobial therapies.
1. Introduction
Infections caused by MDR pathogenic bacteria present a formidable challenge to global public health, as effective treatments remain elusive. Recent projections indicate that antimicrobial resistance was associated with an estimated 4.95 million deaths worldwide in 2019, positioning it as the third leading cause of mortality on a global scale [1]. Furthermore, these projections imply a potential surpassing of the latest estimations provided by the WHO, which anticipate 10 million annual deaths attributable to antimicrobial resistance by 2050. Without significant advancements in antibiotherapy and the development of innovative therapeutic strategies to counter bacterial resistance mechanisms, these escalating concerns are expected to persist.
Bacteria employ four primary mechanisms of resistance that can be split into two types: those that use specific mechanisms for selected antibiotic families or those that use less-specific pathways within a broader spectrum of antibiotics. On the one hand, enzymatic inactivation represents one of these specific mechanisms, typified by AmpC β-lactamase, which degrades the β-lactam core of antibiotics within the β-lactam family [2]. Target alteration constitutes another mechanism observed for specific antibiotic families, disrupting the bacterial replication process and impacting quinolones in various strains, leading to a poorly targeted molecule [3]. On the other hand, membrane mechanisms serve as formidable primary defenses to counteract and reduce intracellular broad-spectrum antibiotic accumulation during exposure. While membrane impermeability restricts the influx of antimicrobials [4], efflux pumps facilitate the expulsion of compounds considered toxic to bacteria, thereby maintaining intracellular concentrations below therapeutic thresholds [5]. Membrane impermeability and efflux pumps are often co-regulated and can confer resistance across multiple antibiotic families, contributing to the emergence of MDR pathogens. Even more concerning, various studies have demonstrated that the overexpression of efflux pumps in bacteria is implicated in the selection of mutations within entire genomes, including genes encoding antibiotic targets [6,7].
It is imperative to distinguish between innate and acquired resistance when focusing on antibiotic resistance. Innate resistance refers to the natural resistance of bacterial species to specific antibiotics, as seen in Escherichia coli, with the intrinsic expression of AmpC, and of the AcrAB-TolC multi-drug efflux system [8]. Acquired resistance enables strains to enhance their resistance levels through mutations [9] or acquisition of genetic material from other bacteria [10]. Moreover, adaptive or induced resistance involves the occasional or excessive activation of previously described mechanisms in response to stress or resistance-inducing molecules. Exposure of specific bacterial species to trigger factors can result in resistance development either by selecting mutant strains or inducing phenotypic adaptations leading to cross-resistance to antibiotics. This form of resistance is transient, with bacteria returning to a basal resistance state once the inducer dissipates.
This review focuses on the second type of resistance mechanism involving efflux pumps and aims to provide a comprehensive overview of the diverse molecules influencing RND efflux pump expression in Gram-negative bacteria. While many of these molecules are antimicrobial agents, others are compounds present in the human body, natural substances, additives, or non-antibiotic drugs. Given that some obscure pathways are associated with each induction mechanism, elucidated induction mechanisms are described.
2. RND Multi-Drug Efflux Pumps and Their Regulation
The polyspecific efflux transporters expressed in Gram-negative bacteria exhibit remarkable diversity and are classified into six distinct families: the RND superfamily, the ATP-binding cassette (ABC) superfamily, the major facilitator superfamily (MFS), the multi-drug and toxic compound extrusion (MATE) family, the small multi-drug resistance (SMR) family, and the proteobacterial antimicrobial compound efflux (PACE) transporter family [11]. Among these, the RND superfamily constitutes the primary player in multi-drug efflux pumps within Gram-negative bacteria, highlighting this family’s significance within the present review context. In Enterobacteriaceae, AcrAB-TolC stands out as the principal and most prevalent efflux pump across various species, including Escherichia coli, Salmonella enterica, and Klebsiella pneumoniae [12]. Pseudomonas aeruginosa, an opportunistic pathogen, exhibits the most abundant efflux system identified, featuring MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY-OprM as the clinically relevant RND efflux pumps [13]. Conversely, in Pseudomonas putida, the role of RND efflux pumps in antibiotic resistance remains incompletely understood, though TtgABC is implicated [14]. Stenotrophomonas maltophilia harbors numerous efflux pumps, with SmeABC, SmeDEF, SmeJK, SmeVWX, and SmeYZ particularly relevant from a clinical standpoint [15,16,17]. Campylobacter jejuni, a gastrointestinal pathogen, relies on CmeABC as its primary efflux pump contributing to antibiotic resistance [18]. Burkholderia cenocepacia possesses several efflux pumps, including CeoAB-OpcM, conferring resistance to clinically significant antibiotics [19].
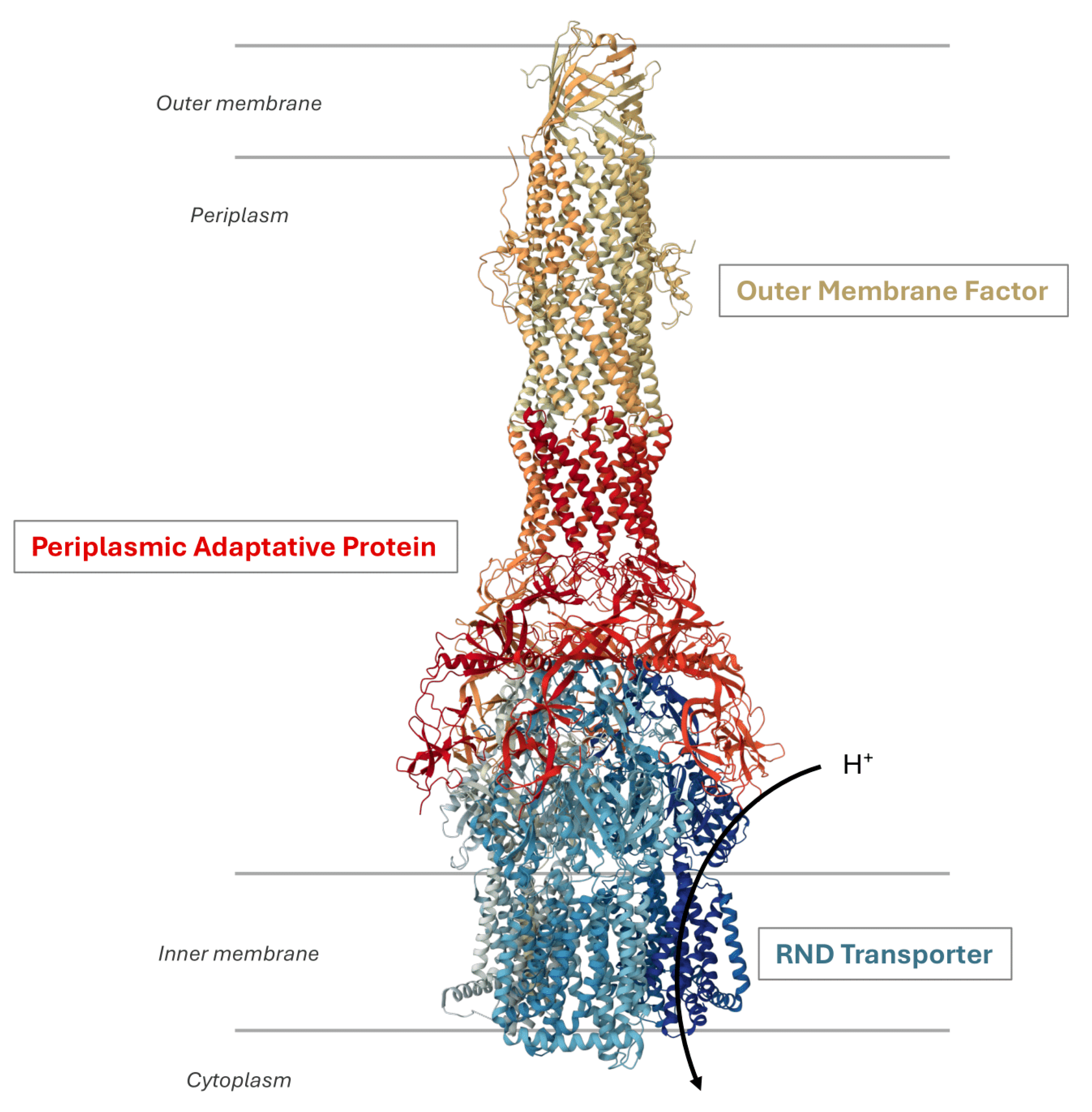
RND efflux pumps possess a tripartite architecture (Figure 1) consisting of an active RND transporter in the inner membrane as a homo- or heterotrimer using the proton motive force for substrate extrusion. This architecture also involves an outer membrane factor (OMF) and a periplasmic adaptor protein (PAP) that bridges the proteins across both membranes [20].
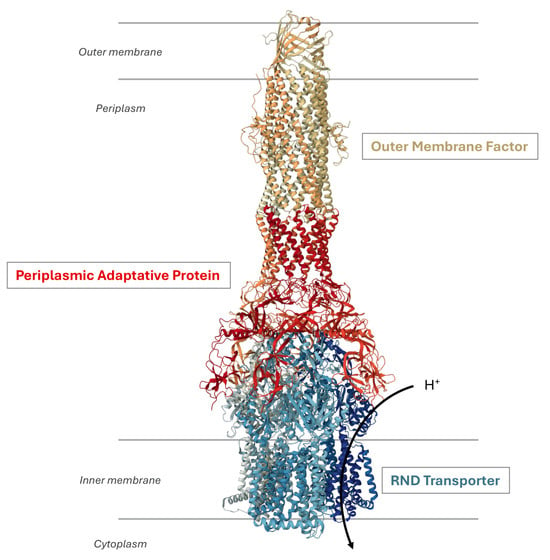
Figure 1.
Organization of an RND efflux pump. The illustration shows the structure of the P. aeruginosa MexAB-OprM system (Protein DataBank entry: 6IOL). It is a tripartite complex composed of the inner membrane RND protein MexB, the outer membrane protein OprM, and the periplasmic adaptative protein MexA. The transport activity is coupled to the translocation of protons in the cytoplasm.
The inner membrane transporter forms an asymmetric trimer where each protomer adopts distinct conformational states designated as loose (L) or access, tight (T) or binding, and open (O) or extrusion [21,22]. This conformational cycle facilitates the sequential binding of substrates, ultimately leading to drug efflux [23,24], whereby substrates are transported by the RND transporter and extruded from the cell through the tripartite complex. Several studies have demonstrated the broad substrate specificity exhibited by these efflux pumps, including structurally diverse molecules such as antibiotics, anticancer agents, dyes, bile salts, detergents, and solvents [20]. Recently, a study based on minimum inhibitory concentration (MIC) values of various efflux-resistant E. coli strains towards distinct classes of antibiotics elucidated the molecular determinants responsible for substrate recognition by AcrAB-TolC [25].
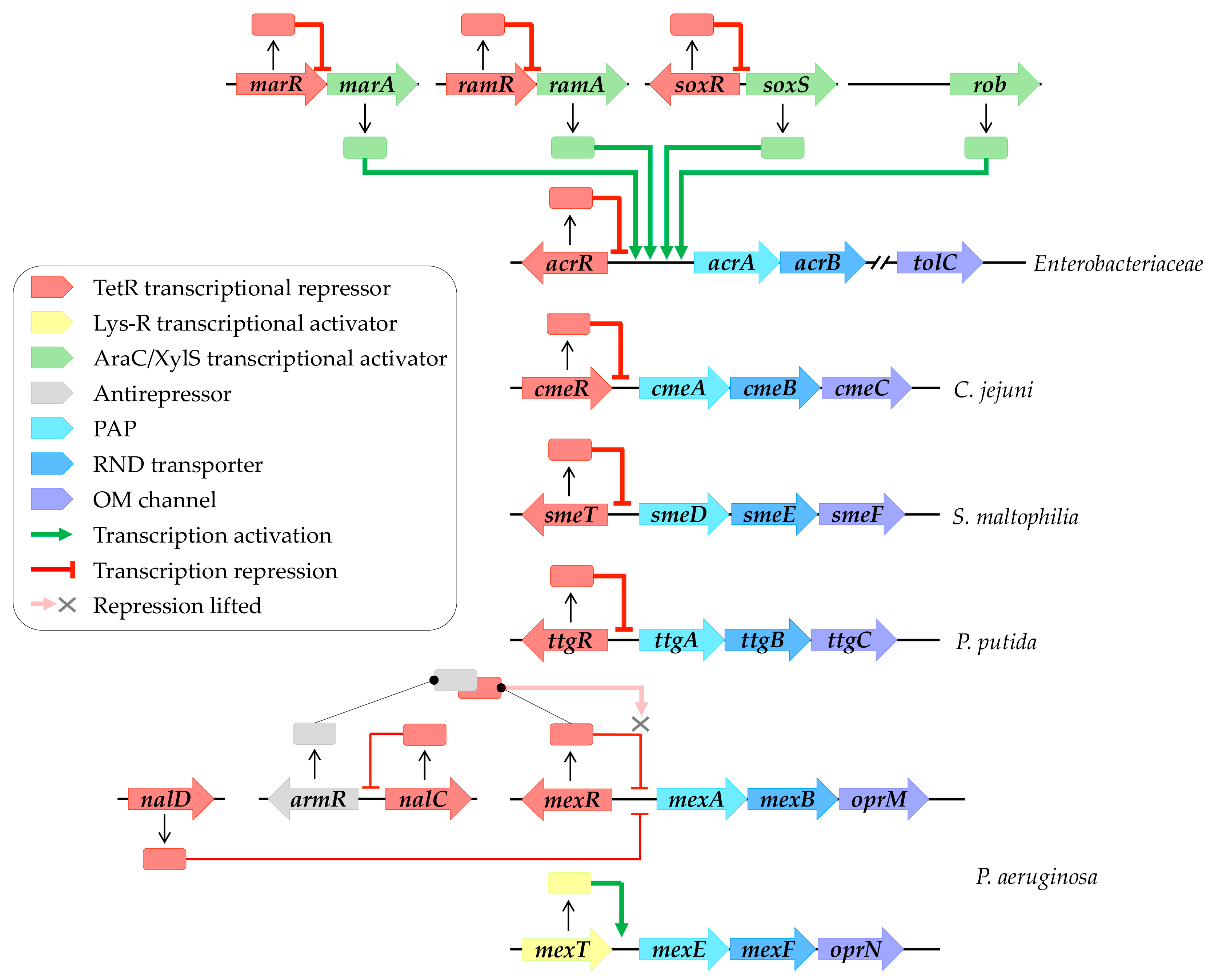
The genes encoding efflux systems are commonly arranged into operons, comprising the RND transporter and the PAP. The third partner may be located within the same operon or elsewhere in the genome. These genes are subject to regulation by local or global regulators (Figure 1). These regulators respond to a diverse array of signals to modulate efflux gene expression [26]. For the sake of simplicity, the following paragraph will provide an overview of the regulatory pathways cited throughout this review (for further details, refer to recent reviews [20,27,28]).
Local regulation commonly involves TetR family transcriptional regulators, which consist of an N-terminal DNA binding domain (NTD) recognizing and binding to a palindromic DNA sequence located in the intergenic region between the regulator and the regulated gene. These regulators also feature a large C-terminal domain (CTD) responsible for ligand binding [29]. For instance, AcrR locally represses and maintains basal levels of AcrAB-TolC in Enterobacteriaceae [30], while CmeR regulates CmeABC in C. jejuni [31]. Furthermore, SmeT controls SmeDEF in S. maltophilia [32], MexR governs MexAB-OprM in P. aeruginosa [33], and TtgR regulates TtgABC in P. putida [34] (Figure 1). Moreover, the repression of the MexAB-OprM system involves NalD and NalC, located elsewhere in the genome [35,36]. NalC indirectly regulates MexAB-OprM expression by repressing ArmR, an antirepressor of MexR [37]. The complex formation between MexR and ArmR prevents MexR attachment to the intergenic promoter region, leading to mexAB-oprM overexpression [38] (Figure 2). In contrast, MexT, a Lys-R family regulator, activates MexEF-OprN expression in P. aeruginosa [39].
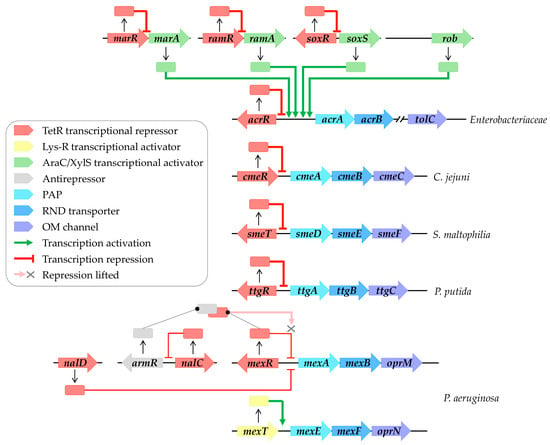
Figure 2.
RND efflux pump transcriptional regulation networks. Local regulation primarily involves TetR family transcriptional regulators (highlighted in red), including AcrR, CmeR, SmeT, TtgR, and MexR, which, respectively, regulate AcrAB-TolC, CmeABC, SmeDEF, TtgABC, and MexAB-OprM systems. Repression of MexAB-OprM systems involves NalD and NalC, located elsewhere in the genome of P. aeruginosa. NalC indirectly modulates expression by inhibiting ArmR, an antirepressor of MexR (highlighted in grey), leading to the alleviation of repression by MexR. MexT (highlighted in yellow) activates MexEF-OprN expression. Global regulation, on the other hand, is orchestrated by AraC/XylS family transcriptional regulators (highlighted in green), including MarA, RamA, SoxS, and Rob, which activate AcrAB-TolC expression. These regulators are subject to local regulation by their own TetR family transcriptional regulators (highlighted in red), such as MarR, RamR, and SoxR. Transcriptional regulatory pathways enabling activation are depicted by green arrows, while those repressing activation are indicated by red arrows.
Global regulation typically involves AraC/XylS family transcriptional regulators, such as MarA, RamA, SoxS, and Rob in Enterobacteriaceae, which activate efflux pump gene expression [40] (Figure 2). These global regulators are themselves locally regulated by their own TetR family transcriptional regulators, including MarR, RamR, and SoxR (Figure 2). External stressors can trigger the release of these repressors, leading to the activation of efflux gene expression, as discussed in subsequent paragraphs.
Furthermore, in addition to transcriptional regulation, many tripartite efflux systems are subject to regulation by two-component systems (TCS) [41]. TCS detect and respond to external stimuli by orchestrating gene expression. The correlation between TCS and antibiotic resistance has been elucidated in numerous pathogens [42,43]; for instance, the AmgRS TCS has been implicated in the development of aminoglycoside resistance in P. aeruginosa through the upregulation of mexXY [44].
3. Induction of Resistance
3.1. Bile
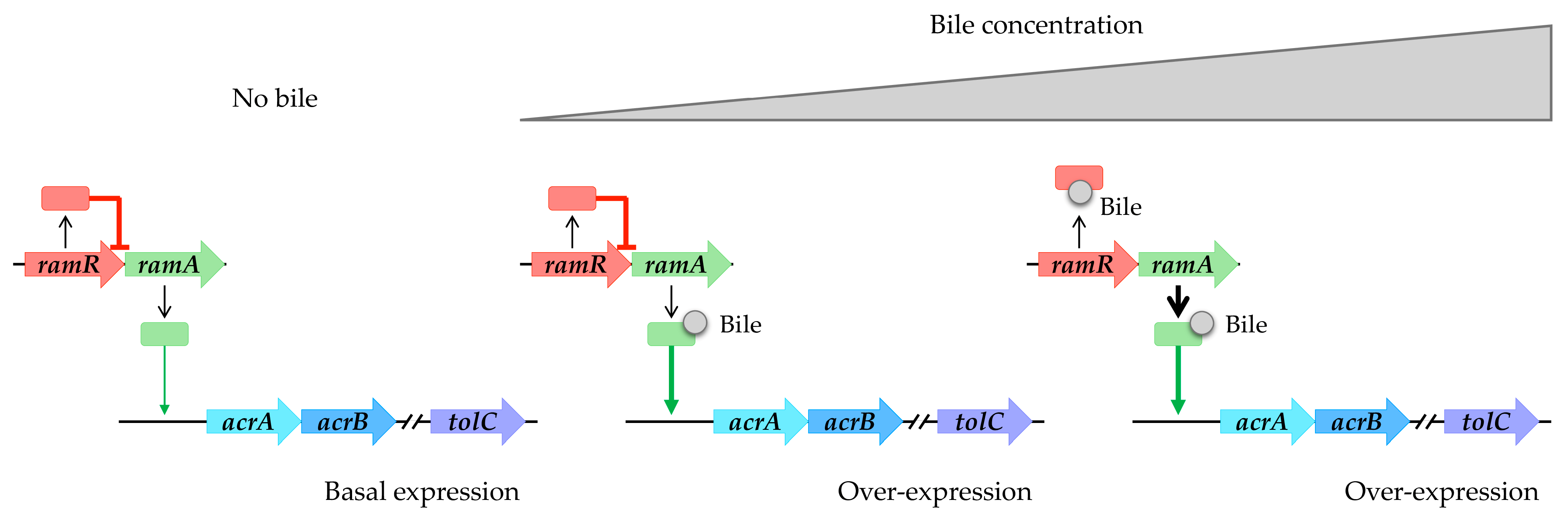
Bile is a complex mixture of organic and inorganic constituents, including fatty acids and bile acids or salts. According to references in the literature, it appears to play a significant role in upregulating the expression of RND efflux pumps (refer to Table 1). Specifically, within the intestinal tract, bile components have been observed to induce the expression of the AcrAB-TolC pump in enterobacteria, including opportunistic pathogens such as S. enterica and E. coli. In the case of S. enterica, bile facilitates the induction of the AcrAB-TolC pump via RamA. This induction occurs through a two-step mechanism: initially, bile binds to RamA, activating it [45]; subsequently, as the bile concentration increases, it binds to RamR. This binding prevents RamR from interacting with the ramA promoter region, leading to the overexpression of ramA and subsequent overproduction of the AcrAB-TolC system [46,47,48] (Figure 3). Structural analyses of RamR complexed with bile components revealed that cholic acid and chenodeoxycholic acid form four hydrogen bonds with Tyr59, Trp85, Ser137, and Asp152 of RamR instead of the typical π-π interaction with Phe155, which is an essential residue for the recognition of many other molecules, inducing conformational changes that are crucial for their operation. It has been challenging to crystallize RamR with deoxycholic acid, likely due to the absence of the 7a-hydroxyl group, which is crucial for forming a hydrogen bond with Asp152 of RamR. This absence also prevents the induction of acrAB-tolC [48].
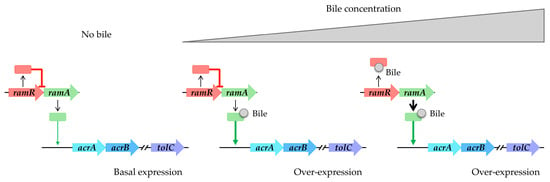
Figure 3.
Bile components induce acrAB-tolC overexpression in S. enterica. Absence of bile results in basal expression of acrAB-tolC. Low bile concentration triggers RamA activation. High bile concentration induces RamR interaction, causing overexpression of ramA and subsequent overproduction of the AcrAB-TolC system.
In E. coli, bile salts induce the overexpression of acrAB while inhibiting the expression of ompF, an outer membrane porin [49,50]. This induction is mediated by Rob. Unlike RamA, the induction by Rob does not involve overexpression but rather a conformational change in existing Rob proteins [51,52]. Shi et al. demonstrated the docking interaction of chenodeoxycholic acid with the ligand binding pocket, which is surrounded by a cluster of aromatic and heterocyclic amino acids. They concluded that the CTD of Rob contains a Gyr-like domain which acts as an environmental sensor interacting with ligands. This interaction structurally stabilizes and activates transcription via allosteric coordination with the NTD [53].
Moreover, bile has been demonstrated to induce overexpression of the cmeABC operon in C. jejuni [54], which encodes for the major RND efflux pump and is regulated by CmeR. The binding of bile salts to the CmeR protein inhibits its interaction with the DNA operon, thereby relieving repression [54,55]. Co-crystallization studies have elucidated the interactions between CmeR protein and bile salts, including taurocholate and cholate, which share a similar chemical structure and charge. These molecules bind to the CmeR-DNA binding region in the same orientation but in an antiparallel mode within the tunnel. Specifically, only two positively charged residues, Lys170 and His175, form essential hydrogen bonds with the steroid backbones of taurocholate and cholate. In the case of taurocholate, CmeR also anchors the molecule by utilizing the positively charged residue His72 to form an additional hydrogen bond with the 3a-hydroxyl group. For cholate, the residue His174 interacts with the non-conjugated 5b-cholanoate tail. These interactions have been corroborated by isothermal titration calorimetry, revealing that the regulator binds to these compounds with dissociation constants (Kd) in the micromolar range [56].
Interestingly, over 80% of cystic fibrosis patients experience increased gastric reflux and aspiration of duodenogastric contents into the lungs [57]. Bile present in the lungs constitutes the primary comorbidity factor for patients with respiratory diseases [58]. In the case of cystic fibrosis, bile has been shown to correlate with a decrease in biodiversity and the emergence of specific pathogens such as P. aeruginosa [59,60,61]. Bile facilitates the induction of genes associated with chronic infections, including the mexAB-oprM operon of P. aeruginosa [61].

Table 1.
Bile components which induce RND efflux pumps.
Table 1.
Bile components which induce RND efflux pumps.
| Molecules | Classification | Pumps | Strains | Mechanisms | References |
|---|---|---|---|---|---|
| Chenodeoxycholate | Bile salt | AcrAB-TolC | E. coli | Rob activation | [54] |
| CmeABC | C. jejuni | CmeR interaction | [52] | ||
| Chenodeoxycholic acid | Bile acid | AcrAB-TolC | E. coli | Rob activation | [53] |
| S. enterica | RamR interaction | [48] | |||
| MexAB-OprM | P. aeruginosa | * | [61] | ||
| Cholate | Bile salt | AcrAB-TolC | E. coli | Rob activation | [52] |
| CmeABC | C. jejuni | CmeR interaction | [54,56] | ||
| Choleate | Bile salt | AcrAB-TolC | S. enterica | RamA activation | [45] |
| CmeABC | C. jejuni | CmeR interaction | [54] | ||
| Cholic acid | Bile acid | AcrAB-TolC | S. enterica | RamA activation and RamR interaction | [45,48] |
| CmeABC | C. jejuni | CmeR interaction | [54] | ||
| Decanoate | Fatty acids | AcrAB-TolC | E. coli | Rob activation | [50,52,53] |
| Deoxycholate | Bile salt | AcrAB-TolC | E. coli | Rob activation | [52] |
| S. enterica | RamR interaction | [46] | |||
| CmeABC | C. jejuni | CmeR interaction | [54] | ||
| Deoxycholic acid | Bile acid | AcrAB-TolC | S. enterica | RamA activation | [45] |
| Glycochenodeoxycholate | Bile salt | AcrAB-TolC | E. coli | Rob activation | [52] |
| Glycocholate | Bile salt | CmeABC | C. jejuni | CmeR interaction | [54] |
| Taurocholate | Bile salt | AcrAB-TolC | E. coli | Rob activation | [52] |
| CmeABC | C. jejuni | CmeR interaction | [54,55,56] | ||
| Taurodeoxycholate | Bile salt | CmeABC | C. jejuni | CmeR interaction | [54] |
* Unknown.
3.2. Antibiotics
Many antibiotics have been described as inducing the expression of RND efflux pumps (refer to Table 2). In 2003, it was demonstrated for the first time that the expression of an RND transporter is directly regulated by antibiotics. Specifically, chloramphenicol, tetracycline, and other plant antimicrobials induce the expression of TtgABC from P. putida by interacting with the regulator TtgR. Upon exposure to these antimicrobial agents, TtgR, capable of binding to various structurally distinct antibiotics, loses its ability to bind to the promoter [62,63,64]. This mechanism has been confirmed through co-crystallizations of TtgR with antibiotics, revealing that most of the characterized ligands bind at a common site parallel to the axis of the dimer and within a hydrophobic binding pocket with few specific interactions. This likely enhances the binding flexibility of the ligand and results in the micromolar affinity of TtgR [63,64,65]. NalD follows a similar mechanism. It interacts with novobiocin, with one NalD dimer binding to two novobiocin molecules with a Kd of 4.65 µM, thereby dissociating it from the promoter and leading to the expression of mexAB-oprM. The involvement of Asn129 and His167 residues in this interaction has been demonstrated [66]. Additionally, aminoglycosides can induce MexAB-OprM expression via the two-component system AmgRS involved in the envelope stress response [67]. This pump can also be induced in the presence of erythromycin, tetracycline, and azithromycin, and this can occur independently of AmgRS activity [67].
MexEF-OprN responds to nitrous stress in P. aeruginosa. The nitroaromatic antibiotic chloramphenicol can induce the expression of mexEF-oprN via the transcriptional regulator MexT [68]. Similarly, chloramphenicol induces CeoAB-OpcM, which is a homologue of MexEF-OprN from B. cenocepacia, by inducing the CeoR regulator, which is a homologue of MexT [69].
The induction of MexXY-OprM is triggered by ribosome-targeting antibiotics, such as chloramphenicol, tetracycline, macrolides, and aminoglycosides, but not by antibiotics acting on other cellular targets [70,71,72]. Similarly, SmeYZ in S. maltophilia is also induced by these ribosome-targeting antibiotics that inhibit protein synthesis. Interestingly, boric acid, an insecticide which prevents tRNA acylation and inhibits protein synthesis, can also induce SmeYZ [73].
Chloramphenicol and tetracycline induce marA and acrB expression in E. coli. Tetracycline, particularly, allows the induction of acrD and acrF [74,75] through the intervention of MarR [76]. The induction mechanism is hypothesized to involve RNA stabilization rather than direct regulation by MarR [77]. Furthermore, carbapenems, representing the final therapeutic option for all Gram-negative bacteria [78], have also been shown to induce efflux [79].

Table 2.
Antibiotics which induce RND efflux pumps.
Table 2.
Antibiotics which induce RND efflux pumps.
| Molecules | Classification | Pumps | Strains | Mechanisms | References |
|---|---|---|---|---|---|
| Amikacin | Aminoglycoside | MexAB-OprM | P. aeruginosa | * | [67,80] |
| Azithromycin | Macrolide | MexAB-OprM | P. aeruginosa | * | [67] |
| MexXY-OprM | Protein synthesis inhibition | [71] | |||
| Azlocillin | Penicillin | MexAB-OprM | P. aeruginosa | * | [80] |
| SmeYZ | S. maltophilia | [73] | |||
| Chloramphenicol | Phenicol | CeoAB-OpcM | B. cenocepacia | ceoR induction | [69] |
| AcrAB-TolC | E. coli | marA induction | [74] | ||
| MexEF-OprN | P. aeruginosa | MexT-dependent (nitrosative stress) | [68] | ||
| MexXY-OprM | Protein synthesis inhibition | [71,72] | |||
| TtgABC | P. putida | TtgR interaction | [62,65] | ||
| SmeYZ | S. maltophilia | Protein synthesis inhibition | [73] | ||
| Chlortetracycline | Tetracycline | SmeVWX | S. maltophilia | * | [73] |
| Cinoxacin | Penicillin | SmeYZ | S. maltophilia | * | [73] |
| SmeVWX | |||||
| Cloxacillin | Penicillin | SmeVWX | S. maltophilia | * | [73] |
| Ethionamide | Antitubercular agent | MexAB-OprM | P. aeruginosa | * | [80] |
| Erythromycin | Macrolide | MexAB-OprM | P. aeruginosa | * | [67] |
| MexXY-OprM | Protein synthesis inhibition | [70,71,72] | |||
| SmeYZ | S. maltophilia | [73] | |||
| SmeVWX | * | ||||
| Fusidic acid | Fusidanine | SmeYZ | S. maltophilia | Protein synthesis inhibition | [73] |
| Gentamicin | Aminoglycoside | MexAB-OprM | P. aeruginosa | AmgRS activation | [67] |
| MexXY-OprM | Protein synthesis inhibition | [70,71] | |||
| Kanamycin | Aminoglycoside | MexAB-OprM | P. aeruginosa | AmgRS activation | [67] |
| MexXY-OprM | Protein synthesis inhibition | [72] | |||
| Lincomycin | Lincosamide | SmeYZ | S. maltophilia | Protein synthesis inhibition | [73] |
| Meropenem | Carbapenem | AcrAB-TolC | E. coli | marA induction | [79] |
| Neomycin | Aminoglycoside | MexAB-OprM | P. aeruginosa | AmgRS activation | [67] |
| Novobiocin | Aminocoumarine | MexAB-OprM | P. aeruginosa | NalD interaction | [66] |
| Oleandomycin | Macrolide | SmeYZ | S. maltophilia | Protein synthesis inhibition | [73] |
| Paromycin | Aminoglycoside | MexAB-OprM | P. aeruginosa | AmgRS activation | [67] |
| Penimepicycline | Tetracycline | SmeYZ | S. maltophilia | Protein synthesis inhibition | [73] |
| SmeVWX | * | ||||
| Puromycin | Aminoglycoside | SmeYZ | S. maltophilia | Protein synthesis inhibition | [73] |
| Rolitetracycline | Tetracycline | SmeYZ | S. maltophilia | Protein synthesis inhibition | [73] |
| Spectinomycin | Aminoglycoside | MexXY-OprM | P. aeruginosa | Protein synthesis inhibition | [71] |
| Spiramycin | Macrolide | SmeYZ | S. maltophilia | Protein synthesis inhibition | [73] |
| SmeVWX | * | ||||
| Sulfadiazine | Sulfonamide | SmeYZ | S. maltophilia | * | [73] |
| Sulfathiazole | Sulfonamide | SmeYZ | S. maltophilia | * | [73] |
| Tetracycline | Tetracycline | AcrAB-TolC | E. coli | marA induction | [74,75] |
| AcrAD-TolC | * | ||||
| AcrEF-TolC | |||||
| MexXY-OprM | P. aeruginosa | Protein synthesis inhibition | [70,71,72] | ||
| TtgABC | P. putida | TtgR interaction | [62,65] | ||
| Tylosin | Macrolide | SmeYZ | S. maltophilia | Protein synthesis inhibition | [73] |
| SmeVWX | * | ||||
| Vancomycin | Glycopeptide | SmeVWX | S. maltophilia | * | [73] |
* Unknown.
3.3. Biocides
While the upregulation of RND efflux pumps in response to antibiotic exposure is well-documented, emerging evidence suggests that biocides, commonly used in disinfection and sanitation, may also induce the expression of these efflux systems (refer to Table 3).
Triclosan, a widely used biocide found in numerous products, such as toothpaste and liquid hand soap, modulates the expression of SmeDEF in S. maltophilia by disrupting the interaction between the transcriptional repressor SmeT and its operator site. This disruption leads to an increase in smeDEF expression, consequently reducing the susceptibility of S. maltophilia to antibiotics such as ciprofloxacin, as evidenced by an increased MIC: from 0.75 µg/mL to 2 µg/mL [81]. Triclosan exerts its effect by binding two molecules to SmeT, with a Kd of 0.63 µM. One triclosan molecule binds to the bottom of the ligand-binding pocket, adopting a conformation reminiscent of the interaction between the plant antimicrobial molecule phloretin and TtgR in P. putida [65], where it is parallel to the ⍺6 helix and stacks against the phenolic ring of Phe70, a residue crucial for ligand binding. The second molecule binds near the dimer interface, interacting with the α6 helix via its phenolic ring. This binding event stabilizes the NTD of each subunit of the homodimer, preventing DNA binding [81].
Biocides are capable of interacting with bacterial membranes, such as benzalkonium chloride, chlorhexidine, and dequalinium chloride, and disrupting them [82]. Biocides can trigger the upregulation of the RND MexCD-OprJ efflux pump in P. aeruginosa, thereby decreasing its susceptibility to certain antibiotics [80,83,84,85]. These findings hold clinical relevance, given that these biocidal agents are commonly employed in antiseptic and disinfectant protocols in clinical context. For instance, exposure to 10 µg/mL of dequalinium chloride resulted in a 54-fold increase in mexCD-oprJ expression within 30 min of addition (with no further inducers present in the media), with a sustained 10-fold increase observed even after 120 min, indicating a potential “induction memory” [80]. It has been postulated that the membrane damage caused by these biocides, rather than the biocides themselves, induces the mexCD-oprJ operon. Supporting this notion, it was demonstrated that chlorhexidine induces mexCD-oprJ by interacting with AlgU, which is a sigma factor in P. aeruginosa analogous to RpoE in E. coli, where RpoE plays a pivotal role as a membrane stress response-associated sigma factor [84].
Exposure to chlorinated phenols, such as pentachlorophenol, and to chlorinated phenol-based disinfectants, such as triclosan, results in the development of an antibiotic resistance phenotype in P. aeruginosa by inducing mexAB-oprM [86,87,88,89]. Transcriptional analyses following pentachlorophenol exposure have revealed the overexpression of mexAB, mexR, armR, and nalC genes [86,87]. NalC can reversibly bind to chlorinated phenols and chlorophenol-containing chemicals and be dissociated from the promoter when linked with it. This binding can facilitate the upregulation of the NalC regulon [87]. Overproduction of ArmR and formation of MexR-ArmR complexes contribute to mexAB-oprM overexpression [87,88]. Evidence of overexpression in an armR-depleted strain suggests the involvement of other mechanisms that still require MexR [88]. Although pentachlorophenol does not directly affect MexR binding to DNA, it is hypothesized that oxidative stress induced by this molecule affects MexR, a redox-sensitive regulator. The oxidation of two cysteines in MexR leads to conformational changes in the protein, hindering its binding to the promoter DNA region [90,91].
Furthermore, in E. coli, it has been demonstrated that compounds with a chlorinated phenol structure can enhance resistance to various antibiotics by repressing ompF in a micF-dependent manner and inducing marRAB, leading to overexpression of acrAB-tolC. This induction likely occurs through interaction with MarR, as the mechanism is not dependent on SoxS and thus does not result from oxidative stress generation [92]. In contrast, paraquat induces acrAB via SoxS in S. enterica [93]. In these bacteria, as observed with bile, co-crystallization of the dequalinium–RamR complex revealed that binding increases the distance between the NTD of the helix–turn–helix motifs in the RamR dimer. The binding of this compound to RamR reduces its DNA-binding affinity, leading to the increased expression of ramA and, subsequently, acrAB [94]. Additionally, in E. coli, treatment with the iron chelator dipyridyl leads to increased transcription of the Rob regulon. The low-activity form of Rob undergoes post-translational conversion to a high-activity form [51,53]. Studies of enterobacteria such as E. coli and S. enterica have shown that responses to different herbicides may vary depending on the species exposed, considering that pre-exposure is not necessary. This suggests that induction due to herbicide exposure occurs more promptly than the interaction of antibiotics with their targets [95].

Table 3.
Biocides that induce RND efflux pumps.
Table 3.
Biocides that induce RND efflux pumps.
| Molecules | Classification | Pumps | Strains | Mechanisms | References |
|---|---|---|---|---|---|
| 2,4-Dichlorophenol | Herbicide precursor | MexAB-OprM | P. aeruginosa | NalC interaction | [87,89] |
| AcrAB-TolC | E. coli | MarR interaction | [96] | ||
| 2,4-Dichlorophenoxyacetic acid | Herbicide | AcrAB-TolC | E. coli | marRAB induction | [92,95] |
| S. enterica | * | [95] | |||
| 2,4,6-Trichlorophenol | Fungicide | MexAB-OprM | P. aeruginosa | NalC interaction | [87,89] |
| 4,4′-Dipyridyl | Degradation of the herbicide paraquat | AcrAB-TolC | E. coli | Rob activation | [51,53] |
| Acriflavine | Antiseptic (fungal infections of aquarium fish) | MexAB-OprM | P. aeruginosa | * | [80] |
| MexCD-OprJ | [80,83] | ||||
| Benzethonium chloride | Cationic surfactant; disinfectant; quaternary ammonium | MexCD-OprJ | P. aeruginosa | Membrane stress (AlgU induction) | [80] |
| Benzalkonium chloride | Cationic surfactant; disinfectant; quaternary ammonium | MexCD-OprJ | P. aeruginosa | Membrane stress (AlgU induction) | [83] |
| Boric acid | Insecticide | SmeYZ | S. maltophilia | Protein synthesis inhibition | [73] |
| Cetylpyridinium chloride | Antiseptic (personal care products); topical anti-infective; pharmaceutical preservative; quaternary ammonium | MexCD-OprJ | P. aeruginosa | Membrane stress (AlgU induction) | [80] |
| SmeYZ | S. maltophilia | * | [73] | ||
| SmeVWX | |||||
| Dicamba | Herbicide | AcrAB-TolC | E. coli | * | [95] |
| S. enterica | |||||
| Dodecyltrimethylammonium bromide | Detergent; surface active agent | SmeVWX | S. maltophilia | * | [73] |
| Dodine | Fungicide | MexCD-OprJ | P. aeruginosa | * | [80] |
| Glyphosate | Herbicide | AcrAB-TolC | E. coli | * | [95] |
| S. enterica | |||||
| Ortho-benzyl-parachlorophenol | Disinfectant | MexAB-OprM | P. aeruginosa | * | [89] |
| Paraquat | Herbicide; quaternary ammonium | AcrAB-TolC | E. coli | MarR interaction | [97] |
| S. enterica | SoxS induction | [93] | |||
| SmeVWX | S. maltophilia | * | [73] | ||
| Pentachlorophenol | Herbicide | MexAB-OprM | P. aeruginosa | NalC interaction Oxydative stress (MexR oxidation) | [80,86,87,88,89] |
| MexJKL | * | [86] | |||
| Poly(hexamethylenebiguanide) hydrochloride | Disinfectant | MexCD-OprJ | P. aeruginosa | Membrane stress (AlgU induction) | [84] |
| Sodium cyanate | Briding agent between reagents in the production of herbicides | MexAB-OprM | P. aeruginosa | * | [80] |
| MexCD-OprJ | |||||
| Sodium metaborate | Herbicide | SmeYZ | S. maltophilia | * | [73] |
| Triclosan | Antiseptic; disinfectant | MexAB-OprM | P. aeruginosa | NalC interaction | [81] |
| SmeDEF | S. maltophilia | SmeT interaction | [87,89] |
* Unknown.
3.4. Drugs
Among the drugs cataloged in Table 4, sodium salicylate’s impact on bacterial resistance to antibiotics, particularly in E. coli, has been the most extensively studied. Sodium salicylate and acetyl salicylic acid belong to the class of non-steroidal anti-inflammatory drugs (NSAIDs), which exhibit antipyretic and anti-platelet aggregation properties. They are employed to alleviate fever, pain, and inflammatory rheumatism and in the prevention of stroke and infarction. Salicylic acid and salicylate represent the primary metabolites of aspirin. In the presence of salicylate, E. coli’s resistance level mirrors that of a mar mutant, conferring resistance to quinolones, cephalosporins, ampicillin, tetracycline, and chloramphenicol [40,97,98,99,100,101,102,103]. At the molecular level, the interaction of salicylate with MarR prevents its binding to marO, which constitutes the operator region [76,104]. The de-repression of the marRAB operon increased MarA production [96,97,103,105,106], subsequently reducing antibiotic accumulation. This occurs due to a decrease in influx caused by increased micF transcription, leading to reduced OmpF levels, and due to an increase in efflux through the induction of acrAB transcription by MarA. Acetaminophen and ibuprofen similarly induce marA and acrB, heightening resistance to ciprofloxacin, nalidixic acid, and tetracycline [98,103]. However, acetaminophen-induced resistance is not totally attributable to marA—as evidenced by elevated MICs in marA-depleted strains—unlike ibuprofen-induced resistance, which is entirely dependent on marA [103]. The involvement of Rob in this induction, as described in previous sections, is hypothesized.
Clofibric acid and ethacrynic acid, employed for hypertriglyceridemia and as diuretic, respectively, share a chlorinated phenoxy structure and increase resistance in uropathogenic E. coli strains to various antibiotics in the same way as aspirin: via micF-dependent ompF repression and marRAB induction [92].
In S. enterica, the co-crystallization of the rhodamine 6G-RamR complex exhibits an interaction with a Kd of 26.4 µM, increasing the distance between the NTD helix–turn–helix motifs in the RamR dimer [94].
Procaine and atropine, used as a local anesthetic and for preoperative sedation, respectively, may affect P. aeruginosa’s sensitivity to antibiotics in surgical patients. Despite differing structures, these drugs, with similar pharmacological properties, induce mexCD-oprJ, thereby enhancing P. aeruginosa’s resistance to ciprofloxacin [80].
A distinct induction mechanism is observed for SmeVWX in S. maltophilia. This mechanism involves the thiol reactivity of inducing compounds. Menadione, sodium selenite, and clioquinol, respectively, react with thiol groups, catalyze the oxidation of thiol groups, and interact with thiol and amino groups. All these compounds enable induction of this efflux pump (starting from 4µM for menadione) and reduce S. maltophilia susceptibility to ofloxacin and chloramphenicol [73,107].

Table 4.
Drugs that induce RND efflux pumps.
Table 4.
Drugs that induce RND efflux pumps.
| Molecules | Classification | Pumps | Strains | Mechanisms | References |
|---|---|---|---|---|---|
| 9′-Aminoacridine | Topical antiseptic (eye drops) | MexAB-OprM | P. aeruginosa | * | [80] |
| MexCD-OprJ | |||||
| Acetaminophen (paracetamol) | Antipyretic; non-narcotic analgesic | AcrAB-TolC | E. coli | marA induction | [97,98,103] |
| Acetyl salicyclic acid (aspirin) | NSAID; antipyretic; analgesic; platelet aggregation inhibitors | AcrAB-TolC | E. coli | marA induction | [98,103] |
| Alexidine | Disinfectant (skin and mucous membrane) | MexCD-OprJ | P. aeruginosa | Membrane stress (AlgU induction) | [80,84] |
| Amitriptyline | Non-narcotic analgesic | MexCD-OprJ | P. aeruginosa | * | [80] |
| Atropine | Anesthetic; adjuvant | MexCD-OprJ | P. aeruginosa | * | [80] |
| Cetrimide 1 | Local antiseptic; quaternary ammonium | MexCD-OprJ | P. aeruginosa | Membrane stress (AlgU induction) | [84] |
| Chlorhexidine | Antiseptic (dermatology and dental) | MexCD-OprJ | P. aeruginosa | Membrane stress (AlgU induction) | [83,84] |
| Chloroxylenol | Topical disinfectant | MexAB-OprM | P. aeruginosa | * | [89] |
| Chlorquinaldol | Antiseptic (dermatology) | SmeVWX | S. maltophilia | * | [73] |
| Clofibric acid | Anticholesteremic | AcrAB-TolC | E. coli | marA induction | [92] |
| Clioquinol | Antifungal and antiprotozoal drug | SmeVWX | S. maltophilia | Thiol reactivity | [73] |
| Diamide | Radiation-sensitizing agent (radiation therapy) | MexAB-OprM | P. aeruginosa | AmgRS activation | [67] |
| Dequalinium chloride | Antiseptic; disinfectant; quaternary ammonium | MexCD-OprJ | P. aeruginosa | Membrane stress (AlgU induction) | [80,85] |
| AcrAB-TolC | S. enterica | RamR interaction | [94] | ||
| SmeYZ | S. maltophilia | * | [73] | ||
| Domiphen bromide | Antiseptic; quaternary ammonium | MexCD-OprJ | P. aeruginosa | Membrane stress (AlgU induction) | [80] |
| Ethacrynic acid | Diuretic | AcrAB-TolC | E. coli | marA induction | [92] |
| Ibuprofen | NSAID; antipyretic; non-narcotic analgesic | AcrAB-TolC | E. coli | marA induction | [103] |
| Menadione | Vitamin K3 | AcrAB-TolC | E. coli | MarR interaction | [96,97] |
| SmeVWX | S. maltophilia | Thiol reactivity | [73,107] | ||
| Orphenadrine | Skeletal muscle relaxant (Parkinson’s) | MexCD-OprJ | P. aeruginosa | * | [80] |
| Plumbagin | Antineoplastic agent (chemotherapy); adjuvant; anticoagulant; contraceptive agent; cardiotonic agent | AcrAB-TolC | E. coli | MarR interaction | [96,97] |
| SmeVWX | S. maltophilia | * | [107] | ||
| Procaine | Local anesthetic | MexCD-OprJ | P. aeruginosa | * | [80] |
| Proflavine | Topical antiseptic; acriflavine derivative | AcrAB-TolC | E. coli | AcrR interaction | [108] |
| MexAB-OprM | P. aeruginosa | * | [80] | ||
| MexCD-OprJ | |||||
| Propanolol | -blocker (hypertension, anxiety, panic attacks, etc.) | MexCD-OprJ | P. aeruginosa | * | [80] |
| Protamine sulfate | Anticoagulant | SmeYZ | S. maltophilia | * | [73] |
| SmeVWX | |||||
| Puromycin | Antineoplastic agent (chemotherapy) | SmeYZ | S. maltophilia | * | [73] |
| Rhodamine 6G | Antineoplastic agent (chemotherapy) | AcrAB-TolC | S. enterica | RamR interaction | [94] |
| E. coli | Rob interaction | [53] | |||
| AcrR interaction | [108] | ||||
| MexCD-OprJ | P. aeruginosa | * | [83,109] | ||
| S-nitrosoglutathione | Nitric oxide donors (asthma, CF 2, embolization prevention, or diabetic leg ulcers) | MexEF-OprN | P. aeruginosa | Nitrosative stress | [68] |
| Sodium salicylate | NSAID; antipyretic; analgesic | CeoAB-OpcM | B. cenocepacia | * | [69] |
| CmeABC | C. jejuni | CmeR interaction | [55] | ||
| AcrAB-TolC | E. coli | MarR interaction | [97,98,99,104] | ||
| S. enterica | [99] | ||||
| Sodium selenite | Phase I clinical trial in terminal cancer patients | SmeVWX | S. maltophilia | Thiol reactivity | [73] |
| Tetraphenylphosphonium chloride | Antineoplastic agent (chemotherapy) | MexCD-OprJ | P. aeruginosa | * | [83,109] |
* Unknown. 1 Tetradonium bromide; cetrinomium bromide; laurtrimonium bromide. 2 Cystic fibrosis.
3.5. Food and Cosmetic Additives
The impact of additives on the induction of antibiotic resistance has been investigated (refer to Table 5). In a 2022 study, non-caloric artificial sweeteners, such as saccharin, sucralose, aspartame, and acesulfame-K, were investigated. Introduced nearly a century ago as sugar substitutes with potent sweetness and low caloric content, these sweeteners have garnered attention. Yu and Guo demonstrated that at a concentration of 300 mg/mL, they can induce the upregulation of acrAB-tolC and increase intracellular ROS and cell envelope permeability in both E. coli and K. pneumoniae [110].
Furthermore, sodium benzoate, commonly known as E211 in the context of food additives, serves as a widely employed preservative in food and cosmetics due to its efficacy against yeast, bacteria, and fungi. It exhibits a lower effect on induction of acrAB-tolC in E. coli, with an induction ratio of 2.3 for 5 mM of sodium benzoate compared to 7.1 for 5 mM of salicylate [97,98].

Table 5.
Additives that induce RND efflux pumps.
Table 5.
Additives that induce RND efflux pumps.
| Molecules | Classification | Pumps | Strains | Mechanisms | References |
|---|---|---|---|---|---|
| Acesulfame potassium | Food additive; artificial sweetener | AcrAB-TolC | E. coli | * | [110] |
| K. pneumoniae | |||||
| Aspartame | Food additive; artificial sweetener | AcrAB-TolC | E. coli | * | [110] |
| K. pneumoniae | |||||
| Saccharin | Food additive; artificial sweetener | AcrAB-TolC | E. coli | * | [110] |
| K. pneumoniae | |||||
| Sucralose | Food additive; artificial sweetener | AcrAB-TolC | E. coli | * | [110] |
| K. pneumoniae | |||||
| Sodium benzoate | Food preservative; antifungal agent | AcrAB-TolC | E. coli | * | [97,98] |
* Unknown.
3.6. Natural Compounds
Essential oils and their constituents are increasingly used due to their potential in combating bacterial infections; however, they have been shown to act counterproductively by inducing a mechanism of resistance to antibiotics (refer to Table 6). Cinnamaldehyde, the main component of cinnamon oil, has exhibited promising antimicrobial properties against various pathogens, including P. aeruginosa [111]. Nevertheless, exposure of P. aeruginosa to subinhibitory concentrations of cinnamaldehyde resulted in a robust yet transient upregulation of operons encoding the MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY-OprM efflux systems. This multifaceted activation led to increased resistance to a range of antibiotics, including meropenem, ceftazidime, tobramycin, and ciprofloxacin, with resistance levels escalating from twofold to eightfold [112,113]. The NalC regulator is implicated in the control of the MexAB-OprM system, where it facilitates the production of the ArmR antirepressor [113]. In the case of MexEF-OprN, electrophilic molecules such as cinnamaldehyde and methylglyoxal activate CmrA, thereby inducing mexS and PA2048. This cascade allows for the accumulation of oxidized products, subsequently activating MexT and leading to the overexpression of mexEF-oprN [112]. Additionally, cinnamate induces acrAB-tolC via the induction of marRAB [98].
Moreover, citral demonstrates induction of mexEF-oprN and mexXY-oprM, enhancing resistance to various antibiotics, including imipenem (2-fold), gentamicin (8-fold), tobramycin (8-fold), ciprofloxacin (2-fold), and colistin (over 128-fold). In this case, efflux is not the only factor involved. Citral also impedes the attachment of aminoglycosides and colistin to the cell surface, and Schiff base formation, which can occur between the aldehyde group of citral and the amine group of tobramycin or colistin, that results in decreased antibiotic activity [114].
As described in the previous sections, the co-crystallization of the berberine–RamR complex revealed that binding increases the distance between the NTD helix–turn–helix motifs in the RamR dimer, with a Kd of 17.9 µM, thereby increasing the expression of ramA and, subsequently, acrAB [94].
Heavy metals and metal cations present in the environment have historically been utilized as antimicrobials. These metals represent a class of natural compounds capable of inducing the expression of RND efflux pumps. While metals are essential as cofactors in numerous bacterial processes, their toxicity at elevated concentrations necessitates that bacteria possess systems for maintaining cellular metal homeostasis. In some instances, this regulation involves efflux pumps that expel these toxic substances from the cell [115]. The CusCBA efflux system, for example, confers bacterial tolerance to copper and silver ions. The expression of cusCBA is naturally induced by these substrates and is regulated by the CusRS two-component system found in various Enterobacteriaceae such as E. coli and K. pneumoniae [116,117]. Similarly, in Helicobacter pylori, the expression of the CrdABC efflux system is induced by copper via the CrdRS two-component system [118]. In addition, CzcABC in P. aeruginosa confers resistance to zinc, cadmium, and cobalt, and its regulation is mediated by the metal-inducible CzcRS two-component system that is activated directly by its specific substrates or indirectly in the presence of copper [119,120]. In some cases, the regulation of efflux systems can serve as an environmental signal reflecting the surrounding ecosystem. For instance, the MtrCDE system in Neisseria gonorrhoeae is indirectly regulated by iron availability. Its expression increases under iron-limited conditions, a scenario that bacteria encounter during host infection [121]. Cross-resistance between heavy metals and antibiotics is an important phenomenon in which exposure to one agent induces resistance mechanisms against others. For example, the mdtABC operon is upregulated in response to excess zinc, conferring resistance to the antibiotic novobiocin [122,123,124]. Additionally, P. aeruginosa isolates exposed to zinc demonstrate resistance not only to cadmium and cobalt but also to the antibiotic imipenem. This cross-resistance reveals a co-regulation mechanism in which imipenem influx is coordinated with heavy metal efflux via the CzcRS two-component system [125]. The interaction between metals and antibiotic resistance involves intricate regulatory networks, often mediated by two-component systems, that allow bacteria to survive in hostile environments by expelling toxic compounds and developing resistance to multiple antimicrobial agents.

Table 6.
Drugs that induce RND efflux pumps.
Table 6.
Drugs that induce RND efflux pumps.
| Molecules | Classification | Pumps | Strains | Mechanisms | References |
|---|---|---|---|---|---|
| Berberine | Food supplement | AcrAB-TolC | S. enterica | RamR interaction | [94] |
| Cadmium | Heavy metal | CzcABC | P. aeruginosa | CzcRS activation | [119,120] |
| Cinnamaldehyde | Component of cinnamon oil | MexAB-OprM | P. aeruginosa | NalC interaction | [112,113] |
| MexCD-OprJ | * | ||||
| MexEF-OprN | |||||
| MexXY-OprM | |||||
| Cinnamate | Component of cinnamon oil | AcrAB-TolC | E. coli | marRAB induction | [98] |
| Citral | Component of many commercial oils (lemon glass, verbena, etc.); flavoring agents and fragrance | MexEF-OprN | P. aeruginosa | * | [114] |
| MexXY-OprM | |||||
| Cobalt | Heavy metal | CzcABC | P. aeruginosa | CzcRS activation | [119,120] |
| Copper | Metal cation | CusCBA | E. coli | CusRS activation | [116,117] |
| K. pneumoniae | |||||
| CrdABC | H. pylori | CrdABC activation | [118] | ||
| CzcABC | P. aeruginosa | CzcRS activation | [119,120] | ||
| Iron | Metal cation | MtrCDE | N. gonorrhoeae | Repression by MpeR of the repressor MtrR | [121] |
| Methylglyoxal | Found in honey and soft drinks | MexEF-OprN | P. aeruginosa | * | [112] |
| Sanguinarine | Natural alkaloid; toothpaste, mouthwash | MexAB-OprM | P. aeruginosa | * | [80] |
| MexCD-OprJ | |||||
| Zinc | Metal cation | CzcABC | P. aeruginosa | CzcRS activation | [119,120,125] |
| MdtABC | E. coli | BaeSR activation | [124] |
* Unknown.
4. Conclusions
The escalating threat posed by MDR pathogenic bacteria to global public health necessitates urgent and concerted efforts to address antibiotic resistance. Understanding the diverse mechanisms employed by bacteria to resist antibiotics, particularly the role of RND efflux pumps, is pivotal in this endeavor.
In this review, we developed a comprehensive insight into the interplay between bile, biocides, pharmaceuticals, and various other compounds, shedding light on their roles in the modulation of RND efflux pump expression in bacterial pathogens. Due to the chemical diversity of the inducing molecules, it is impossible to draw conclusions about structure–activity relationships. Herein lies the subtlety of these efflux pumps: they have a wide range of substrates, and their regulators can interact with a wide range of molecules. The clinical implications of efflux pump induction by non-antibiotic compounds warrant further investigation. The impact of environmental factors, food additives, and pharmaceuticals on the emergence and dissemination of antibiotic resistance poses significant challenges for public health. Therefore, comprehensive surveillance programs are essential to monitor the prevalence and dynamics of efflux pump-mediated resistance in clinical settings and the environment.
The prospects for future research in this field are multifaceted. Firstly, there is a need for deeper mechanistic insights into the regulation of efflux pump expression and the interplay between various regulatory pathways. Understanding how environmental cues and stressors modulate efflux pumps’ activity can inform the development of novel therapeutic interventions to combat antibiotic resistance. Additionally, efforts should be directed towards exploring alternative strategies to target efflux pumps, either through the design of efflux pump inhibitors (EPIs) [126] or through the development of new antimicrobial agents that are less susceptible to efflux-mediated resistance [25]. Finally, the diagnosis of infection by bacteria overexpressing an efflux system needs to be developed and improved as a routine in hospitals and the community [9]. Although antibacterial resistance arises through various mechanisms, the increased active efflux of antibiotics is particularly significant. A single type of efflux pump can confer resistance to multiple drugs simultaneously. Furthermore, the overproduction of efflux pumps in bacteria significantly contributes to the selection of target mutations, both of which culminate in a MDR phenotype [6,7]. Despite the first discovery of efflux pumps over 40 years ago, their clinical significance remains challenging to ascertain. This difficulty primarily arises from the absence of reliable methods for detecting efflux levels in bacterial strains isolated from infected patients or animals. Additionally, the current lack of EPIs on the market diminishes the incentive for clinicians to investigate efflux mechanisms in clinical isolates. RND efflux pumps play a crucial role in antimicrobial resistance. Therefore, assessing the efflux capacity of clinical isolates could substantially improve the management of infections, especially if effective EPIs are available. Unfortunately, as of now, no EPI is undergoing clinical trials. This highlights an urgent need for increased research and development in this area to enhance the fight against MDR pathogens.
In summary, addressing antibiotic resistance requires a multidimensional approach that encompasses understanding the molecular mechanisms of resistance, exploring innovative therapeutic strategies, and implementing robust surveillance measures. By elucidating the intricate interplay between bacterial pathogens and their resistance mechanisms, we can strive towards mitigating the threat of antibiotic resistance and safeguarding the efficacy of antimicrobial treatments for future generations.
Author Contributions
Writing—original draft preparation, M.N. and J.-M.B.; writing—review and editing, M.N. and J.-M.B.; funding acquisition, J.-M.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by l’Agence Nationale de la Recherche (ANR, France), ANR “Spice-Up” ANR-19-CE44-0015-01.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We deeply thank Martin Picard (IBPC Paris) and Jean Michel Brunel (MCT Marseille) for their support and encouragement. We wish to thank Alexis Lodé (IBPC Paris) and Johan Revol-Tissot (BSC Strasbourg) for their careful reading and useful comments on the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Abbreviations
ABC: ATP-binding cassette; CF: Cystic fibrosis; CTD: C-terminal domain; MATE: Multi-drug and toxic compound extrusion; MDR: Multi-drug resistant; MFS: Major facilitator superfamily; MIC: Minimum inhibitory concentration; NAIDs: Non-steroidal anti-inflammatory drugs; NTD: N-terminal domain; OMF: Outer membrane factor; PACE: Proteobacterial antimicrobial compound efflux; PAP: Periplasmic adaptor protein; RND: Resistance–nodulation–cell division; SMR: Small multi-drug resistance.
References
- Antimicrobial Resistance Collaborators. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Tooke, C.L.; Hinchliffe, P.; Bragginton, E.C.; Colenso, C.K.; Hirvonen, V.H.A.; Takebayashi, Y.; Spencer, J. β-Lactamases and β-Lactamase Inhibitors in the 21st Century. J. Mol. Biol. 2019, 431, 3472–3500. [Google Scholar] [CrossRef]
- Schaenzer, A.J.; Wright, G.D. Antibiotic Resistance by Enzymatic Modification of Antibiotic Targets. Trends Mol. Med. 2020, 26, 768–782. [Google Scholar] [CrossRef]
- Masi, M.; Winterhalter, M.; Pagès, J.-M. Outer Membrane Porins. Subcell Biochem. 2019, 92, 79–123. [Google Scholar] [CrossRef]
- Vergalli, J.; Bodrenko, I.V.; Masi, M.; Moynié, L.; Acosta-Gutiérrez, S.; Naismith, J.H.; Davin-Regli, A.; Ceccarelli, M.; van den Berg, B.; Winterhalter, M.; et al. Porins and Small-Molecule Translocation across the Outer Membrane of Gram-Negative Bacteria. Nat. Rev. Microbiol. 2020, 18, 164–176. [Google Scholar] [CrossRef]
- Grimsey, E.M.; Weston, N.; Ricci, V.; Stone, J.W.; Piddock, L.J.V. Overexpression of RamA, Which Regulates Production of the Multidrug Resistance Efflux Pump AcrAB-TolC, Increases Mutation Rate and Influences Drug Resistance Phenotype. Antimicrob. Agents Chemother. 2020, 64, e02460-19. [Google Scholar] [CrossRef]
- El Meouche, I.; Dunlop, M.J. Heterogeneity in Efflux Pump Expression Predisposes Antibiotic-Resistant Cells to Mutation. Science 2018, 362, 686–690. [Google Scholar] [CrossRef]
- Mazzariol, A.; Cornaglia, G.; Nikaido, H. Contributions of the AmpC Beta-Lactamase and the AcrAB Multidrug Efflux System in Intrinsic Resistance of Escherichia coli K-12 to Beta-Lactams. Antimicrob. Agents Chemother. 2000, 44, 1387–1390. [Google Scholar] [CrossRef]
- Ferrand, A.; Vergalli, J.; Bosi, C.; Pantel, A.; Pagès, J.-M.; Davin-Regli, A. Contribution of Efflux and Mutations in Fluoroquinolone Susceptibility in MDR Enterobacterial Isolates: A Quantitative and Molecular Study. J. Antimicrob. Chemother. 2023, 78, 1532–1542. [Google Scholar] [CrossRef]
- Poirel, L.; Madec, J.-Y.; Lupo, A.; Schink, A.-K.; Kieffer, N.; Nordmann, P.; Schwarz, S. Antimicrobial Resistance in Escherichia coli. Microbiol. Spectr. 2018, 6, 10–1128. [Google Scholar] [CrossRef]
- Huang, L.; Wu, C.; Gao, H.; Xu, C.; Dai, M.; Huang, L.; Hao, H.; Wang, X.; Cheng, G. Bacterial Multidrug Efflux Pumps at the Frontline of Antimicrobial Resistance: An Overview. Antibiotics 2022, 11, 520. [Google Scholar] [CrossRef]
- Hernando-Amado, S.; Blanco, P.; Alcalde-Rico, M.; Corona, F.; Reales-Calderón, J.A.; Sánchez, M.B.; Martínez, J.L. Multidrug Efflux Pumps as Main Players in Intrinsic and Acquired Resistance to Antimicrobials. Drug Resist. Updat. 2016, 28, 13–27. [Google Scholar] [CrossRef]
- Bialvaei, A.Z.; Rahbar, M.; Hamidi-Farahani, R.; Asgari, A.; Esmailkhani, A.; Dashti, Y.M.; Soleiman-Meigooni, S. Expression of RND Efflux Pumps Mediated Antibiotic Resistance in Pseudomonas Aeruginosa Clinical Strains. Microb. Pathog. 2021, 153, 104789. [Google Scholar] [CrossRef]
- Puja, H.; Comment, G.; Chassagne, S.; Plésiat, P.; Jeannot, K. Coordinate Overexpression of Two RND Efflux Systems, ParXY and TtgABC, Is Responsible for Multidrug Resistance in Pseudomonas Putida. Environ. Microbiol. 2020, 22, 5222–5231. [Google Scholar] [CrossRef]
- Chang, L.-L.; Chen, H.-F.; Chang, C.-Y.; Lee, T.-M.; Wu, W.-J. Contribution of Integrons, and SmeABC and SmeDEF Efflux Pumps to Multidrug Resistance in Clinical Isolates of Stenotrophomonas Maltophilia. J. Antimicrob. Chemother. 2004, 53, 518–521. [Google Scholar] [CrossRef]
- García-León, G.; Ruiz de Alegría Puig, C.; García de la Fuente, C.; Martínez-Martínez, L.; Martínez, J.L.; Sánchez, M.B. High-Level Quinolone Resistance Is Associated with the Overexpression of smeVWX in Stenotrophomonas Maltophilia Clinical Isolates. Clin. Microbiol. Infect. 2015, 21, 464–467. [Google Scholar] [CrossRef]
- Lin, Y.-T.; Huang, Y.-W.; Chen, S.-J.; Chang, C.-W.; Yang, T.-C. The SmeYZ Efflux Pump of Stenotrophomonas Maltophilia Contributes to Drug Resistance, Virulence-Related Characteristics, and Virulence in Mice. Antimicrob. Agents Chemother. 2015, 59, 4067–4073. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, M.; Yang, W.; Fang, Y.; Wang, G.; Hou, F. A Seventeen-Year Observation of the Antimicrobial Susceptibility of Clinical Campylobacter Jejuni and the Molecular Mechanisms of Erythromycin-Resistant Isolates in Beijing, China. Int. J. Infect. Dis. 2016, 42, 28–33. [Google Scholar] [CrossRef]
- Podnecky, N.L.; Rhodes, K.A.; Schweizer, H.P. Efflux Pump-Mediated Drug Resistance in Burkholderia. Front. Microbiol. 2015, 6, 305. [Google Scholar] [CrossRef]
- Alav, I.; Kobylka, J.; Kuth, M.S.; Pos, K.M.; Picard, M.; Blair, J.M.A.; Bavro, V.N. Structure, Assembly, and Function of Tripartite Efflux and Type 1 Secretion Systems in Gram-Negative Bacteria. Chem. Rev. 2021, 121, 5479–5596. [Google Scholar] [CrossRef]
- Murakami, S.; Nakashima, R.; Yamashita, E.; Matsumoto, T.; Yamaguchi, A. Crystal Structures of a Multidrug Transporter Reveal a Functionally Rotating Mechanism. Nature 2006, 443, 173–179. [Google Scholar] [CrossRef]
- Seeger, M.A.; Schiefner, A.; Eicher, T.; Verrey, F.; Diederichs, K.; Pos, K.M. Structural Asymmetry of AcrB Trimer Suggests a Peristaltic Pump Mechanism. Science 2006, 313, 1295–1298. [Google Scholar] [CrossRef]
- Seeger, M.A.; Diederichs, K.; Eicher, T.; Brandstätter, L.; Schiefner, A.; Verrey, F.; Pos, K.M. The AcrB Efflux Pump: Conformational Cycling and Peristalsis Lead to Multidrug Resistance. Curr. Drug Targets 2008, 9, 729–749. [Google Scholar] [CrossRef]
- Takatsuka, Y.; Nikaido, H. Covalently Linked Trimer of the AcrB Multidrug Efflux Pump Provides Support for the Functional Rotating Mechanism. J. Bacteriol. 2009, 191, 1729–1737. [Google Scholar] [CrossRef]
- Revol-Tissot, J.; Boyer, G.; Alibert, S. Molecular Determinant Deciphering of MIC-Guided RND Efflux Substrates in E. Coli. Front. Drug Discov. 2024, 4, 1326121. [Google Scholar] [CrossRef]
- Blair, J.M.A.; Richmond, G.E.; Piddock, L.J.V. Multidrug Efflux Pumps in Gram-Negative Bacteria and Their Role in Antibiotic Resistance. Future Microbiol. 2014, 9, 1165–1177. [Google Scholar] [CrossRef]
- Ferrand, A.; Vergalli, J.; Pagès, J.-M.; Davin-Regli, A. An Intertwined Network of Regulation Controls Membrane Permeability Including Drug Influx and Efflux in Enterobacteriaceae. Microorganisms 2020, 8, 833. [Google Scholar] [CrossRef]
- Du, D.; Wang-Kan, X.; Neuberger, A.; van Veen, H.W.; Pos, K.M.; Piddock, L.J.V.; Luisi, B.F. Multidrug Efflux Pumps: Structure, Function and Regulation. Nat. Rev. Microbiol. 2018, 16, 523–539. [Google Scholar] [CrossRef]
- Colclough, A.L.; Scadden, J.; Blair, J.M.A. TetR-Family Transcription Factors in Gram-Negative Bacteria: Conservation, Variation and Implications for Efflux-Mediated Antimicrobial Resistance. BMC Genom. 2019, 20, 731. [Google Scholar] [CrossRef]
- Harmon, D.E.; Ruiz, C. The Multidrug Efflux Regulator AcrR of Escherichia coli Responds to Exogenous and Endogenous Ligands To Regulate Efflux and Detoxification. mSphere 2022, 7, e0047422. [Google Scholar] [CrossRef]
- Grinnage-Pulley, T.; Mu, Y.; Dai, L.; Zhang, Q. Dual Repression of the Multidrug Efflux Pump CmeABC by CosR and CmeR in Campylobacter Jejuni. Front. Microbiol. 2016, 7, 1097. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, P.; Alonso, A.; Martinez, J.L. Regulatory Regions of smeDEF in Stenotrophomonas Maltophilia Strains Expressing Different Amounts of the Multidrug Efflux Pump SmeDEF. Antimicrob. Agents Chemother. 2004, 48, 2274–2276. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, P.; Rojo, F.; Martínez, J.L. Transcriptional Regulation of mexR, the Repressor of Pseudomonas Aeruginosa mexAB-oprM Multidrug Efflux Pump. FEMS Microbiol. Lett. 2002, 207, 63–68. [Google Scholar] [CrossRef]
- Duque, E.; Segura, A.; Mosqueda, G.; Ramos, J.L. Global and Cognate Regulators Control the Expression of the Organic Solvent Efflux Pumps TtgABC and TtgDEF of Pseudomonas Putida. Mol. Microbiol. 2001, 39, 1100–1106. [Google Scholar] [CrossRef]
- Morita, Y.; Cao, L.; Gould, V.C.; Avison, M.B.; Poole, K. nalD Encodes a Second Repressor of the mexAB-oprM Multidrug Efflux Operon of Pseudomonas Aeruginosa. J. Bacteriol. 2006, 188, 8649–8654. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Srikumar, R.; Poole, K. MexAB-OprM Hyperexpression in NalC-Type Multidrug-Resistant Pseudomonas Aeruginosa: Identification and Characterization of the nalC Gene Encoding a Repressor of PA3720-PA3719. Mol. Microbiol. 2004, 53, 1423–1436. [Google Scholar] [CrossRef]
- Daigle, D.M.; Cao, L.; Fraud, S.; Wilke, M.S.; Pacey, A.; Klinoski, R.; Strynadka, N.C.; Dean, C.R.; Poole, K. Protein Modulator of Multidrug Efflux Gene Expression in Pseudomonas Aeruginosa. J. Bacteriol. 2007, 189, 5441–5451. [Google Scholar] [CrossRef]
- Wilke, M.S.; Heller, M.; Creagh, A.L.; Haynes, C.A.; McIntosh, L.P.; Poole, K.; Strynadka, N.C.J. The Crystal Structure of MexR from Pseudomonas Aeruginosa in Complex with Its Antirepressor ArmR. Proc. Natl. Acad. Sci. USA 2008, 105, 14832–14837. [Google Scholar] [CrossRef]
- Köhler, T.; Epp, S.F.; Curty, L.K.; Pechère, J.C. Characterization of MexT, the Regulator of the MexE-MexF-OprN Multidrug Efflux System of Pseudomonas Aeruginosa. J. Bacteriol. 1999, 181, 6300–6305. [Google Scholar] [CrossRef]
- Duval, V.; Lister, I.M. MarA, SoxS and Rob of Escherichia coli—Global Regulators of Multidrug Resistance, Virulence and Stress Response. Int. J. Biotechnol. Wellness Ind. 2013, 2, 101–124. [Google Scholar] [CrossRef]
- De Gaetano, G.V.; Lentini, G.; Famà, A.; Coppolino, F.; Beninati, C. Antimicrobial Resistance: Two-Component Regulatory Systems and Multidrug Efflux Pumps. Antibiotics 2023, 12, 965. [Google Scholar] [CrossRef] [PubMed]
- Bhagirath, A.Y.; Li, Y.; Patidar, R.; Yerex, K.; Ma, X.; Kumar, A.; Duan, K. Two Component Regulatory Systems and Antibiotic Resistance in Gram-Negative Pathogens. Int. J. Mol. Sci. 2019, 20, 1781. [Google Scholar] [CrossRef] [PubMed]
- Tierney, A.R.; Rather, P.N. Roles of Two-Component Regulatory Systems in Antibiotic Resistance. Future Microbiol. 2019, 14, 533–552. [Google Scholar] [CrossRef] [PubMed]
- Lau, C.H.-F.; Fraud, S.; Jones, M.; Peterson, S.N.; Poole, K. Mutational Activation of the AmgRS Two-Component System in Aminoglycoside-Resistant Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 2013, 57, 2243–2251. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, E.; Yamaguchi, A.; Nishino, K. AcrAB Multidrug Efflux Pump Regulation in Salmonella enterica Serovar Typhimurium by RamA in Response to Environmental Signals. J. Biol. Chem. 2008, 283, 24245–24253. [Google Scholar] [CrossRef] [PubMed]
- Prouty, A.M.; Brodsky, I.E.; Falkow, S.; Gunn, J.S. Bile-Salt-Mediated Induction of Antimicrobial and Bile Resistance in Salmonella Typhimurium. Microbiology 2004, 150, 775–783. [Google Scholar] [CrossRef] [PubMed]
- Baucheron, S.; Nishino, K.; Monchaux, I.; Canepa, S.; Maurel, M.-C.; Coste, F.; Roussel, A.; Cloeckaert, A.; Giraud, E. Bile-Mediated Activation of the acrAB and tolC Multidrug Efflux Genes Occurs Mainly through Transcriptional Derepression of ramA in Salmonella enterica Serovar Typhimurium. J. Antimicrob. Chemother. 2014, 69, 2400–2406. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, S.; Nakashima, R.; Sakurai, K.; Baucheron, S.; Giraud, E.; Doublet, B.; Cloeckaert, A.; Nishino, K. Crystal Structure of the Multidrug Resistance Regulator RamR Complexed with Bile Acids. Sci. Rep. 2019, 9, 177. [Google Scholar] [CrossRef] [PubMed]
- Kus, J.V.; Gebremedhin, A.; Dang, V.; Tran, S.-L.; Serbanescu, A.; Barnett Foster, D. Bile Salts Induce Resistance to Polymyxin in Enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 2011, 193, 4509–4515. [Google Scholar] [CrossRef]
- Ma, D.; Cook, D.N.; Alberti, M.; Pon, N.G.; Nikaido, H.; Hearst, J.E. Genes acrA and acrB Encode a Stress-Induced Efflux System of Escherichia coli. Mol. Microbiol. 1995, 16, 45–55. [Google Scholar] [CrossRef]
- Rosner, J.L.; Dangi, B.; Gronenborn, A.M.; Martin, R.G. Posttranscriptional Activation of the Transcriptional Activator Rob by Dipyridyl in Escherichia coli. J. Bacteriol. 2002, 184, 1407–1416. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, E.Y.; Bertenthal, D.; Nilles, M.L.; Bertrand, K.P.; Nikaido, H. Bile Salts and Fatty Acids Induce the Expression of Escherichia coli AcrAB Multidrug Efflux Pump through Their Interaction with Rob Regulatory Protein. Mol. Microbiol. 2003, 48, 1609–1619. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Wang, F.; Li, F.; Wang, L.; Xiong, Y.; Wen, A.; Jin, Y.; Jin, S.; Gao, F.; Feng, Z.; et al. Structural Basis of Transcription Activation by Rob, a Pleiotropic AraC/XylS Family Regulator. Nucleic Acids Res. 2022, 50, 5974–5987. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Cagliero, C.; Guo, B.; Barton, Y.-W.; Maurel, M.-C.; Payot, S.; Zhang, Q. Bile Salts Modulate Expression of the CmeABC Multidrug Efflux Pump in Campylobacter Jejuni. J. Bacteriol. 2005, 187, 7417–7424. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Pu, X.-Y.; Zhang, Q. Salicylate Functions as an Efflux Pump Inducer and Promotes the Emergence of Fluoroquinolone-Resistant Campylobacter Jejuni Mutants. Appl. Environ. Microbiol. 2011, 77, 7128–7133. [Google Scholar] [CrossRef] [PubMed]
- Lei, H.-T.; Shen, Z.; Surana, P.; Routh, M.D.; Su, C.-C.; Zhang, Q.; Yu, E.W. Crystal Structures of CmeR-Bile Acid Complexes from Campylobacter Jejuni. Protein. Sci. 2011, 20, 712–723. [Google Scholar] [CrossRef] [PubMed]
- Benson, V.S.; Müllerová, H.; Vestbo, J.; Wedzicha, J.A.; Patel, A.; Hurst, J.R. Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints (ECLIPSE) Investigators Associations between Gastro-Oesophageal Reflux, Its Management and Exacerbations of Chronic Obstructive Pulmonary Disease. Respir. Med. 2015, 109, 1147–1154. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rosen, R.; Lurie, M.; Kane, M.; DiFilippo, C.; Cohen, A.; Freiberger, D.; Boyer, D.; Visner, G.; Narvaez-Rivas, M.; Liu, E.; et al. Risk Factors for Bile Aspiration and Its Impact on Clinical Outcomes. Clin. Transl. Gastroenterol. 2021, 12, e00434. [Google Scholar] [CrossRef] [PubMed]
- Reen, F.J.; Woods, D.F.; Mooij, M.J.; Adams, C.; O’Gara, F. Respiratory Pathogens Adopt a Chronic Lifestyle in Response to Bile. PLoS ONE 2012, 7, e45978. [Google Scholar] [CrossRef]
- Reen, F.J.; Woods, D.F.; Mooij, M.J.; Chróinín, M.N.; Mullane, D.; Zhou, L.; Quille, J.; Fitzpatrick, D.; Glennon, J.D.; McGlacken, G.P.; et al. Aspirated Bile: A Major Host Trigger Modulating Respiratory Pathogen Colonisation in Cystic Fibrosis Patients. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 1763–1771. [Google Scholar] [CrossRef]
- Reen, F.J.; Flynn, S.; Woods, D.F.; Dunphy, N.; Chróinín, M.N.; Mullane, D.; Stick, S.; Adams, C.; O’Gara, F. Bile Signalling Promotes Chronic Respiratory Infections and Antibiotic Tolerance. Sci. Rep. 2016, 6, 29768. [Google Scholar] [CrossRef] [PubMed]
- Terán, W.; Felipe, A.; Segura, A.; Rojas, A.; Ramos, J.-L.; Gallegos, M.-T. Antibiotic-Dependent Induction of Pseudomonas Putida DOT-T1E TtgABC Efflux Pump Is Mediated by the Drug Binding Repressor TtgR. Antimicrob. Agents Chemother. 2003, 47, 3067–3072. [Google Scholar] [CrossRef] [PubMed]
- Daniels, C.; Daddaoua, A.; Lu, D.; Zhang, X.; Ramos, J.-L. Domain Cross-Talk during Effector Binding to the Multidrug Binding TTGR Regulator. J. Biol. Chem. 2010, 285, 21372–21381. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Escamilla, A.M.; Fernandez-Ballester, G.; Morel, B.; Casares-Atienza, S.; Ramos, J.L. Molecular Binding Mechanism of TtgR Repressor to Antibiotics and Antimicrobials. PLoS ONE 2015, 10, e0138469. [Google Scholar] [CrossRef] [PubMed]
- Alguel, Y.; Meng, C.; Terán, W.; Krell, T.; Ramos, J.L.; Gallegos, M.-T.; Zhang, X. Crystal Structures of Multidrug Binding Protein TtgR in Complex with Antibiotics and Plant Antimicrobials. J. Mol. Biol. 2007, 369, 829–840. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Wang, D.; Zhou, W.; Sang, H.; Liu, X.; Ge, Z.; Zhang, J.; Lan, L.; Yang, C.-G.; Chen, H. Novobiocin Binding to NalD Induces the Expression of the MexAB-OprM Pump in Pseudomonas Aeruginosa. Mol. Microbiol. 2016, 100, 749–758. [Google Scholar] [CrossRef]
- Fruci, M.; Poole, K. Aminoglycoside-Inducible Expression of the mexAB-oprM Multidrug Efflux Operon in Pseudomonas Aeruginosa: Involvement of the Envelope Stress-Responsive AmgRS Two-Component System. PLoS ONE 2018, 13, e0205036. [Google Scholar] [CrossRef] [PubMed]
- Fetar, H.; Gilmour, C.; Klinoski, R.; Daigle, D.M.; Dean, C.R.; Poole, K. mexEF-oprN Multidrug Efflux Operon of Pseudomonas Aeruginosa: Regulation by the MexT Activator in Response to Nitrosative Stress and Chloramphenicol. Antimicrob. Agents Chemother. 2011, 55, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Nair, B.M.; Cheung, K.-J.; Griffith, A.; Burns, J.L. Salicylate Induces an Antibiotic Efflux Pump in Burkholderia Cepacia Complex Genomovar III (B. Cenocepacia). J. Clin. Investig. 2004, 113, 464–473. [Google Scholar] [CrossRef]
- Masuda, N.; Sakagawa, E.; Ohya, S.; Gotoh, N.; Tsujimoto, H.; Nishino, T. Contribution of the MexX-MexY-oprM Efflux System to Intrinsic Resistance in Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 2000, 44, 2242–2246. [Google Scholar] [CrossRef]
- Jeannot, K.; Sobel, M.L.; El Garch, F.; Poole, K.; Plésiat, P. Induction of the MexXY Efflux Pump in Pseudomonas Aeruginosa Is Dependent on Drug-Ribosome Interaction. J. Bacteriol. 2005, 187, 5341–5346. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Sobel, M.L.; Poole, K. Antibiotic Inducibility of the MexXY Multidrug Efflux System of Pseudomonas Aeruginosa: Involvement of the Antibiotic-Inducible PA5471 Gene Product. J. Bacteriol. 2006, 188, 1847–1855. [Google Scholar] [CrossRef] [PubMed]
- Blanco, P.; Corona, F.; Martínez, J.L. Biolog Phenotype Microarray Is a Tool for the Identification of Multidrug Resistance Efflux Pump Inducers. Antimicrob. Agents Chemother. 2018, 62, e01263-18. [Google Scholar] [CrossRef]
- Hächler, H.; Cohen, S.P.; Levy, S.B. marA, a Regulated Locus Which Controls Expression of Chromosomal Multiple Antibiotic Resistance in Escherichia coli. J. Bacteriol. 1991, 173, 5532–5538. [Google Scholar] [CrossRef] [PubMed]
- Viveiros, M.; Jesus, A.; Brito, M.; Leandro, C.; Martins, M.; Ordway, D.; Molnar, A.M.; Molnar, J.; Amaral, L. Inducement and Reversal of Tetracycline Resistance in Escherichia coli K-12 and Expression of Proton Gradient-Dependent Multidrug Efflux Pump Genes. Antimicrob. Agents Chemother. 2005, 49, 3578–3582. [Google Scholar] [CrossRef]
- Alekshun, M.N.; Levy, S.B.; Mealy, T.R.; Seaton, B.A.; Head, J.F. The Crystal Structure of MarR, a Regulator of Multiple Antibiotic Resistance, at 2.3 A Resolution. Nat. Struct. Biol. 2001, 8, 710–714. [Google Scholar] [CrossRef]
- Lopez, P.J.; Marchand, I.; Yarchuk, O.; Dreyfus, M. Translation Inhibitors Stabilize Escherichia coli mRNAs Independently of Ribosome Protection. Proc. Natl. Acad. Sci. USA 1998, 95, 6067–6072. [Google Scholar] [CrossRef] [PubMed]
- Hsueh, P.-R.; Hoban, D.J.; Carmeli, Y.; Chen, S.-Y.; Desikan, S.; Alejandria, M.; Ko, W.-C.; Binh, T.Q. Consensus Review of the Epidemiology and Appropriate Antimicrobial Therapy of Complicated Urinary Tract Infections in Asia-Pacific Region. J. Infect. 2011, 63, 114–123. [Google Scholar] [CrossRef]
- Chetri, S.; Das, B.J.; Bhowmik, D.; Chanda, D.D.; Chakravarty, A.; Bhattacharjee, A. Transcriptional Response of Mar, Sox and Rob Regulon against Concentration Gradient Carbapenem Stress within Escherichia coli Isolated from Hospital Acquired Infection. BMC Res. Notes 2020, 13, 168. [Google Scholar] [CrossRef]
- Laborda, P.; Alcalde-Rico, M.; Blanco, P.; Martínez, J.L.; Hernando-Amado, S. Novel Inducers of the Expression of Multidrug Efflux Pumps That Trigger Pseudomonas Aeruginosa Transient Antibiotic Resistance. Antimicrob. Agents Chemother. 2019, 63, e01095-19. [Google Scholar] [CrossRef]
- Hernández, A.; Ruiz, F.M.; Romero, A.; Martínez, J.L. The Binding of Triclosan to SmeT, the Repressor of the Multidrug Efflux Pump SmeDEF, Induces Antibiotic Resistance in Stenotrophomonas Maltophilia. PLoS Pathog. 2011, 7, e1002103. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, P.; Moore, L.E. Cationic Antiseptics: Diversity of Action under a Common Epithet. J. Appl. Microbiol. 2005, 99, 703–715. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Murata, T.; Mima, T.; Shiota, S.; Kuroda, T.; Mizushima, T.; Gotoh, N.; Nishino, T.; Tsuchiya, T. Induction of mexCD-oprJ Operon for a Multidrug Efflux Pump by Disinfectants in Wild-Type Pseudomonas Aeruginosa PAO1. J. Antimicrob. Chemother. 2003, 51, 991–994. [Google Scholar] [CrossRef] [PubMed]
- Fraud, S.; Campigotto, A.J.; Chen, Z.; Poole, K. MexCD-OprJ Multidrug Efflux System of Pseudomonas Aeruginosa: Involvement in Chlorhexidine Resistance and Induction by Membrane-Damaging Agents Dependent upon the AlgU Stress Response Sigma Factor. Antimicrob. Agents Chemother. 2008, 52, 4478–4482. [Google Scholar] [CrossRef] [PubMed]
- Hernando-Amado, S.; Laborda, P.; Martínez, J.L. Tackling Antibiotic Resistance by Inducing Transient and Robust Collateral Sensitivity. Nat. Commun. 2023, 14, 1723. [Google Scholar] [CrossRef] [PubMed]
- Muller, J.F.; Stevens, A.M.; Craig, J.; Love, N.G. Transcriptome Analysis Reveals That Multidrug Efflux Genes Are Upregulated to Protect Pseudomonas Aeruginosa from Pentachlorophenol Stress. Appl. Environ. Microbiol. 2007, 73, 4550–4558. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Cremers, C.M.; Jakob, U.; Love, N.G. Chlorinated Phenols Control the Expression of the Multidrug Resistance Efflux Pump MexAB-OprM in Pseudomonas Aeruginosa by Interacting with NalC. Mol. Microbiol. 2011, 79, 1547–1556. [Google Scholar] [CrossRef] [PubMed]
- Starr, L.M.; Fruci, M.; Poole, K. Pentachlorophenol Induction of the Pseudomonas Aeruginosa mexAB-oprM Efflux Operon: Involvement of Repressors NalC and MexR and the Antirepressor ArmR. PLoS ONE 2012, 7, e32684. [Google Scholar] [CrossRef] [PubMed]
- Muller, J.F.; Ghosh, S.; Ikuma, K.; Stevens, A.M.; Love, N.G. Chlorinated Phenol-Induced Physiological Antibiotic Resistance in Pseudomonas Aeruginosa. FEMS Microbiol. Lett. 2015, 362, fnv172. [Google Scholar] [CrossRef]
- Chen, H.; Hu, J.; Chen, P.R.; Lan, L.; Li, Z.; Hicks, L.M.; Dinner, A.R.; He, C. The Pseudomonas Aeruginosa Multidrug Efflux Regulator MexR Uses an Oxidation-Sensing Mechanism. Proc. Natl. Acad. Sci. USA 2008, 105, 13586–13591. [Google Scholar] [CrossRef]
- Chen, H.; Yi, C.; Zhang, J.; Zhang, W.; Ge, Z.; Yang, C.-G.; He, C. Structural Insight into the Oxidation-Sensing Mechanism of the Antibiotic Resistance of Regulator MexR. EMBO Rep. 2010, 11, 685–690. [Google Scholar] [CrossRef] [PubMed]
- Balagué, C.; Véscovi, E.G. Activation of Multiple Antibiotic Resistance in Uropathogenic Escherichia coli Strains by Aryloxoalcanoic Acid Compounds. Antimicrob. Agents Chemother. 2001, 45, 1815–1822. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, E.; Shirosaka, I.; Yamaguchi, A.; Nishino, K. Regulation of the AcrAB Multidrug Efflux Pump in Salmonella enterica Serovar Typhimurium in Response to Indole and Paraquat. Microbiology 2011, 157, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, S.; Nikaido, E.; Nakashima, R.; Sakurai, K.; Fujiwara, D.; Fujii, I.; Nishino, K. The Crystal Structure of Multidrug-Resistance Regulator RamR with Multiple Drugs. Nat. Commun. 2013, 4, 2078. [Google Scholar] [CrossRef]
- Kurenbach, B.; Marjoshi, D.; Amábile-Cuevas, C.F.; Ferguson, G.C.; Godsoe, W.; Gibson, P.; Heinemann, J.A. Sublethal Exposure to Commercial Formulations of the Herbicides Dicamba, 2,4-Dichlorophenoxyacetic Acid, and Glyphosate Cause Changes in Antibiotic Susceptibility in Escherichia coli and Salmonella enterica Serovar Typhimurium. mBio 2015, 6, e00009-15. [Google Scholar] [CrossRef]
- Alekshun, M.N.; Levy, S.B. Alteration of the Repressor Activity of MarR, the Negative Regulator of the Escherichia coli marRAB Locus, by Multiple Chemicals in Vitro. J. Bacteriol. 1999, 181, 4669–4672. [Google Scholar] [CrossRef]
- Seoane, A.S.; Levy, S.B. Characterization of MarR, the Repressor of the Multiple Antibiotic Resistance (Mar) Operon in Escherichia coli. J. Bacteriol. 1995, 177, 3414–3419. [Google Scholar] [CrossRef]
- Cohen, S.P.; Levy, S.B.; Foulds, J.; Rosner, J.L. Salicylate Induction of Antibiotic Resistance in Escherichia coli: Activation of the Mar Operon and a Mar-Independent Pathway. J. Bacteriol. 1993, 175, 7856–7862. [Google Scholar] [CrossRef]
- Sulavik, M.C.; Dazer, M.; Miller, P.F. The Salmonella Typhimurium Mar Locus: Molecular and Genetic Analyses and Assessment of Its Role in Virulence. J. Bacteriol. 1997, 179, 1857–1866. [Google Scholar] [CrossRef]
- Randall, L.P.; Woodward, M.J. Multiple Antibiotic Resistance (Mar) Locus in Salmonella enterica Serovar Typhimurium DT104. Appl. Environ. Microbiol. 2001, 67, 1190–1197. [Google Scholar] [CrossRef]
- Chubiz, L.M.; Rao, C.V. Aromatic Acid Metabolites of Escherichia coli K-12 Can Induce the marRAB Operon. J. Bacteriol. 2010, 192, 4786–4789. [Google Scholar] [CrossRef] [PubMed]
- Hartog, E.; Menashe, O.; Kler, E.; Yaron, S. Salicylate Reduces the Antimicrobial Activity of Ciprofloxacin against Extracellular Salmonella enterica Serovar Typhimurium, but Not against Salmonella in Macrophages. J. Antimicrob. Chemother. 2010, 65, 888–896. [Google Scholar] [CrossRef] [PubMed]
- Verma, T.; Bhaskarla, C.; Sadhir, I.; Sreedharan, S.; Nandi, D. Non-Steroidal Anti-Inflammatory Drugs, Acetaminophen and Ibuprofen, Induce Phenotypic Antibiotic Resistance in Escherichia coli: Roles of marA and acrB. FEMS Microbiol. Lett. 2018, 365, fny251. [Google Scholar] [CrossRef] [PubMed]
- Duval, V.; McMurry, L.M.; Foster, K.; Head, J.F.; Levy, S.B. Mutational Analysis of the Multiple-Antibiotic Resistance Regulator MarR Reveals a Ligand Binding Pocket at the Interface between the Dimerization and DNA Binding Domains. J. Bacteriol. 2013, 195, 3341–3351. [Google Scholar] [CrossRef]
- Martin, R.G.; Rosner, J.L. Binding of Purified Multiple Antibiotic-Resistance Repressor Protein (MarR) to Mar Operator Sequences. Proc. Natl. Acad. Sci. USA 1995, 92, 5456–5460. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Kunze, C.; Dunlop, M.J. Salicylate Increases Fitness Cost Associated with MarA-Mediated Antibiotic Resistance. Biophys. J. 2019, 117, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Blanco, P.; Corona, F.; Sánchez, M.B.; Martínez, J.L. Vitamin K3 Induces the Expression of the Stenotrophomonas Maltophilia SmeVWX Multidrug Efflux Pump. Antimicrob. Agents Chemother. 2017, 61, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Su, C.-C.; Rutherford, D.J.; Yu, E.W. Characterization of the Multidrug Efflux Regulator AcrR from Escherichia coli. Biochem. Biophys. Res. Commun. 2007, 361, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Komori, Y.; Mima, T.; Kuroda, T.; Mizushima, T.; Tsuchiya, T. Construction of a Series of Mutants Lacking All of the Four Major Mex Operons for Multidrug Efflux Pumps or Possessing Each One of the Operons from Pseudomonas Aeruginosa PAO1: MexCD-OprJ Is an Inducible Pump. FEMS Microbiol. Lett. 2001, 202, 139–143. [Google Scholar] [CrossRef]
- Yu, Z.; Guo, J. Non-Caloric Artificial Sweeteners Exhibit Antimicrobial Activity against Bacteria and Promote Bacterial Evolution of Antibiotic Tolerance. J. Hazard. Mater. 2022, 433, 128840. [Google Scholar] [CrossRef]
- Elcocks, E.R.; Spencer-Phillips, P.T.N.; Adukwu, E.C. Rapid Bactericidal Effect of Cinnamon Bark Essential Oil against Pseudomonas Aeruginosa. J. Appl. Microbiol. 2020, 128, 1025–1037. [Google Scholar] [CrossRef] [PubMed]
- Juarez, P.; Jeannot, K.; Plésiat, P.; Llanes, C. Toxic Electrophiles Induce Expression of the Multidrug Efflux Pump MexEF-OprN in Pseudomonas Aeruginosa through a Novel Transcriptional Regulator, CmrA. Antimicrob. Agents Chemother. 2017, 61, e00585-17. [Google Scholar] [CrossRef] [PubMed]
- Tetard, A.; Zedet, A.; Girard, C.; Plésiat, P.; Llanes, C. Cinnamaldehyde Induces Expression of Efflux Pumps and Multidrug Resistance in Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 2019, 63, e01081-19. [Google Scholar] [CrossRef] [PubMed]
- Tetard, A.; Foley, S.; Mislin, G.L.A.; Brunel, J.-M.; Oliva, E.; Torrealba Anzola, F.; Zedet, A.; Cardey, B.; Pellequer, Y.; Ramseyer, C.; et al. Negative Impact of Citral on Susceptibility of Pseudomonas Aeruginosa to Antibiotics. Front. Microbiol. 2021, 12, 709838. [Google Scholar] [CrossRef] [PubMed]
- Nies, D.H. Efflux-Mediated Heavy Metal Resistance in Prokaryotes. FEMS Microbiol. Rev. 2003, 27, 313–339. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Ishihama, A. Transcriptional Response of Escherichia coli to External Copper. Mol. Microbiol. 2005, 56, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Pontel, L.B.; Soncini, F.C. Alternative Periplasmic Copper-Resistance Mechanisms in Gram Negative Bacteria. Mol. Microbiol. 2009, 73, 212–225. [Google Scholar] [CrossRef] [PubMed]
- Waidner, B.; Melchers, K.; Stähler, F.N.; Kist, M.; Bereswill, S. The Helicobacter Pylori CrdRS Two-Component Regulation System (HP1364/HP1365) Is Required for Copper-Mediated Induction of the Copper Resistance Determinant CrdA. J. Bacteriol. 2005, 187, 4683–4688. [Google Scholar] [CrossRef] [PubMed]
- Caille, O.; Rossier, C.; Perron, K. A Copper-Activated Two-Component System Interacts with Zinc and Imipenem Resistance in Pseudomonas Aeruginosa. J. Bacteriol. 2007, 189, 4561–4568. [Google Scholar] [CrossRef]
- Dieppois, G.; Ducret, V.; Caille, O.; Perron, K. The Transcriptional Regulator CzcR Modulates Antibiotic Resistance and Quorum Sensing in Pseudomonas Aeruginosa. PLoS ONE 2012, 7, e38148. [Google Scholar] [CrossRef]
- Mercante, A.D.; Jackson, L.; Johnson, P.J.T.; Stringer, V.A.; Dyer, D.W.; Shafer, W.M. MpeR Regulates the Mtr Efflux Locus in Neisseria Gonorrhoeae and Modulates Antimicrobial Resistance by an Iron-Responsive Mechanism. Antimicrob. Agents Chemother. 2012, 56, 1491–1501. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.; Mao, W.; Warren, M.S.; Mistry, A.; Hoshino, K.; Okumura, R.; Ishida, H.; Lomovskaya, O. Interplay between Efflux Pumps May Provide Either Additive or Multiplicative Effects on Drug Resistance. J. Bacteriol. 2000, 182, 3142–3150. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.J.; Barrett, J.A.; Poole, R.K. Genome-Wide Transcriptional Response of Chemostat-Cultured Escherichia coli to Zinc. J. Bacteriol. 2005, 187, 1124–1134. [Google Scholar] [CrossRef] [PubMed]
- Baranova, N.; Nikaido, H. The baeSR Two-Component Regulatory System Activates Transcription of the yegMNOB (mdtABCD) Transporter Gene Cluster in Escherichia coli and Increases Its Resistance to Novobiocin and Deoxycholate. J. Bacteriol. 2002, 184, 4168–4176. [Google Scholar] [CrossRef]
- Perron, K.; Caille, O.; Rossier, C.; van Delden, C.; Dumas, J.-L.; Köhler, T. CzcR-CzcS, a Two-Component System Involved in Heavy Metal and Carbapenem Resistance in Pseudomonas Aeruginosa. J. Biol. Chem. 2004, 279, 8761–8768. [Google Scholar] [CrossRef]
- Compagne, N.; Vieira Da Cruz, A.; Müller, R.T.; Hartkoorn, R.C.; Flipo, M.; Pos, K.M. Update on the Discovery of Efflux Pump Inhibitors against Critical Priority Gram-Negative Bacteria. Antibiotics 2023, 12, 180. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).