Broad-Spectrum Antibacterial Activity of Antioxidant Octyl Gallate and Its Impact on Gut Microbiome
Abstract
:1. Introduction
2. Results
2.1. OG Possesses Potent Antibacterial Activity against Gram-Positive Bacteria
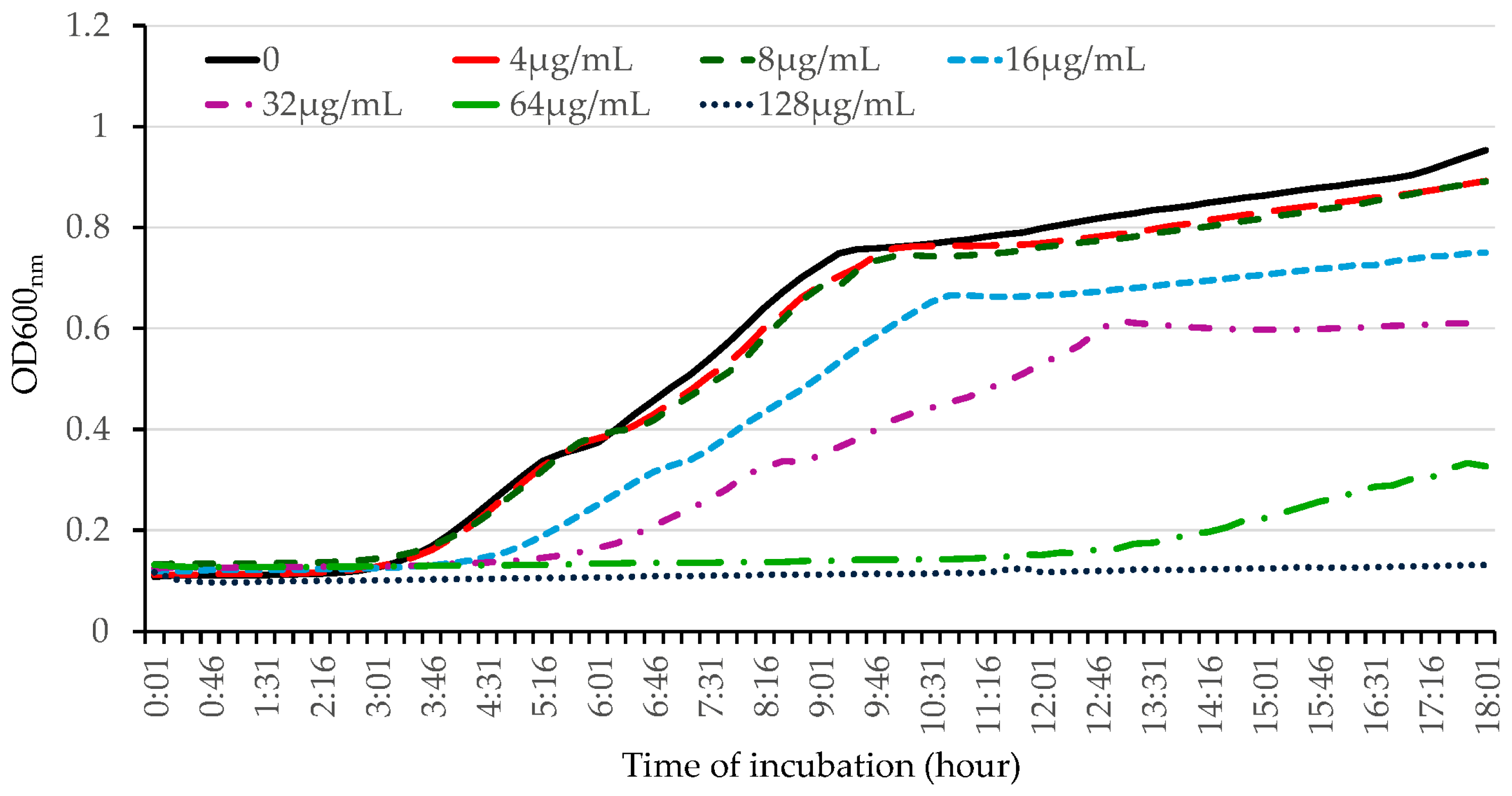
2.2. OG Possesses Robust Bactericidal Activity against HA-MRSA
2.3. OG Enhances the Cell Permeability of HA-MRSA and E. coli
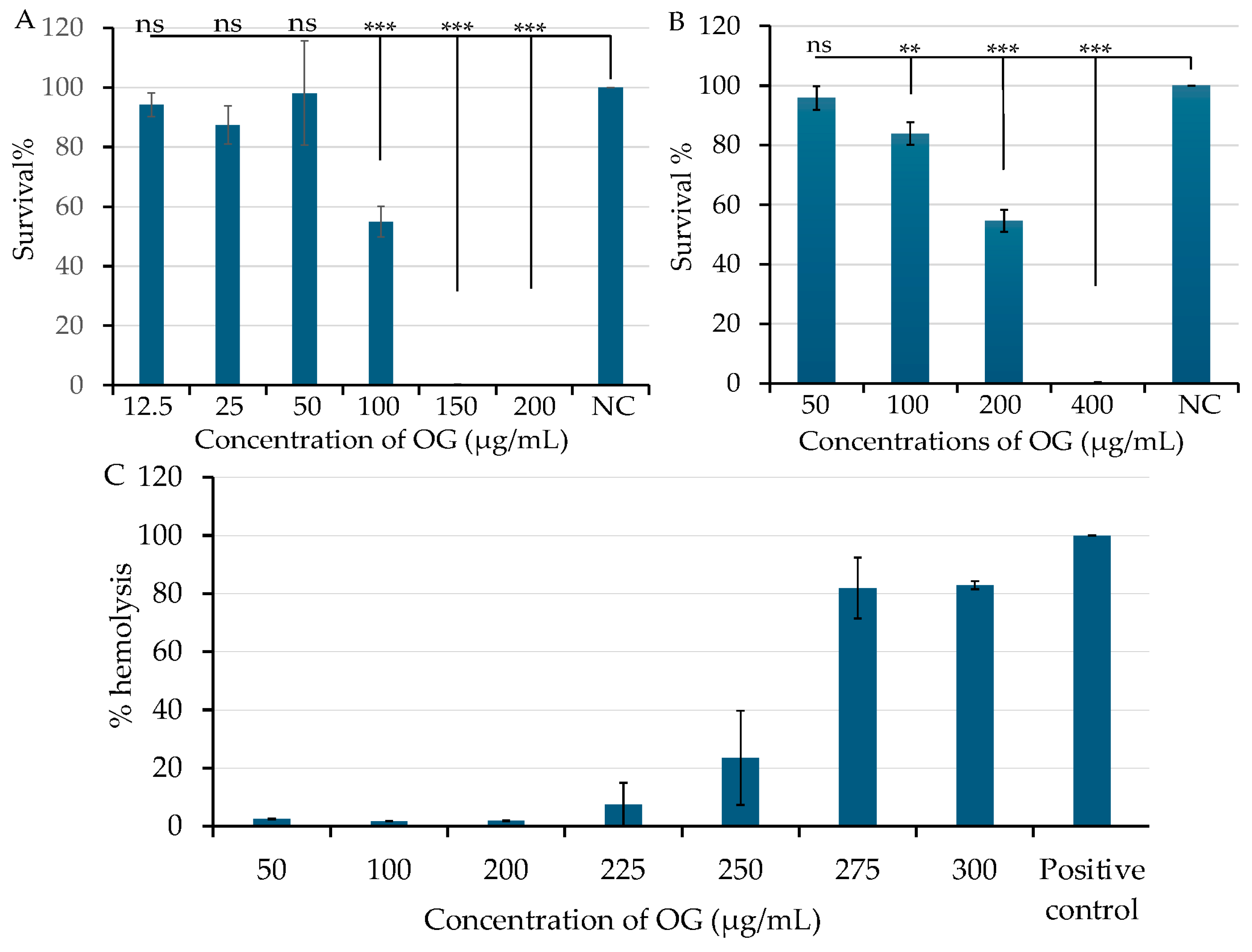
2.4. OG Has Low Cytotoxicity and Hemolytic Activity
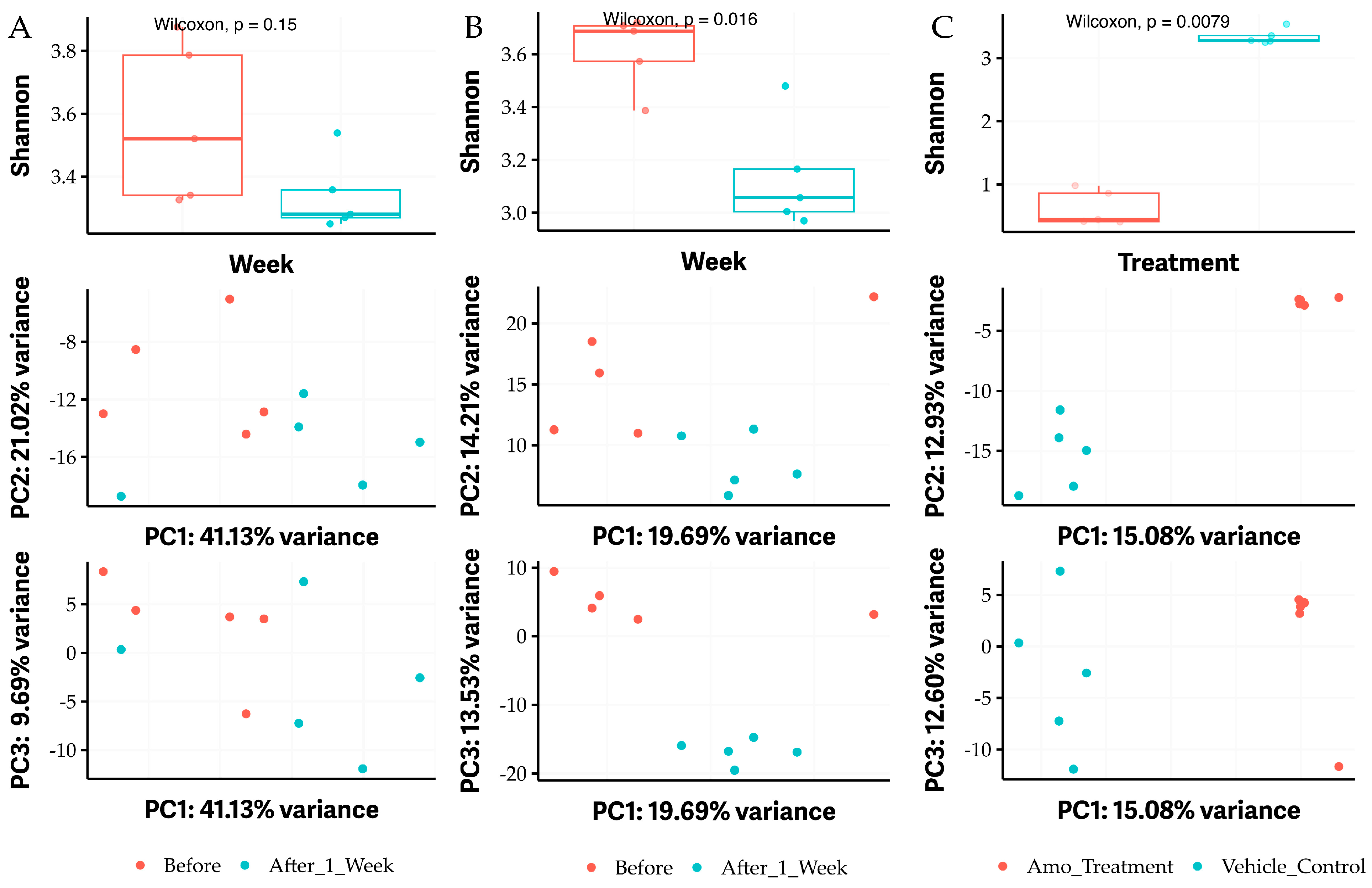
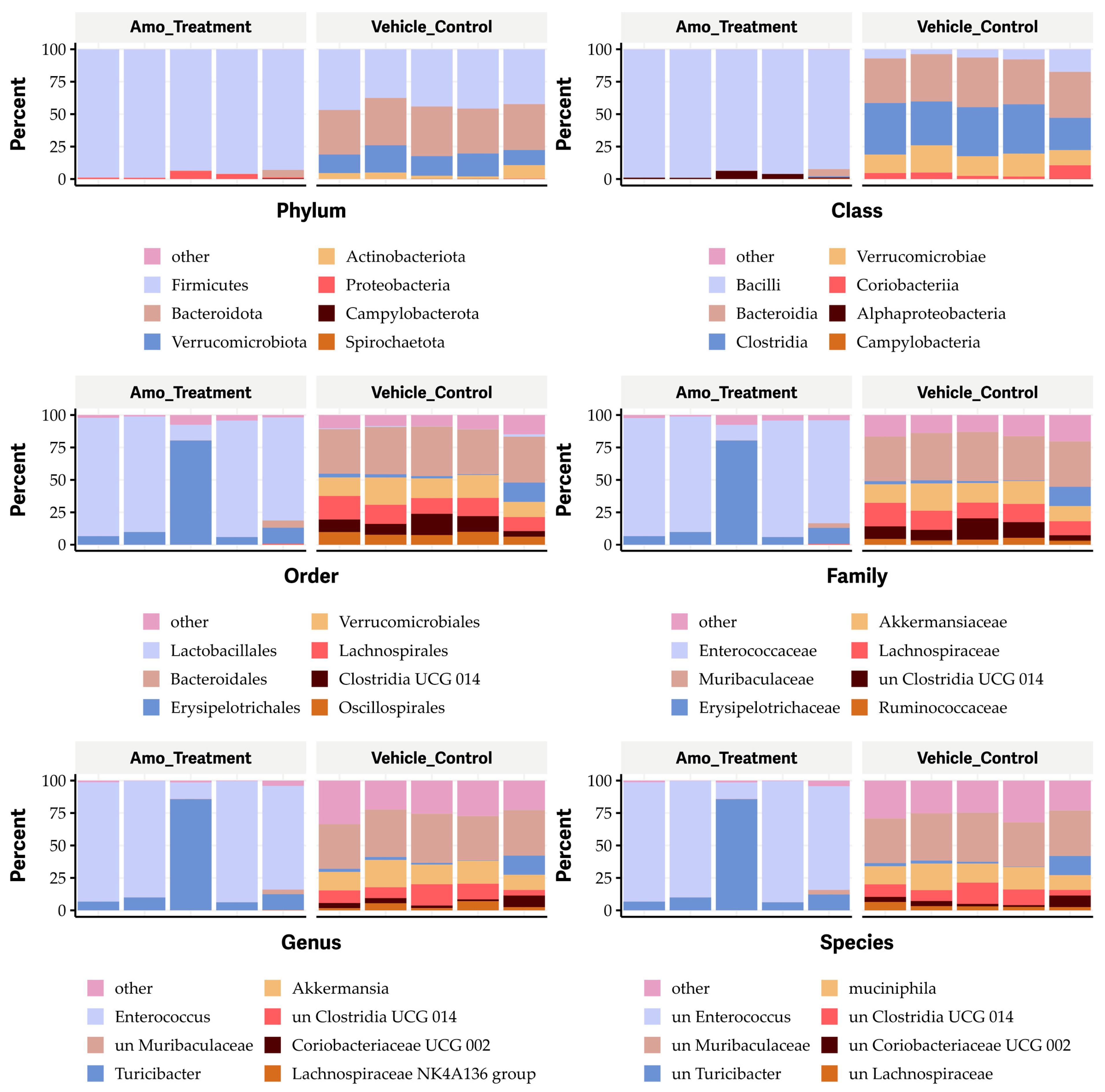
2.5. Feeding OG Affects the Diversity of Gut Microbiome of C57BL/6 Mice
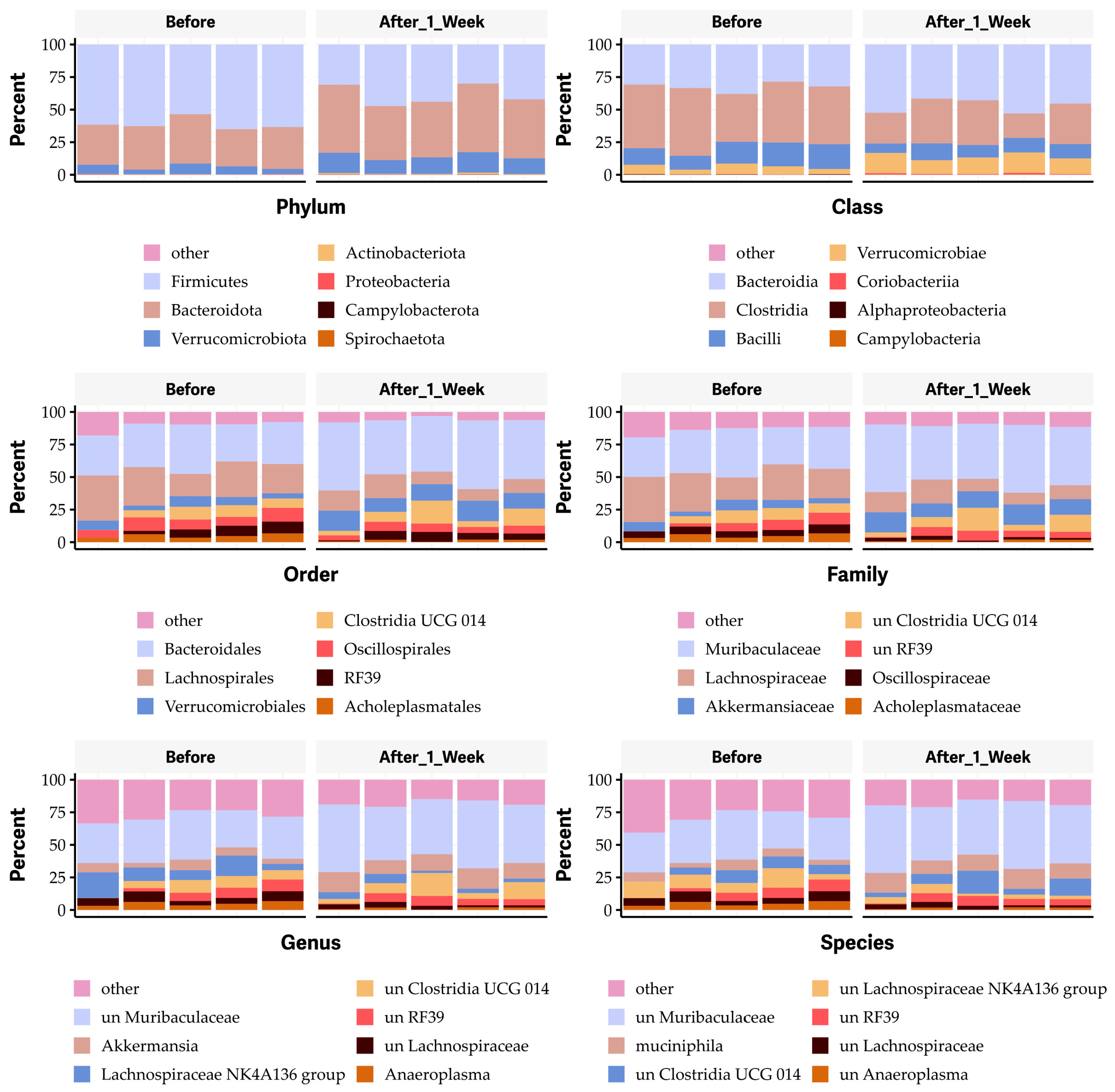
2.6. Oral Administration of OG Alters the Bacterial Species of Gut Microbiome in Mice
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Antibiotics and Chemical Compounds
4.3. Eukaryotic Cell Culture
4.4. MIC and MBC Assays
4.5. Kinetic Time-Killing Assays
4.6. Bacterial Cell Permeability Assays
4.7. Cytotoxicity Assay with Human Foreskin Fibroblast (HFF) and Lung A549 Epithelial Cells
4.8. Hemolytic Activity Assay with Sheep Red Blood Cells
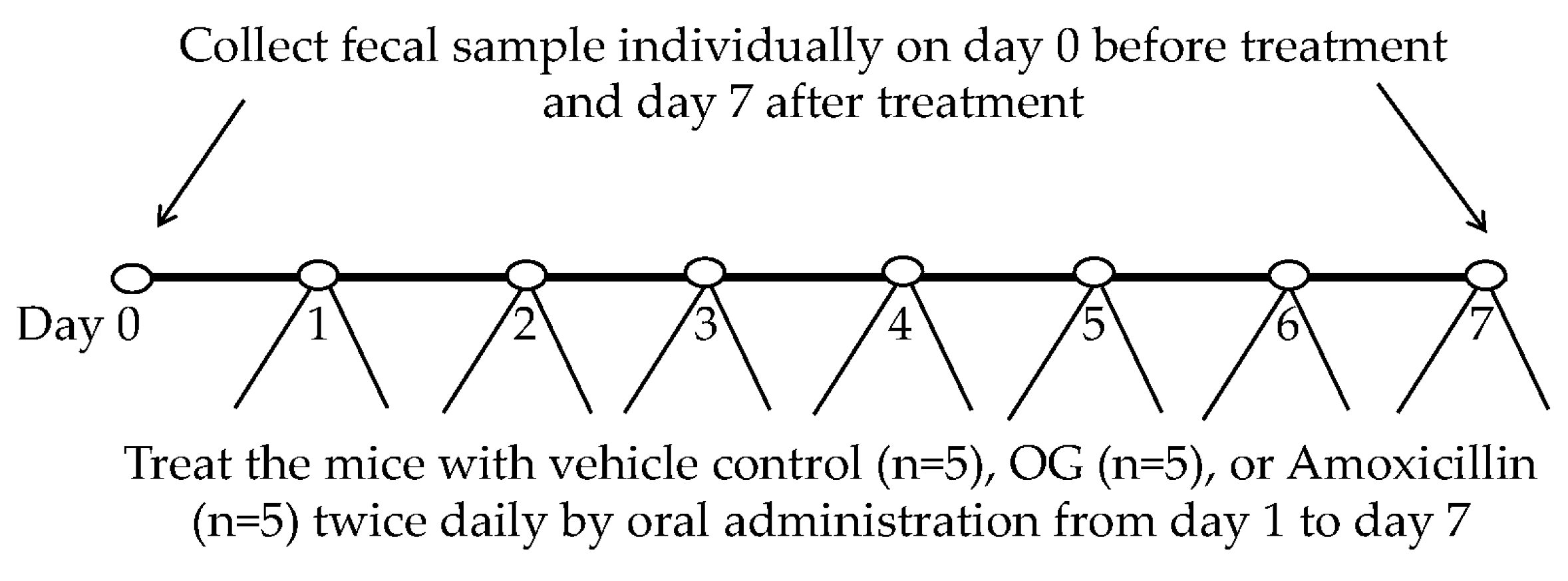
4.9. Animal Experiments and Collection of Fecal Samples from Mice
4.10. Total DNA Purification from Individual Fecal Sample and 16s RNA Sequencing and Microbiome Analysis
4.11. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kadariya, J.; Smith, T.C.; Thapaliya, D. Staphylococcus aureus and staphylococcal food-borne disease: An ongoing challenge in public health. Biomed Res. Int. 2014, 2014, 827965. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.A.; Sharma-Kuinkel, B.K.; Maskarinec, S.A.; Eichenberger, E.M.; Shah, P.P.; Carugati, M.; Holland, T.L.; Fowler, V.G., Jr. Methicillin-resistant Staphylococcus aureus: An overview of basic and clinical research. Nat. Rev. Microbiol. 2019, 17, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed]
- CDC. Antibiotic Resistance Threats in the United States; CDC: Atlanta, GA, USA, 2019.
- WHO. Global Shortage of Innovative Antibiotics Fuels Emergency and Spread of Drug-Resistance; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- Kubo, I.; Fujita, K.; Nihei, K. Anti-Salmonella activity of alkyl gallates. J. Agric. Food Chem. 2002, 50, 6692–6696. [Google Scholar] [CrossRef] [PubMed]
- Kubo, I.; Fujita, K.; Nihei, K.; Nihei, A. Antibacterial activity of alkyl gallates against Bacillus subtilis. J. Agric. Food Chem. 2004, 52, 1072–1076. [Google Scholar] [CrossRef] [PubMed]
- Stapleton, P.D.; Shah, S.; Anderson, J.C.; Hara, Y.; Hamilton-Miller, J.M.; Taylor, P.W. Modulation of beta-lactam resistance in Staphylococcus aureus by catechins and gallates. Int. J. Antimicrob. Agents 2004, 23, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Rúa, J.; Fernández-Álvarez, L.; de Castro, C.; Del Valle, P.; de Arriaga, D.; García-Armesto, M.R. Antibacterial activity against foodborne Staphylococcus aureus and antioxidant capacity of various pure phenolic compounds. Foodborne Pathog. Dis. 2011, 8, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Jeon, S. Synergistic antibacterial effect of octyl gallate and antibiotics against resistant bacterial strains. J. Appl. Microbiol. 2016, 120, 1175–1182. [Google Scholar]
- Oh, Y.; Park, S.; Lee, H. Synergistic effect of octyl gallate and gentamicin against MRSA. Antimicrob. Agents Chemother. 2018, 62, e01034-18. [Google Scholar]
- Santativongchai, P.; Tulayakul, P.; Ji, Y.; Jeon, B. Synergistic Potentiation of Antimicrobial and Antibiofilm Activities of Penicillin and Bacitracin by Octyl Gallate, a Food-Grade Antioxidant, in Staphylococcus epidermidis. Antibiotics 2022, 11, 1775. [Google Scholar] [CrossRef]
- Silva, R.T.C.; Guidotti-Takeuchi, M.; Peixoto, J.L.M.; Demarqui, F.M.; Mori, A.P.; Dumont, C.F.; Ferreira, G.R.A.; Pereira, G.M.; Rossi, D.A.; Corbi, P.P.; et al. New Palladium(II) Complexes Containing Methyl Gallate and Octyl Gallate: Effect against Mycobacterium tuberculosis and Campylobacter jejuni. Molecules 2023, 28, 3887. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Park, J.S.; Choi, H.Y.; Kwak, J.H.; Kim, W.G. Differential effects of alkyl gallates on quorum sensing in Pseudomonas aeruginosa. Sci. Rep. 2019, 9, 7741. [Google Scholar] [CrossRef] [PubMed]
- Gabe, V.; Kacergius, T.; Abu-Lafi, S.; Zeidan, M.; Abu-Farich, B.; Austys, D.; Masalha, M.; Rayan, A. Suppressive Effects of Octyl Gallate on Streptococcus mutans Biofilm Formation, Acidogenicity, and Gene Expression. Molecules 2019, 24, 3170. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.G.; Jiang, L.; Lin, S.; Jin, W.G.; Gu, Q.; Chen, Y.W.; Zhang, K.; Ettelaie, R. Ultra-efficient antimicrobial photodynamic inactivation system based on blue light and octyl gallate for ablation of planktonic bacteria and biofilms of Pseudomonas fluorescens. Food Chem. 2022, 374, 131585. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.Z.; Chen, W.X.; Zhao, Y.X.; Fang, Q.; Wang, L.G.; Tian, S.Y.; Shi, Y.G.; Chen, J.S. Ascorbic acid potentiates photodynamic inactivation mediated by octyl gallate and blue light for rapid eradication of planktonic bacteria and biofilms. Food Chem. 2024, 448, 139073. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.C.; Jeon, B. Novel adjuvant strategy to potentiate bacitracin against MDR MRSA. J. Antimicrob. Chemother. 2016, 71, 1260–1263. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Takeuchi, F.; Kuroda, M. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 2002, 359, 1819–1827. [Google Scholar] [CrossRef]
- Gill, S.R.; Fouts, D.E.; Archer, G.L. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain. J. Bacteriol. 2005, 187, 2426–2438. [Google Scholar] [CrossRef]
- Montgomery, C.P.; Boyle-Vavra, S.; Daum, R.S. Importance of the global regulators Agr and SaeRS in the pathogenesis of CA-MRSA USA300 infection. PLoS ONE 2010, 5, e15177. [Google Scholar] [CrossRef]
- Yang, J.; Brown, C.; Noland, W.; Johnson, T.J.; Ji, Y. Identification and Validation of a Novel Antibacterial Compound MZ-01 against Methicillin-Resistant Staphylococcus aureus. Antibiotics 2022, 11, 1550. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, J.; Ji, M.; Phillips, J.; Wylam, M.; Ji, Y. Identification of cbiO Gene Critical for Biofilm Formation by MRSA CFSa36 Strain Isolated from Pediatric Patient with Cystic Fibrosis. Pathogens 2021, 10, 1363. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Yang, M.; Sreevatsan, S.; Davies, P.R. Prevalence and Characterization of Staphylococcus aureus in Growing Pigs in the USA. PLoS ONE 2015, 10, e0143670. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.X.; Kasper, K.J.; Zeppa, J.J.; McCormick, J.K. Superantigens Modulate Bacterial Density during Staphylococcus aureus Nasal Colonization. Toxins 2015, 7, 1821–1836. [Google Scholar] [CrossRef] [PubMed]
- Wilson, G.J.; Tuffs, S.W.; Wee, B.A.; Seo, K.S.; Park, N.; Connelley, T.; Guinane, C.M.; Morrison, W.I.; Fitzgerald, J.R. Bovine Staphylococcus aureus Superantigens Stimulate the Entire T Cell Repertoire of Cattle. Infect. Immun. 2018, 86, e00505-18. [Google Scholar] [CrossRef] [PubMed]
- Schindler, B.D.; Kaatz, G.W. Multidrug efflux pumps of Gram-positive bacteria. Drug Resist. Updates 2016, 27, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lubelski, J.; Konings, W.N.; Driessen, A.J. Distribution and physiology of ABC-type transporters contributing to multidrug resistance in bacteria. Microbiol. Mol. Biol. Rev. 2007, 71, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, H.; Zgurskaya, H.I. AcrAB and related multidrug efflux pumps of Escherichia coli. J. Mol. Microbiol. Biotechnol. 2001, 3, 215–218. [Google Scholar]
- Lomovskaya, O.; Lewis, K. Emr, an Escherichia coli locus for multidrug resistance. Proc. Natl. Acad. Sci. USA 1992, 89, 8938–8942. [Google Scholar] [CrossRef] [PubMed]
- Tikhonova, E.B.; Wang, Q.; Zgurskaya, H.I. Chimeric analysis of the multicomponent multidrug efflux transporters from gram-negative bacteria. J. Bacteriol. 2007, 189, 2633–2643. [Google Scholar] [CrossRef]
- FDA. CFR (Code of Federal Regulations), Title 21: Food and Drugs. Chapter I—Food and Drug Administration, Department of Health and Human Services, Part 170—Food Additives; Office of the Federal Register: Washington, DC, USA, 2008.
- FDA. CFR (Code of Federal Regulations), Title 21: Food and Drugs. Chapter I—Food and Drug Administration, Department of Health and Human Services, Part 184—Direct Food Substances Affirmed as Generally Recognized as Safe (GRAS); Office of the Federal Register: Washington, DC, USA, 2008.
- Available online: https://www.efsa.europa.eu/en/efsajournal/pub/4248 (accessed on 18 July 2024).
- Binda, C.; Lopetuso, L.R.; Rizzatti, G.; Gibiino, G.; Cennamo, V.; Gasbarrini, A. Actinobacteria: A relevant minority for the maintenance of gut homeostasis. Dig. Liver Dis. 2018, 50, 421–428. [Google Scholar] [CrossRef]
- Feng, Q.; Chen, W.D.; Wang, Y.D. Gut Microbiota: An Integral Moderator in Health and Disease. Front. Microbiol. 2018, 9, 151. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.J.; Wan, L.; Gao, A.; Yu, Y.X.; Zhou, S.Y.; He, Q.; Li, G.; Ren, H.; Lian, X.L.; Zhao, D.H.; et al. Targeted inhibition of gut bacterial β-glucuronidases by octyl gallate alleviates mycophenolate mofetil-induced gastrointestinal toxicity. Int. J. Biol. Macromol. 2024, 264 Pt 1, 130145. [Google Scholar] [CrossRef] [PubMed]
- Modi, S.R.; Collins, J.J.; Relman, D.A. Antibiotics and the gut microbiota. J. Clin. Investig. 2014, 124, 4212–4218. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.; Doe, A.; Brown, B. Changes in Gut Microbiome Composition and their Implications. J. Microb. Ecol. 2023, 12, 456–470. [Google Scholar]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; Vaughan, E.E.; Plugge, C.M.; de Vos, W.M. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 1469–1476. [Google Scholar] [CrossRef] [PubMed]
- Lagkouvardos, I.; Lesker, T.R.; Hitch, T.C.A.; Gálvez, E.J.C.; Smit, N.; Neuhaus, K.; Wang, J.; Baines, J.F.; Abt, B.; Stecher, B.; et al. Sequence and cultivation study of Muribaculaceae reveals novel species, host preference, and functional potential of this yet undescribed family. Microbiome 2019, 7, 28. [Google Scholar] [CrossRef] [PubMed]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef]
- Zhu, B.; Wang, X.; Li, L. Human gut microbiome: The second genome of human body. Protein Cell 2010, 1, 718–725. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Dethlefsen, L.; Huse, S.; Sogin, M.L.; Relman, D.A. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008, 6, e280. [Google Scholar] [CrossRef] [PubMed]
- Mruk-Mazurkiewicz, H.; Kulaszyńska, M.; Czarnecka, W.; Podkówka, A.; Ekstedt, N.; Zawodny, P.; Wierzbicka-Woś, A.; Marlicz, W.; Skupin, B.; Stachowska, E.; et al. Insights into the Mechanisms of Action of Akkermansia muciniphila in the Treatment of Non-Communicable Diseases. Nutrients 2024, 16, 1695. [Google Scholar] [CrossRef] [PubMed]
- Pellegrino, A.; Coppola, G.; Santopaolo, F.; Gasbarrini, A.; Ponziani, F.R. Role of Akkermansia in Human Diseases: From Causation to Therapeutic Properties. Nutrients 2023, 15, 1815. [Google Scholar] [CrossRef] [PubMed]
- Png, C.W.; Lindén, S.K.; Gilshenan, K.S.; Zoetendal, E.G.; McSweeney, C.S.; Sly, L.I.; McGuckin, M.A.; Florin, T.H. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 2010, 105, 2420–2428. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.C.; Han, F.; Song, M.R.; Chen, S.; Li, Q.; Zhang, Q.; Zhu, K.; Shen, J.Z. Natural flavones from Morus alba against methicillin-resistant Staphylococcus aureus via targeting the proton motive force and membrane permeability. J. Agric. Food Chem. 2019, 67, 10222–10234. [Google Scholar] [CrossRef] [PubMed]
- Espinasse, A.; Goswami, M.; Yang, J.; Vorasin, O.; Ji, Y.; Carlson, E.E. Targeting Multidrug Resistant Staphylococcus Infections with Bacterial Histidine Kinase Inhibitors. Chem. Sci. 2023, 14, 5028–5037. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Ramette, A. Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 2007, 62, 142–160. [Google Scholar] [CrossRef] [PubMed]



| Strain/Species | Discerption | OG MIC µg/mL | OG MBC µg/mL | Source or Ref. |
|---|---|---|---|---|
| S. aureus | MRSA | |||
| WCUH29 | HA-MRSA | 8 | 16 | NCIMB40771 |
| NRS383 | USA200 HA-MRSA | 8 | 16 | NARSA |
| MW2 | USA400 HA-MRSA | 8 | 16 | [19] |
| NRS386 | USA700 MRSA | 8 | 16 | Human isolate, NARSA |
| NRS483 | USA1000 MRSA | 4 | 8 | Human isolate, NARSA |
| NRS484 | USA1100 MRSA | 16 | 32 | Human isolate, NARSA |
| NRS194 | CA-MRSA | 8 | 16 | NARSA |
| NRS248 | CA-MRSA | 8 | 16 | NARSA |
| COL | CA-MRSA | 8 | 16 | [20] |
| JE2 | USA300 CA-MRSA | 8 | 16 | BEI |
| 923 | USA300 CA-MRSA | 8 | 16 | [21] |
| 1371 | USA300 CA-MRSA | 8 | 16 | [22] |
| NRS123 | USA400 CA-MRSA | 8 | 16 | NARSA |
| CFSa36 | MRSA from CF patient | 8 | 16 | [23] |
| ST398 | ST398 LA-MRSA | 8 | 16 | [24] |
| S. aureus | MSSA | |||
| MSA553 | ST30 MSSA | 8 | 16 | Dr. Patrick Schlievert |
| Newman | MSSA | 8 | 16 | [25] |
| RF122 | ET3 ST151 bovine isolate | NA | 16 | [26] |
| Group A Streptococci | ||||
| S. pyogenes | Sore throat isolate | 8 | 16 | Dr. Jeffrey Hall |
| Bacillus subtilis | 8 | 8 | ATCC | |
| Gram-negative bacteria | ||||
| E. coli ATCC 25922 | >128 | >128 | ATCC | |
| A. baumannii ATCC 19606 | 64 | >64 | ATCC | |
| K. pneumoniae ATCC 13883 | >128 | >128 | ATCC | |
| P. aeruginosa ATCC 27853 | >128 | >128 | ATCC | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.; Gould, T.J.; Jeon, B.; Ji, Y. Broad-Spectrum Antibacterial Activity of Antioxidant Octyl Gallate and Its Impact on Gut Microbiome. Antibiotics 2024, 13, 731. https://doi.org/10.3390/antibiotics13080731
Yang J, Gould TJ, Jeon B, Ji Y. Broad-Spectrum Antibacterial Activity of Antioxidant Octyl Gallate and Its Impact on Gut Microbiome. Antibiotics. 2024; 13(8):731. https://doi.org/10.3390/antibiotics13080731
Chicago/Turabian StyleYang, Junshu, Trevor J. Gould, Byeonghwa Jeon, and Yinduo Ji. 2024. "Broad-Spectrum Antibacterial Activity of Antioxidant Octyl Gallate and Its Impact on Gut Microbiome" Antibiotics 13, no. 8: 731. https://doi.org/10.3390/antibiotics13080731







