Abstract
Background/Objectives: Salmonella is a major cause of foodborne illnesses, with multidrug-resistant (MDR) strains posing significant threats to public health worldwide. This study investigated the prevalence and antimicrobial resistance (AMR) of Salmonella, focusing on extended-spectrum β-lactamase (ESBL)-producing Salmonella in retail poultry meat in Korea. Methods: A total of 300 poultry meat samples were collected nationwide from retail markets. Multi-locus sequence typing, serotyping, and antimicrobial susceptibility testing were performed. Whole-genome sequencing (WGS) analysis was conducted against 28 representative ESBL-producing S. Infantis isolates to identify the genetic characteristics and phylogenetic relationship. Results: Salmonella was detected in 81.3% of raw poultry meat samples, with S. Infantis ST32 being the dominant serotype in chicken (53.0%) and S. Typhimurium ST19 predominant in duck (39.0%). MDR was identified in 58.2% of samples, with a significantly higher rate in chicken isolates than in duck isolates (p < 0.001). Notably, 75.3% of chicken MDR isolates were ESBL-producing S. Infantis carrying blaCTX-M-65. WGS of 28 geographically and phenotypically representative ESBL-producing S. Infantis revealed five clonal clusters, suggesting the widespread dissemination of ESBL-producing S. Infantis across Korea’s poultry supply chain. All 28 ESBL-producing S. Infantis isolates contained a pESI-like megaplasmid, carrying multiple resistance and virulence genes, with sequences highly identical to plasmids reported in the United States, indicating potential international transmission. Conclusions: This study emphasizes the urgent need for continuous surveillance and responsible antibiotic use in livestock under a One Health framework. WGS can provide an effective tool for tracking AMR evolution and clonal spread within and across regions.
1. Introduction
Salmonella is a major zoonotic foodborne pathogen with significant public health and economic impacts worldwide [1]. It causes more than 90 million cases of gastroenteritis, with an estimated annual salmonellosis incidence ranging from 200 million to more than 1 billion [2]. According to the Ministry of Food and Drug Safety (MFDS, Korea), Salmonella was responsible for 10,888 reported infections between 2013 and 2023, making it the third most common cause of foodborne illness in Korea after pathogenic Escherichia coli (15,715 cases) and norovirus (10,947 cases) [3]. Salmonella is typically found as a normal component of animal gut microbiota and serves as a source of transmission from animals to humans. Approximately 85% of Salmonella-associated foodborne illnesses are associated with the consumption of contaminated foods, including meat, eggs, and dairy products [4,5]. To date, approximately 2600 Salmonella serovars have been identified and classified as typhoidal or non-typhoidal based on their pathogenicity in humans and animals [4]. Non-typhoidal Salmonella (NTS) is associated with various serotypes and is a leading cause of foodborne diarrhea [6]. Among the NTS serotypes, S. Typhimurium and S. Enteritidis are the most common causes of zoonotic infection of humans and are usually associated with poultry and poultry products [7]. The most prevalent serotypes related to human infections include Enteritidis, Newport, Typhimurium, Javiana, and monophasic Typhimurium 4,[5],12:i:- in the United States and Enteritidis, Typhimurium, monophasic Typhimurium, Infantis, and Newport in the European Union [7].
Antimicrobials have been widely used for prophylaxis and treatment in livestock industries for several decades [8,9]. Moreover, antimicrobials have also been used at subtherapeutic doses to promote growth by regulating intestinal microbiota [10]. However, the emergence of bacteria with antimicrobial resistance (AMR) has led several countries to ban their use for growth promoters [10,11]. Despite these measures, the prolonged and often inappropriate use of antimicrobials places selective pressure on bacteria in animals and the environment, facilitating resistance [12]. The emergence of multidrug-resistant (MDR) Salmonella strains has gradually increased, leading to significant health concerns owing to the potential for treatment failure of salmonellosis [9,13]. Resistance to extended-spectrum β-lactams, particularly third- and fourth-generation cephalosporins and carbapenems, poses a significant threat, as they are critically important antimicrobials in human and veterinary medicine [9].
A particularly concerning development is the global emergence of MDR-emergent S. Infantis (ESI), representing a challenge for the poultry industry [14]. These strains carry a large plasmid, known as the pESI or pESI-like plasmid, which encodes multiple AMR genes, including the extended-spectrum β-lactamase (ESBL) gene [15]. The rising prevalence of ESI in poultry farms and meat products has led to a rise in S. Infantis infections in the United States, South America, and Europe, primarily linked to contaminated chicken products, posing serious risks to both animals and humans [14,16,17,18].
In Korea, the emergence of ESBL-producing MDR S. Infantis was first reported in chicken and duck samples collected from slaughterhouses in 2020 and 2021, respectively [19]. Although not characterized in detail, these isolates were presumed to be ESI strains carrying pESI or pESI-like plasmids. Recently, the presence of ESI carrying pESI-like plasmids was confirmed in broiler farms and egg grading and packing plants through PCR-based detection [20,21]. However, the molecular characterization of these ESI strains was limited due to the methodologies used, such as pulsed-field gel electrophoresis (PFGE) and multiplex PCRs, which provide limited genetic information and make cross-comparisons with international ESI strains challenging. Therefore, we investigated the nationwide distribution of ESBL-producing Salmonella spp., as well as ESI carrying pESI-like plasmids, in retail chicken and duck meat in South Korea. We employed the advanced technique of whole-genome sequencing (WGS) to thoroughly characterize the genetic elements and clonal relationships of ESI isolates and to trace the origins of pESI-like plasmids based on sequence identities.
2. Results
2.1. Prevalence, Serotype, and Sequence Type Distribution of Salmonella in Poultry Meats
The overall prevalence, serotypes, and sequence types (STs) of Salmonella spp. in 300 retail poultry meats are shown in Table 1 and Supplementary Table S1. Salmonella contamination was found in 81.3% (244 out of 300 samples) of retail poultry meat samples. There was no statistically significant difference between the prevalence of Salmonella in chicken (79.0%, 158 out of 200 samples) and duck (86.0%, 86 out of 100 samples) meat. Serotyping of the 244 isolates identified 11 serotypes, with an additional 34 isolates being non-typable. These isolates were associated with 18 STs, with most serotypes belonging to distinct STs. The distribution of serotypes and STs in chickens and ducks showed clear differences. The most prevalent serotype was S. Infantis (53.0%), followed by S. Agona (10.5%) in chicken, S. Typhimurium (39.0%), and S. Infantis and S. Brandenburg (7.0%) in duck meat. All the S. Infantis isolates belonged to ST32, whereas S. Typhimurium was classified as ST19. Additionally, significant differences were observed in the isolation rates of the eight STs between chicken and duck meats (p < 0.05).

Table 1.
Prevalence, serotype, and sequence types (STs) of Salmonella in poultry meats.
Notably, based on the Korean Animal Product Traceability system, we found that all S. Infantis-positive duck meat was processed in slaughterhouses processing both chickens and ducks, whereas all duck meat processed in slaughterhouses that only handle ducks was negative for S. Infantis.
2.2. Comparisons of AMR Profiles of Salmonella enterica Isolates from Chicken and Duck Meat
Overall, 93.7% (n = 148) of Salmonella isolates from chicken meat samples were resistant to at least one antimicrobial agent, and 79.7% (n = 126) of the isolates exhibited an MDR phenotype. In contrast, 58.1% (n = 50) of duck meat isolates were resistant to at least one antimicrobial agent, and 18.6% (n = 16) were MDR (Table 2). Among 244 Salmonella isolates, the highest resistance was observed against NAL (68.0%), followed by TET (54.9%), STR, and FIS (54.5%). Chicken isolates exhibited significantly higher resistance to AMP, CHL, CTX, FIS, GEN, NAL, STR, STX, and TET than that of duck isolates (p < 0.001). Moreover, resistance to AUG2, FEP, FOX, GEN, and CAZ was detected only in the chicken meat isolates. Notably, all the 244 isolates were susceptible to MERO.

Table 2.
Antimicrobial resistance (AMR) profiles of Salmonella in chicken and duck meats.
2.3. ESBL-Producing Salmonella
According to the results of the antimicrobial susceptibility test, 63.3% (n = 100) and 8.1% (n = 7) of chicken and duck meat isolates, respectively, were ESBL-producing Salmonella, which showed resistance against CTX (≥16 µg/mL). Interestingly, all ESBL-producing Salmonella isolates were S. Infantis (Table 2). PCR screening of ESBL-encoding genes revealed that all ESBL-producing S. Infantis harbored the blaCTX-M-65 gene, belonging to the CTX-M-9 group. All ESBL-producing isolates tested negative for blaTEM and blaSHV, besides one isolate from chicken meat that harbored blaTEM-98. All ESBL-producing isolates were MDR and resistant to at least five classes of antimicrobials (Table 3). Most ESBL-producing isolates were resistant to CTX, NAL, AMP (all 100.0%), CHL (99.1%), TET (95.3%), and FIS (92.5%). Twenty-two combinations of AMR patterns were identified. The most common combination of AMR patterns was resistance to AMP-CHL-CTX-FIS-STR-SXT-NAL-TET (41.1%), followed by resistance to AMP-CHL-CTX-FIS-GEN-STR-SXT-NAL-TET and AMP-CHL-CTX-FIS-NAL-STR-TET (15.9%).

Table 3.
AMR patterns of 107 extended-spectrum β-lactamase (ESBL)-producing S. Infantis.
2.4. Genomic Characteristics of ESBL-Producing S. Infantis
To further investigate the genetic characteristics and clonal relationships of ESBL-producing Salmonella, representative ESBL-producing S. Infantis isolates were selected for WGS. For chicken isolates, the three most common AMR patterns (Table 3) were detected across all five provinces. The AMP-CHL-CTX-NAL-TET resistance pattern was detected in three provinces (Gyeonggi, Chungcheong, and Gangwon provinces), while AMP-CHL-CTX-FIS-GEN-NAL-STR-TET resistance was detected in two provinces (Gyeonggi and Chungcheong provinces). One isolate with each AMR pattern was selected from each province, resulting in the selection of twenty isolates. The two isolates that showed resistance to the highest numbers of antimicrobial agents (12 and 11 antimicrobials, Table 3) were included. Three isolates from different retail markets were additionally selected to represent all ESBL-producing S. Infantis-positive retail markets (19 retail markets).
For duck isolates, ESBL-producing S. Infantis was only detected in three markets, all located in Chungcheong Province; all of them showed the same AMR pattern (Table 3). One from each market was included in the WGS analysis. Taken together, 25 chicken-derived and 3 duck-derived ESBL-producing S. Infantis isolates were subjected to WGS.
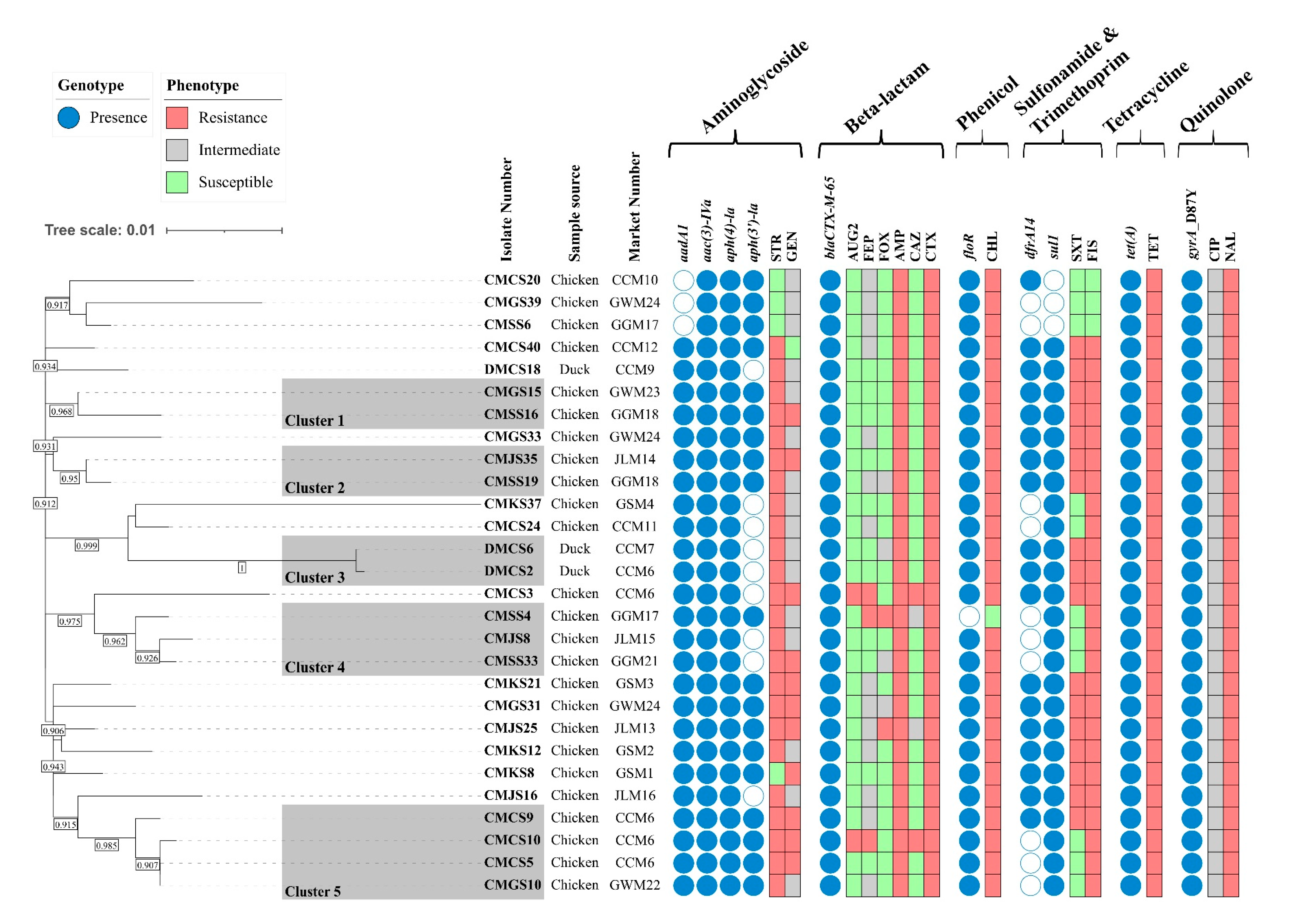
The in silico serotyping revealed that all 28 ESBL-producing isolates were Salmonella enterica serovar Infantis (antigenic formula: 7:r:1,5). To assess the genetic relatedness among the 28 ESBL-producing isolates, an ML phylogenetic tree was constructed based on 1422 SNPs in the core gene alignment. Five clusters were generated, and most isolates clustered together in Clusters 1–5, regardless of their geographical region. The isolates in Cluster 1 (10 SNP differences), Cluster 2 (3 SNP differences), Cluster 4 (median 8.7, range 6–11 SNP differences), and Cluster 5 (median 4, range 0–8 SNP differences) were clustered together, even though they were collected from different geographical regions. Moreover, two isolates collected from the Chungcheong (CMCS5) and Gangwon (CMGS10) provinces in Cluster 5 were identical, with no SNP differences. Among the three duck isolates, DMCS18 belonged to a singleton, but the remaining two isolates (DMCS2 and DMCS6) clustered together in Cluster 3, with only one SNP difference. These two isolates were collected from duck meat purchased from different markets but processed in a slaughterhouse that processed chicken and ducks together, suggesting possible cross-contamination between chicken and duck meat during slaughter. Taken together, the average SNP differences among the isolates within the same clusters were ≤10, indicating close genetic relatedness among the S. Infantis isolates (Figure 1).
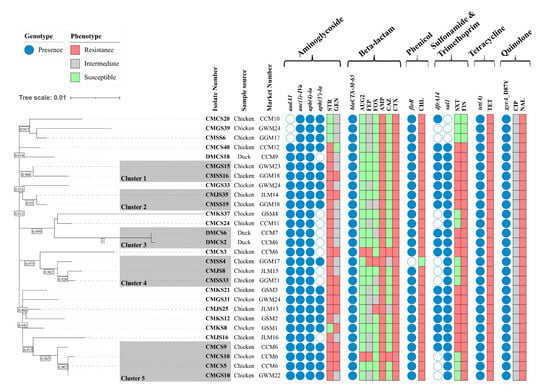
Figure 1.
Phylogenetic tree and AMR profiles of 28 ESBL-producing S. Infantis samples. A maximum-likelihood phylogenetic tree of 28 ESBL-producing S. Infantis samples was constructed using the FDA-CFSAN SNP pipeline and FastTree using S. Infantis SINFA (N649235) as a reference strain. Genetically related isolates were clustered by the framework provided by the FDA. Letters beside the isolate number indicate the sample sources. The bootstraps higher than 0.90 are shown in the box between the separated tree nodes. The presence and absence of AMR genes are indicated in blue and white circles, respectively. The AMR profiles are indicated in red (resistance), gray (intermediate), and green (susceptible) boxes, respectively. Market number: GSM, Gyeongsang Market; CCM, Chungcheong Market; JLM, Jeolla Market; GGM, Gyeonggi Market; GWM, Gangwon Market. AMP, ampicillin; AUG2, amoxicillin/clavulanic acid; CAZ, ceftazidime; CHL, chloramphenicol; CIP, ciprofloxacin; CTX, cefotaxime; FEP, cefepime; FIS, sulfisoxazole; FOX, cefoxitin; GEN, gentamicin; NAL, nalidixic acid; STR, streptomycin; SXT, trimethoprim/sulfamethoxazole; TET, tetracycline.
The AMR genotypes and phenotypes of the 28 ESBL-producing S. Infantis isolates are shown in Figure 1. A total of nine AMR genes and one point mutation were detected in this study. Genes conferring resistance to aminoglycoside (aadA1, aph(4)-Ia, aac(3)-IVa, aph(3′)-Ia), β-lactam (blaCTX-M-65), phenicol, (floR), quinolone (gyrA_D87Y point mutation), sulfonamide (sul1), trimethoprim (dfrA14), and tetracycline (tet(A)) were identified. Plasmid-mediated quinolone resistance (PMQR) genes were not detected in this study. All 28 isolates harbored aac(3)-IVa, aph(4)-Ia, tet(A), blaCTX-M-65, and D87Y point mutations in gyrA (100% prevalence). Most of the AMR genotypes of the ESBL-producing Salmonella isolates corresponded to AMR phenotypes.
2.5. Characterization of pESI-like Plasmids
A plasmid replicon search revealed that only one type of plasmid replicon, IncFIB (pN55391), was present in all 28 S. Infantis isolates.
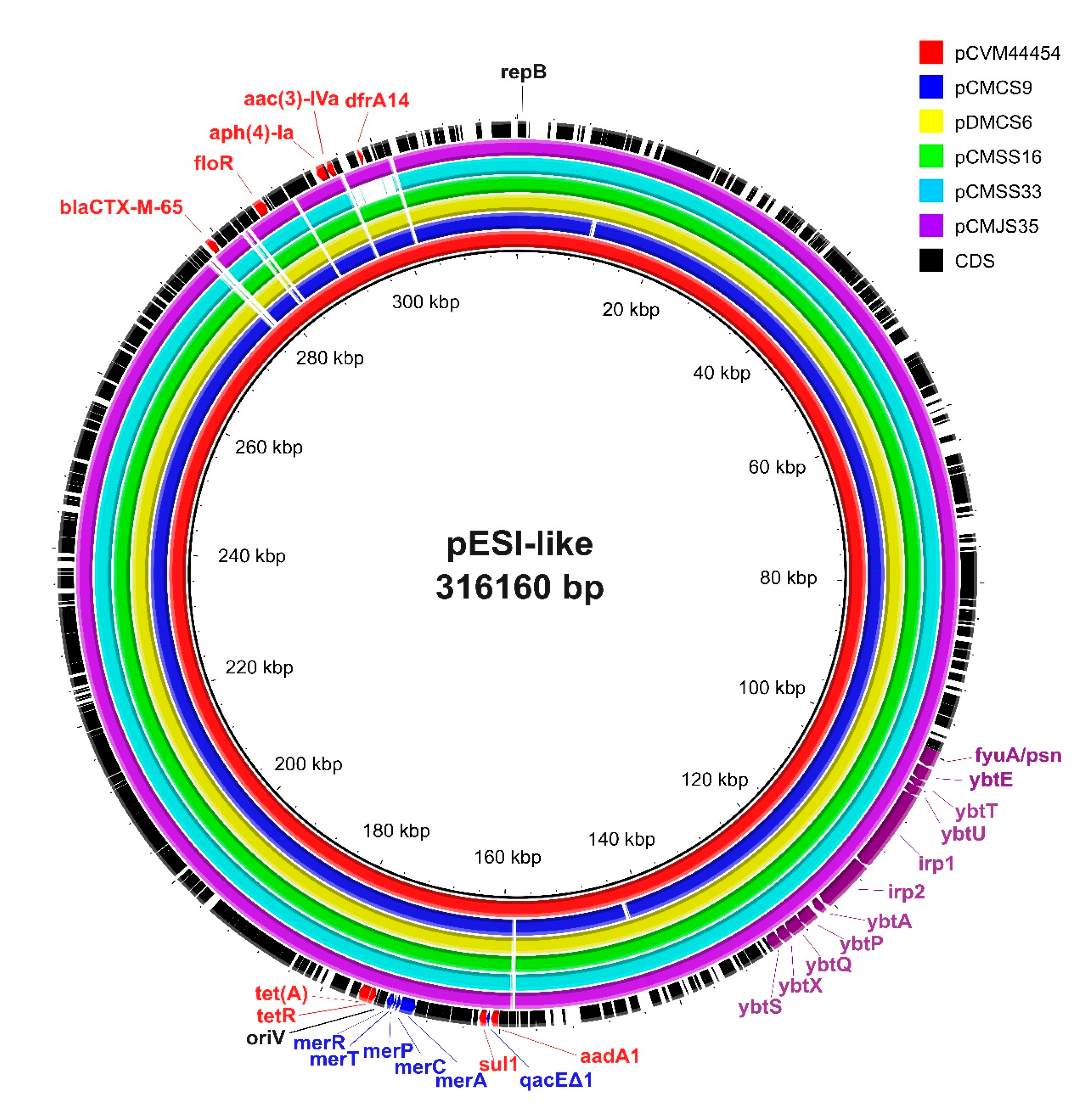
Plasmids were reconstructed using the mob-suite tool to further analyze the characteristics of the pESI-like megaplasmid in ESBL-producing S. Infantis isolates. All 28 ESBL-producing S. Infantis isolates carried a conjugative plasmid with predicted sizes ranging from 300,523 bp to 312,318 bp (Supplementary Table S2). IncI1 pMLST of the plasmids was positive for four genes (ardA, pilL, sogS, trbA), whereas all isolates were negative for the replicase gene repl1. Among the plasmid allele profiles, 26 of the 28 isolates displayed an allele combination of ardA_11, pilL_3, sogS_14, and trbA_8. Two plasmids (pCMKS37 and pCMGS31) showed a one-allele difference in ardA (ardA_3). Genes conferring resistance to aminoglycoside (aadA1, aph(4)-Ia, aac(3)-IVa, aph(3′)-Ia), sulfonamide (sul1), tetracycline (tet(A)), phenicol (floR), trimethoprim (dfrA14), and β-lactam (blaCTX-M-65) were detected in the plasmids, except for one isolate (CMCS20), in which dfrA14 was found in the chromosome. Virulence genes associated with yersiniabactin biosynthesis (irp1, irp2, fyuA/psn, fyuA, ybtA, ybtE, ybtP, ybtQ, ybtS, ybtT, ybtU, ybtX), along with genes conferring resistance to mercury (merC, merP, merT, merR) and quaternary ammonium compounds (qacEΔ1), which are frequently identified in pESI-like plasmids, were detected in all 28 plasmids.
The mob-typer results revealed that all plasmids identified in this study were identical to the pESI plasmid carrying blaCTX-M-65, specifically pCVM44454 (GenBank accession no. CP016413.1) and pN55391 (CP016411.1), which were previously reported in the United States.
To place these findings in a global context, we compared the plasmid sequences from our isolates with representative pESI-like plasmids reported from other countries, including Israel (119944_pESI), Italy (12037823-11_Italy_pESI_CTX-M-1), and Slovenia (pS19). The pESI-like plasmids in this study showed the highest average nucleotide identity (ANI) values when compared to those from the United States (99.99–100%). In contrast, ANI values for the plasmids from Israel, Italy, and Slovenia ranged from 99.12% to 99.64% (Supplementary Table S3).
For further structural comparison, one isolate from each of the five phylogenetic clusters was selected and aligned with pCVM44454. All five representative plasmids exhibited nearly identical structures to pCVM44454, except for pCMSS33A (a chicken isolate), which exhibited a deletion in the region containing dfrA14 (Figure 2).
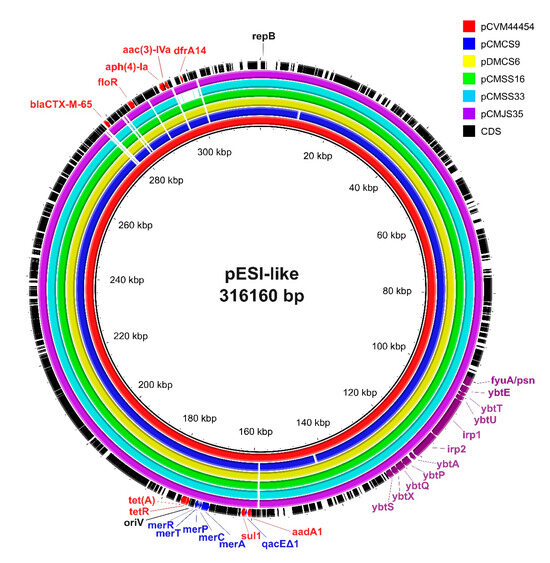
Figure 2.
Comparative genomic analysis of emerging Salmonella enterica Infantis (pESI)-like plasmids. Sequence alignment of pESI-like plasmids in extended-spectrum β-lactamase (ESBL)-producing S. Infantis was performed using BRIG software (v.0.95). The plasmid pCVM44454 (CP016413.1) was used as the reference plasmid. The respective plasmids corresponded with the colors shown in the legend on the right. Structurally similar regions are shown with colors, whereas the missing regions are shown as blank. Antimicrobial resistance-, stress resistance-, virulence-, and replication-related genes are indicated in red, blue, purple, and black letters, respectively.
3. Discussion
Non-typhoid salmonellosis is a major cause of foodborne illness worldwide. Salmonella contamination of poultry meat products occurs during slaughtering and processing, primarily due to exposure to bacteria-containing intestinal contents or improper handling [22]. In the present study, a high rate of Salmonella contamination was detected in poultry meat collected from retail markets across Korea. The prevalence of Salmonella spp. in retail chicken (79.0%) and duck meat (86.0%) was much higher than previously reported figures from Korea (21.2–42.3% in chicken meat and 51.3% in duck meat) [23,24,25,26]. Moreover, the proportion of Salmonella spp. in poultry meat was higher than that reported in China [1], Brazil [27], and the USA [28]. The higher frequency of Salmonella infections may be attributed to the isolation methods used in these studies. Most studies have isolated Salmonella spp. using xylose lysine deoxycholate (XLD) agar, a common isolation medium, whereas we used CHROMagarTM Salmonella for isolation. Maddocks et al. [29] demonstrated the sensitivity and specificity of CHROMagarTM Salmonella compared with other selective media. A clear difference was detected in the distribution of the predominant serotypes and STs of Salmonella between chicken and duck meat (Table 1). The predominant serovars and STs of Salmonella in chicken and duck meat were S. Infantis ST32 and S. Typhimurium ST19, respectively. S. Infantis recently became the most predominant serovar in broiler farms in Korea and is frequently identified as the causative agent of human salmonellosis worldwide [5,20]. In duck meat, S. Infantis ST32 was the third most frequently isolated serotype, with S. Brandenburg ST1954 and non-typable ST17, followed by non-typable ST33. Notably, S. Infantis duck isolates were detected only in duck meat from slaughterhouses that process both chickens and ducks. This suggests that cross-contamination of S. Infantis in duck meat may have occurred during the co-slaughtering process. Considering the serotype distribution in chicken and duck meat, S. Infantis isolated from duck meat may originate from chicken. This finding highlights the potential for S. Infantis transmission through duck meat and underscores the need for surveillance and proper management practices in poultry slaughterhouses.
The extensive use of antimicrobials in human and veterinary medicine for therapeutic or preventive purposes has promoted the emergence of AMR Salmonella strains that pose a threat to both animal and human health [30]. In the present study, we conducted an antimicrobial susceptibility test for the antimicrobials specified by the Korea Disease Control and Prevention Agency and the World Health Organization [31,32]. In results, high resistance rates were observed in poultry meat against NAL (68.0%), TET (54.9%), STR (54.5%), FIS (54.5%), and CHL (54.1%), with AMR profiles consistent with those of findings from previous studies [33,34,35]. Moreover, the resistance rates against nine antimicrobial agents, AMP, CTX, CHL, GEN, NAL, STR, FIS, TET, and SXT, were significantly higher in isolates from chicken meat than in those from duck meat (p < 0.001). A significantly higher proportion of MDR Salmonella was observed in chicken meat (79.7%) than in duck meat (18.6%). These results suggest that the selective pressure of antimicrobials may be higher in the chicken meat supply chain. The prevalence of MDR Salmonella is attributed to the overuse and misuse of antimicrobial agents for the prevention and treatment of diseases, which is consistent with a report that various antimicrobial agents are widely used in the poultry industry in Korea [36]. Moreover, 63.3% of the isolates from chickens and 8.1% of those from ducks were ESBL-producing Salmonella with MDR phenotypes. Consistent with the results reported by Kang et al. [19], the most prevalent MDR pattern among the ESBL-producing S. Infantis isolates was resistance to eight antimicrobial agents (Table 3). Despite the wide range of ESBLs, including TEM, SHV, CTX-M, and OXA β-lactamases, only blaCTX-M-65 was found in S. Infantis ST32. ESBL-producing S. Infantis strains have been reported in several food isolates, raising concerns regarding the significant risk of zoonotic transmission to humans [14]. However, there is limited information on the genetic characteristics of ESBL-producing S. Infantis from poultry sources in Korea, particularly using recent advanced WGS techniques [19]. Therefore, WGS was conducted to better understand the genetic characteristics and relatedness of ESBL-producing S. Infantis isolates from retail poultry meat.
Phylogenetic analysis based on SNP alignment revealed genetic relatedness among some S. Infantis isolates irrespective of their geographical regions (Figure 1). Isolates from different provinces showed a significant clonal relationship, indicating the clonal spread of S. Infantis in retail poultry meat in Korea. Detection of AMR genes in ESBL-producing S. Infantis isolates revealed the presence of multiple resistance genes (Figure 1). The presence of AMR genes in the representative 28 isolates was not consistent with the phylogenetic clusters, indicating that the acquisition or loss of AMR genes adapted to the environment. Most of the AMR genes detected corresponded to the resistant phenotypes observed in this study. All isolates resistant to TET, NAL, and CTX harbored tet(A), gyrA point mutations (D87Y), and blaCTX-M-65, respectively. In addition, 27 isolates were resistant to CHL and carried floR. Isolates carrying both trimethoprim (dfrA14) and sulfonamide (sul1) resistance genes were consistent with the SXT resistance phenotype. These findings demonstrate a clear correlation between the AMR genotype and phenotype. However, discrepancies were observed in aminoglycoside resistance, wherein the aminoglycoside genotype and phenotype did not match in some isolates. For instance, aadA1 and aac(3)-IVa are known to confer resistance against STR and GEN, respectively [37]. However, consistent with Alzahrani et al. [33], false positive results for aminoglycoside resistance have been reported for susceptible isolates.
Several researchers have noted that the S. Infantis strain carries a megaplasmid, termed the “plasmid of emerging S. Infantis” (pESI), which contains multiple AMR, metal resistance, and virulence genes conferring significant adaptive advantages to the bacteria both within environments and hosts [38]. The pESI megaplasmid was first identified in Israel in 2008. Subsequently, numerous studies have shown the presence of pESI-carrying S. Infantis strains in food-producing animals, meats, and humans worldwide, raising concerns about the potential dissemination of MDR S. Infantis strains [14]. In Korea, based on the report of the Korean Veterinary Antimicrobial Resistance Monitoring System (KVARS), ESBL-producing MDR S. Infantis was first detected in S. Infantis isolates from slaughterhouses in 2020, and the detected isolates suddenly increased the following year. These ESBL-producing MDR S. Infantis strains were ST32 and typically carried blaCTX-M-65, which were strongly linked to ESI carrying pESI plasmid [19]. Soon after, blaCTX-M-65 carrying a pESI-like plasmid in S. Infantis was reported in samples collected from egg packing facilities and broiler farms in 2022, through detection of repA, ipf, and K88-like genes using PCR [20,21]. However, in these studies, the clonal relatedness was analyzed using PFGE, which limits the ability to compare with strains reported by other research groups, as well as us. Moreover, some genetic components of pESI-like plasmids were screened by PCR. In the study by Kang et al. [19], ESBL-producing MDR S. Infantis isolated from slaughterhouses harbored blaCTX-M-65 and IncFIB replicon, which is consistent with our findings. However, all ESI carrying a pESI plasmid isolated from egg packing facilities harbored two ESBL-producing genes (blaCTX-M-65 and blaTEM-1), and their plasmid replicon type was IncP [21], which is a different type of pESI plasmid from those found in slaughterhouses [19] and meat in this study. These findings may indicate that ESI strains in broiler and layer farms carry different types of pESI-like plasmids in Korea. It is plausible that the previous study mistyped the plasmid replicon, as they used PCR-based replicon typing (PBRT) developed in 2005 [39]. The PBRT method was initially designed to detect the origin of replication (oriV) for IncP plasmids and the RepFIB replication protein A (repB) gene for IncFIB replicon-type plasmids. Several studies have demonstrated that pESI plasmids possess an oriV region of IncP-1α, which is typed as an IncP replicon when using the PBRT method [15,40]. Xu et al. [41] have demonstrated the sequence variability of the repB genes in the IncFIB replicon type, encompassing 7 primary types and 70 subtypes, which may account for the failure of targeting IncFIB in pESI-like plasmids with PBRT. Supporting this, the pESI-like plasmids detected in this study were found to possess IncFIB (pN55391), repB, and oriV (Figure 2, Supplementary Table S2), which is consistent with the findings of García-Soto et al. [42]. Based on the in silico PCR analysis, the replicon type of the pESI-like plasmids in this study was identified as IncP when applied with PBRT primers. Therefore, the PBRT method developed two decades ago should be replaced with a new method reflecting recent genetic information about Salmonella.
All 28 S. Infantis isolates analyzed in this study carried pESI-like plasmids harboring multiple resistance genes, including blaCTX-M-65. Previous studies have demonstrated that pESI-like megaplasmids commonly contain a diverse combination of AMR genes, particularly those conferring resistance to aminoglycosides, sulfonamides, tetracyclines, trimethoprim, and phenicols [14,16,21,37]. All 28 plasmids harbored yersiniabactin-encoding genes, which confer a siderophore-dependent iron uptake system that enhances Salmonella survival and virulence [38].
Globally, similar pESI-like megaplasmids have been reported in S. Infantis isolates from poultry and clinical samples in the United States, Israel, and several European countries. These plasmids commonly carry many genes for AMR, heavy metal resistance, oxidative stress, and virulence, in line with our findings [14,18,43]. The close genetic similarity (ANI > 99.9%) between Korean pESI-like plasmids and those from the United States suggests either a shared evolutionary origin or potential international transmission. As Alba et al. [44] noted, the structural variability of pESI plasmids likely reflects adaptation to environmental selective pressures. Recombination and rearrangement events may be evolutionarily favored, allowing plasmids to maintain core genetic determinants while acquiring additional resistance or virulence genes.
Notably, pESI-like plasmids are not exclusive to S. Infantis. Several studies have documented their horizontal transfer into non-Infantis Salmonella serotypes across different countries [45,46,47]. This highlights the potential for broader dissemination of MDR traits across serotypes throughout the poultry production chain.
Taken together, these results provide comprehensive WGS-based molecular characterization of S. Infantis in Korea, supporting the urgent need for ongoing surveillance across all sectors of poultry production. A nationally integrated monitoring system grounded in the One Health approach is essential to mitigate the risk of emerging MDR Salmonella lineages and to control their spread both within Korea and beyond its borders.
4. Materials and Methods
4.1. Sample Collection
In 2023, 300 poultry meat samples were collected from 20 retail markets across five provinces in Korea: Gyeonggi, Gyeongsang, Jeolla, Chungcheong, and Gangwon. Four retail markets were arbitrarily selected in each province, and ten chicken and five duck samples were collected from each retail market. Samples were stored in a cooled icebox at 4 °C and transported to the laboratory within 24 h of collection for isolation of Salmonella spp.
4.2. Isolation and Identification of Salmonella spp.
Each whole-body chicken or duck meat sample was rinsed in a sterile plastic bag containing 400 mL of buffered peptone water (BPW; BD, Sparks, MD, USA). Each rinsate was incubated at 37 °C for 24 h. Following that, 100 µL of the enriched BPW cultures were transferred into 10 mL of Rappaport–Vassiliadis (RV; BD) broth and incubated at 42 °C for 24 h [26]. A loopful of each RV culture was streaked onto CHROMagarTM Salmonella (Paris, France) and incubated at 37 °C for 24 h. Up to three presumptive isolates were selected and subcultured on blood agar plates (Asan Pharm Co., Seoul, Korea) at 37 °C for 24 h. Identification of presumptive Salmonella isolates was determined by a TaqMan-based real-time PCR assay targeting the invA gene, as previously described [48].
4.3. Serotyping and Multi-Locus Sequence Typing
The serotypes of the Salmonella isolates were determined using multiplex PCR as described previously [49]. Multi-locus sequence typing (MLST) was conducted against seven housekeeping genes (aroC, dnaN, hemD, hisD, purE, sucA, and thrA), and STs were identified using the MLST database (https://pubmlst.org/organisms/salmonella-spp, accessed on 1 August 2023).
4.4. Antimicrobial Susceptibility Test
Antimicrobial susceptibility testing of Salmonella isolates against 16 antimicrobial agents was performed following the standardized protocols established by MFDS and the Animal and Plant Quarantine Agency [19,50,51]. The minimum inhibitory concentration (MIC) values were determined according to the broth microdilution method using the SensititreTM KRNV6F kit (Thermo Fisher Scientific, Waltham, MA, USA). The antimicrobial agents used were amoxicillin/clavulanic acid at a 2:1 ratio (AUG2), ampicillin (AMP), cefepime (FEP), cefotaxime (CTX), cefoxitin (FOX), ceftazidime (CAZ), chloramphenicol (CHL), ciprofloxacin (CIP), colistin (COL), gentamicin (GEN), meropenem (MERO), nalidixic acid (NAL), streptomycin (STR), sulfisoxazole (FIS), tetracycline (TET), and trimethoprim/sulfamethoxazole (SXT). Escherichia coli ATCC 25922 was used as the quality control reference strain in all tests. The MIC values of the isolates, except for STR, were interpreted according to the guidelines of the Clinical and Laboratory Standards Institute [52]. The breakpoint of STR was interpreted according to the National Antimicrobial Resistance Monitoring System [53]. Isolates resistant to CTX or CAZ or both were considered ESBL-producing Salmonella spp. for further identification as described below. When a strain was resistant to three or more antimicrobial classes, it was categorized as exhibiting MDR.
4.5. Characterization of the β-Lactamase-Encoding Genes
Detection of the β-lactamase-encoding genes was performed against the isolates resistant to CTX or CAZ or both. The presence of genes encoding different types of β-lactamases [Temoniera (TEM), sulphyldryl-variable (SHV), and cefotaxime (CTX-M)] was confirmed by PCR. Subtypes were determined by sequencing analysis as previously described [54,55]. For CTX-M-related genes, specific primer sets were used to identify each CTX-M group: CTX-M-1, CTX-M-2, CTX-M-8, CTX-M-9, and CTX-M-25. Sequence analyses were performed using BLAST at the National Center for Biotechnology Information (NCBI, https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 23 August 2023).
4.6. Whole-Genome Sequencing Analysis
A total of 28 representative ESBL-producing S. Infantis isolates were subjected to WGS to determine their genetic characteristics and phylogenetic relationships. WGS was performed by Macrogen (Seoul, Korea) following previously described [56].
To predict Salmonella serotype, in silico serotyping was performed using SeqSero2 (v.1.2.1) [57]. Single-nucleotide polymorphism (SNP) analysis was performed using the FDA-CFSAN SNP Pipeline. S. Infantis SINFA strain (GenBank accession number: N649235) was used as the reference genome for analysis [58]. A maximum likelihood (ML) tree was created using FastTree [59]. According to the framework for interpreting SNP analysis by the FDA-CFSAN, a cluster was defined when (1) there were 20 or fewer SNP differences, (2) the phylogenetic analysis showed a monophyletic relationship, and (3) there was a bootstrap support of 0.90 or higher [60].
Point mutations and genes leading to AMR were detected using AMRFinder Plus (version 3.11.26) [61]. PlasmidFinder (v. 2.1.6) was used to identify plasmid replicons [62]. Default parameters were used for all bioinformatics tools. The resulting data were visualized using iTOL (version 6.0; https://itol.embl.de/, accessed on 12 July 2024).
4.7. Detection and Reconstruction of pESI-like Plasmids
Plasmid sequences from the draft assemblies were typed and reconstructed using the MOB-Suite tool (v.3.1.8) with default parameters [63]. The reconstructed plasmids were annotated using Prokka (v.1.14.6). Plasmid MLST (pMLST) of the reconstructed plasmids was performed using pMLST (v.2.0) according to the IncI1 pMLST scheme on the Center for Genomic Epidemiology website (CGE, https://www.genomicepidemiology.org/, accessed on 13 August 2024). To determine the organization of AMR and virulence genes in the plasmids, AMRFinderPlus (v.3.11.26) and the Virulence Factor Database (VFDB; https://www.mgc.ac.cn/VFs/main.htm, accessed on 30 August 2024) were used, respectively [64]. Comparison and visualization of the characteristics of the plasmids were compared with those of the pESI-like plasmids available in the NCBI database using the OrthoANI algorithm [65] and visualized using the BLAST Ring Image Generator (BRIG, v.0.95) [66]. The WGS data of the pESI or pESI-like plasmids, pCVM44454 (GenBank accession number: CP016413.1), pN55391 (CP016411.1), 12037823-11_pESI-CTX-M-1 (OW849779.2), 119944_pESI (CP047882.1) and pS19 (CP092041.1), were used for the analysis.
4.8. Statistical Analysis
Chi-square tests were conducted to analyze significant differences in the isolation rates and AMR rates among Salmonella isolates from different samples using the SPSS program (v.26, Chicago, IL, USA). Statistical significance was set at a p-value of <0.05.
5. Conclusions
This study identified a high prevalence of Salmonella in retail poultry meat across Korea, with S. Infantis being the most dominant serotype in chicken meat. Notably, most S. Infantis isolates were ESBL-producing strains carrying blaCTX-M-65 gene. WGS of 28 representative ESBL-producing S. Infantis isolates revealed close genetic relatedness among isolates from geographically distinct regions, suggesting clonal dissemination of these MDR strains throughout the poultry supply chain.
All 28 isolates harbored a pESI-like megaplasmid encoding multiple AMR, virulence, and stress resistance genes, which is consistent with global reports of emerging S. Infantis lineages. Importantly, our findings highlight previously underemphasized aspects, such as potential cross-contamination between chicken and duck meat during processing at shared slaughter facilities. This underscores the necessity of implementing stricter hygiene measures and targeted interventions in mixed processing environments.
Taken together, these results provide a comprehensive molecular characterization of S. infantis in Korea, underscoring the urgent need for continuous nationwide monitoring, expanded genomic surveillance, and improved biosecurity measures in poultry production. Future studies incorporating isolates from farms, slaughterhouses, and clinical sources are essential to build a more integrated One Health framework to tackle the dissemination of MDR Salmonella and mitigate risks to public health.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics14040366/s1, Table S1: Characteristics of Salmonella spp. isolated from retail poultry meats in this study; Table S2: Metadata of pESI-like plasmids in 28 ESBL-producing S. Infantis strains; Table S3: Pairwise ANI values for the pESI-like plasmids.
Author Contributions
Conceptualization, Y.K., H.C., S.-J.Y., J.-C.C. and K.T.P.; methodology, Y.K., H.C. and M.L.; software, Y.K. and M.L.; validation, K.T.P.; formal analysis, Y.K., H.C., A.H. and K.T.P.; investigation, Y.K., H.C. and M.L.; resources, S.-J.Y., J.-C.C. and K.T.P.; data curation, Y.K. and H.C.; writing—original draft preparation, Y.K; writing—review and editing, Y.K., H.C., M.L., A.H., S.-J.Y., J.-C.C. and K.T.P.; visualization, Y.K. and M.L.; supervision, K.T.P.; project administration, K.T.P.; funding acquisition, K.T.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Ministry of Food and Drug Safety, grant number 23194MFDS012.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Genome sequence data of S. Infantis isolates have been deposited in the NCBI under BioProject accession number PRJNA1237260.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Zeng, Y.B.; Xiong, L.G.; Tan, M.F.; Li, H.Q.; Yan, H.; Zhang, L.; Yin, D.F.; Kang, Z.F.; Wei, Q.P.; Luo, L.G. Prevalence and Antimicrobial Resistance of Salmonella in Pork, Chicken, and Duck from Retail Markets of China. Foodborne Pathog. Dis. 2019, 16, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Chlebicz, A.; Śliżewska, K. Campylobacteriosis, Salmonellosis, Yersiniosis, and Listeriosis as Zoonotic Foodborne Diseases: A Review. Int. J. Environ. Res. Public Health 2018, 15, 863. [Google Scholar] [CrossRef] [PubMed]
- Ministry of Food and Drug Administration. Food poisoning statistics. Available online: https://www.foodsafetykorea.go.kr/portal/healthyfoodlife/foodPoisoningStat.do?menu_no=4425&menu_grp=MENU_NEW02 (accessed on 19 January 2024).
- Lamichhane, B.; Mawad, A.M.M.; Saleh, M.; Kelley, W.G.; Harrington, P.J., 2nd; Lovestad, C.W.; Amezcua, J.; Sarhan, M.M.; El Zowalaty, M.E.; Ramadan, H.; et al. Salmonellosis: An Overview of Epidemiology, Pathogenesis, and Innovative Approaches to Mitigate the Antimicrobial Resistant Infections. Antibiotics 2024, 13, 76. [Google Scholar] [CrossRef]
- Naushad, S.; Huang, H.; Ogunremi, D. Salmonella: A Brief Review. In Salmonella—Perspectives for Low-Cost Prevention, Control and Treatment; Huang, H., Naushad, S., Eds.; IntechOpen: Rijeka, Croatia, 2023. [Google Scholar]
- Cheng, R.A.; Eade, C.R.; Wiedmann, M. Embracing Diversity: Differences in Virulence Mechanisms, Disease Severity, and Host Adaptations Contribute to the Success of Nontyphoidal Salmonella as a Foodborne Pathogen. Front. Microbiol. 2019, 10, 1368. [Google Scholar] [CrossRef]
- Ferrari, R.G.; Rosario, D.K.A.; Cunha-Neto, A.; Mano, S.B.; Figueiredo, E.E.S.; Conte-Junior, C.A. Worldwide Epidemiology of Salmonella Serovars in Animal-Based Foods: A Meta-analysis. Appl. Environ. Microbiol. 2019, 85, e00591-19. [Google Scholar] [CrossRef]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef]
- Hur, J.; Jawale, C.; Lee, J.H. Antimicrobial resistance of Salmonella isolated from food animals: A review. Food Res. Int. 2012, 45, 819–830. [Google Scholar] [CrossRef]
- Nazeer, N.; Uribe-Diaz, S.; Rodriguez-Lecompte, J.C.; Ahmed, M. Antimicrobial peptides as an alternative to relieve antimicrobial growth promoters in poultry. Br. Poult. Sci. 2021, 62, 672–685. [Google Scholar] [CrossRef]
- Salim, H.M.; Huque, K.S.; Kamaruddin, K.M.; Beg, M. Global restriction of using antibiotic growth promoters and alternative strategies in poultry production. Sci. Prog. 2018, 101, 52–75. [Google Scholar] [CrossRef]
- Almansour, A.M.; Alhadlaq, M.A.; Alzahrani, K.O.; Mukhtar, L.E.; Alharbi, A.L.; Alajel, S.M. The Silent Threat: Antimicrobial-Resistant Pathogens in Food-Producing Animals and Their Impact on Public Health. Microorganisms 2023, 11, 2127. [Google Scholar] [CrossRef]
- World Health Organization. Critically Important Antimicrobials for Human Medicine: 6th Revision; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Alvarez, D.M.; Barrón-Montenegro, R.; Conejeros, J.; Rivera, D.; Undurraga, E.A.; Moreno-Switt, A.I. A review of the global emergence of multidrug-resistant Salmonella enterica subsp. enterica Serovar Infantis. Int. J. Food Microbiol. 2023, 403, 110297. [Google Scholar] [CrossRef]
- McMillan, E.A.; Wasilenko, J.L.; Tagg, K.A.; Chen, J.C.; Simmons, M.; Gupta, S.K.; Tillman, G.E.; Folster, J.; Jackson, C.R.; Frye, J.G. Carriage and Gene Content Variability of the pESI-Like Plasmid Associated with Salmonella Infantis Recently Established in United States Poultry Production. Genes 2020, 11, 1516. [Google Scholar] [CrossRef] [PubMed]
- Franco, A.; Leekitcharoenphon, P.; Feltrin, F.; Alba, P.; Cordaro, G.; Iurescia, M.; Tolli, R.; D’Incau, M.; Staffolani, M.; Di Giannatale, E.; et al. Emergence of a Clonal Lineage of Multidrug-Resistant ESBL-Producing Salmonella Infantis Transmitted from Broilers and Broiler Meat to Humans in Italy between 2011 and 2014. PLoS ONE 2015, 10, e0144802. [Google Scholar] [CrossRef] [PubMed]
- McMillan, E.A.; Weinroth, M.D.; Frye, J.G. Increased Prevalence of Salmonella Infantis Isolated from Raw Chicken and Turkey Products in the United States Is Due to a Single Clonal Lineage Carrying the pESI Plasmid. Microorganisms 2022, 10, 1478. [Google Scholar] [CrossRef]
- Tyson, G.H.; Li, C.; Harrison, L.B.; Martin, G.; Hsu, C.H.; Tate, H.; Tran, T.T.; Strain, E.; Zhao, S. A Multidrug-Resistant Salmonella Infantis Clone is Spreading and Recombining in the United States. Microb. Drug Resist. 2021, 27, 792–799. [Google Scholar] [CrossRef]
- Kang, H.S.; Ali, M.S.; Na, S.H.; Moon, B.Y.; Kim, J.I.; Hwang, Y.J.; Yoon, S.S.; Park, S.C.; Lim, S.K. Nationwide surveillance and characterization of the third-generation cephalosporin-resistant Salmonella enterica serovar infantis isolated from chickens in South Korea between 2010 and 2022. Heliyon 2024, 10, e37124. [Google Scholar] [CrossRef]
- Kim, M.B.; Jung, H.R.; Lee, Y.J. Emergence of Salmonella Infantis carrying the pESI megaplasmid in commercial farms of five major integrated broiler operations in Korea. Poult. Sci. 2024, 103, 103516. [Google Scholar] [CrossRef]
- Kim, M.B.; Lee, Y.J. Emergence of Salmonella Infantis carrying the pESI-like plasmid from eggs in egg grading and packing plants in Korea. Food Microbiol. 2024, 122, 104568. [Google Scholar] [CrossRef]
- Yeh, Y.; Purushothaman, P.; Gupta, N.; Ragnone, M.; Verma, S.C.; de Mello, A.S. Bacteriophage application on red meats and poultry: Effects on Salmonella population in final ground products. Meat Sci. 2017, 127, 30–34. [Google Scholar] [CrossRef]
- Hyeon, J.Y.; Chon, J.W.; Hwang, I.G.; Kwak, H.S.; Kim, M.S.; Kim, S.K.; Choi, I.S.; Song, C.S.; Park, C.; Seo, K.H. Prevalence, antibiotic resistance, and molecular characterization of Salmonella serovars in retail meat products. J. Food Prot. 2011, 74, 161–166. [Google Scholar] [CrossRef]
- Kim, M.S.; Lim, T.H.; Jang, J.H.; Lee, D.H.; Kim, B.Y.; Kwon, J.H.; Choi, S.W.; Noh, J.Y.; Hong, Y.H.; Lee, S.B.; et al. Prevalence and antimicrobial resistance of Salmonella species isolated from chicken meats produced by different integrated broiler operations in Korea. Poult. Sci. 2012, 91, 2370–2375. [Google Scholar] [CrossRef] [PubMed]
- Koh, Y.; Bae, Y.; Lee, Y.S.; Kang, D.H.; Kim, S.H. Prevalence and Characteristics of Salmonella spp. Isolated from Raw Chicken Meat in the Republic of Korea. J. Microbiol. Biotechnol. 2022, 32, 1307–1314. [Google Scholar] [CrossRef] [PubMed]
- Yoon, R.H.; Cha, S.Y.; Wei, B.; Roh, J.H.; Seo, H.S.; Oh, J.Y.; Jang, H.K. Prevalence of Salmonella isolates and antimicrobial resistance in poultry meat from South Korea. J. Food Prot. 2014, 77, 1579–1582. [Google Scholar] [CrossRef]
- Lunara Santos Pavelquesi, S.; Carolina Almeida de Oliveira Ferreira, A.; Fernandes Silva Rodrigues, L.; Maria de Souza Silva, C.; Cristina Rodrigues da Silva, I.; Castilho Orsi, D. Prevalence and Antimicrobial Resistance of Salmonella spp. Isolated From Chilled Chicken Meat Commercialized at Retail in Federal District, Brazil. J. Food Prot. 2023, 86, 100130. [Google Scholar] [CrossRef]
- Bolkenov, B.; Lee, K.Y.; Atwill, E.R.; Pitesky, M.; Rickard, M.; Hung-Fan, M.; Shafii, M.; Lavelle, K.; Huang, A.; Sebti, J.; et al. Phenotypic and genotypic characterization of antimicrobial resistance of non-typhoidal Salmonella from retail meat in California. Int. J. Food Microbiol. 2024, 421, 110785. [Google Scholar] [CrossRef]
- Maddocks, S.; Olma, T.; Chen, S. Comparison of CHROMagar Salmonella medium and xylose-lysine-desoxycholate and Salmonella-Shigella agars for isolation of Salmonella strains from stool samples. J. Clin. Microbiol. 2002, 40, 2999–3003. [Google Scholar] [CrossRef]
- Su, L.H.; Chiu, C.H.; Chu, C.; Ou, J.T. Antimicrobial resistance in nontyphoid Salmonella serotypes: A global challenge. Clin. Infect. Dis. 2004, 39, 546–551. [Google Scholar] [CrossRef]
- Korea Disease Control and Prevention Agency. One Health AMR. Available online: https://nih.go.kr/nohas/common/main.do (accessed on 20 March 2025).
- World Health Organization. Integrated Surveillance of Antimicrobial Resistance in Foodborne Bacteria: Application of a One Health Approach: Guidance from the WHO Advisory Group on Integrated Surveillanec of Antimicrobial Resistance (AGISAR); World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Alzahrani, K.O.; Al-Reshoodi, F.M.; Alshdokhi, E.A.; Alhamed, A.S.; Al Hadlaq, M.A.; Mujallad, M.I.; Mukhtar, L.E.; Alsufyani, A.T.; Alajlan, A.A.; Al Rashidy, M.S.; et al. Antimicrobial resistance and genomic characterization of Salmonella enterica isolates from chicken meat. Front. Microbiol. 2023, 14, 1104164. [Google Scholar] [CrossRef]
- Castello, A.; Piraino, C.; Butera, G.; Alio, V.; Cardamone, C.; Oliveri, G.; Cascone, G.; Ciravolo, C.; Costa, A. Prevalence and antimicrobial resistance profiles of Salmonella spp. in poultry meat. Ital. J. Food Saf. 2023, 12, 11135. [Google Scholar] [CrossRef]
- Perin, A.P.; Martins, B.T.F.; Barreiros, M.A.B.; Yamatogi, R.S.; Nero, L.A.; Dos Santos Bersot, L. Occurrence, quantification, pulse types, and antimicrobial susceptibility of Salmonella sp. isolated from chicken meat in the state of Paraná, Brazil. Braz. J. Microbiol. 2020, 51, 335–345. [Google Scholar] [CrossRef]
- Jeon, H.Y.; Kim, Y.B.; Lim, S.K.; Lee, Y.J.; Seo, K.W. Characteristics of cephalosporin-resistant Salmonella isolates from poultry in Korea, 2010-2017. Poult. Sci. 2019, 98, 957–965. [Google Scholar] [CrossRef] [PubMed]
- Tate, H.; Folster, J.P.; Hsu, C.H.; Chen, J.; Hoffmann, M.; Li, C.; Morales, C.; Tyson, G.H.; Mukherjee, S.; Brown, A.C.; et al. Comparative Analysis of Extended-Spectrum-β-Lactamase CTX-M-65-Producing Salmonella enterica Serovar Infantis Isolates from Humans, Food Animals, and Retail Chickens in the United States. Antimicrob. Agents Chemother. 2017, 61, e00488-17. [Google Scholar] [CrossRef] [PubMed]
- Aviv, G.; Tsyba, K.; Steck, N.; Salmon-Divon, M.; Cornelius, A.; Rahav, G.; Grassl, G.A.; Gal-Mor, O. A unique megaplasmid contributes to stress tolerance and pathogenicity of an emergent Salmonella enterica serovar Infantis strain. Environ. Microbiol. 2014, 16, 977–994. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Bertini, A.; Villa, L.; Falbo, V.; Hopkins, K.L.; Threlfall, E.J. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 2005, 63, 219–228. [Google Scholar] [CrossRef]
- Vázquez, X.; Fernández, J.; Rodríguez-Lozano, J.; Calvo, J.; Rodicio, R.; Rodicio, M.R. Genomic Analysis of Two MDR Isolates of Salmonella enterica Serovar Infantis from a Spanish Hospital Bearing the blaCTX-M-65 Gene with or without fosA3 in pESI-like Plasmids. Antibiotics 2022, 11, 786. [Google Scholar] [CrossRef]
- Xu, Y.; Jing, Y.; Hu, L.; Cheng, Q.; Gao, H.; Zhang, Z.; Yang, H.; Zhao, Y.; Zhou, D.; Yin, Z.; et al. IncFIB-4.1 and IncFIB-4.2 Single-Replicon Plasmids: Small Backbones with Large Accessory Regions. Infect. Drug Resist. 2022, 15, 1191–1203. [Google Scholar] [CrossRef]
- García-Soto, S.; Abdel-Glil, M.Y.; Tomaso, H.; Linde, J.; Methner, U. Emergence of Multidrug-Resistant Salmonella enterica Subspecies enterica Serovar Infantis of Multilocus Sequence Type 2283 in German Broiler Farms. Front. Microbiol. 2020, 11, 1741. [Google Scholar] [CrossRef]
- Diamant, I.; Adani, B.; Sylman, M.; Rahav, G.; Gal-Mor, O. The transcriptional regulation of the horizontally acquired iron uptake system, yersiniabactin and its contribution to oxidative stress tolerance and pathogenicity of globally emerging salmonella strains. Gut Microbes 2024, 16, 2369339. [Google Scholar] [CrossRef]
- Alba, P.; Carfora, V.; Feltrin, F.; Diaconu, E.L.; Sorbara, L.; Dell’Aira, E.; Cerci, T.; Ianzano, A.; Donati, V.; Franco, A.; et al. Evidence of structural rearrangements in ESBL-positive pESI(like) megaplasmids of S. Infantis. FEMS Microbiol. Lett. 2023, 370, fnad014. [Google Scholar] [CrossRef]
- Cohen, E.; Kriger, O.; Amit, S.; Davidovich, M.; Rahav, G.; Gal-Mor, O. The emergence of a multidrug resistant Salmonella Muenchen in Israel is associated with horizontal acquisition of the epidemic pESI plasmid. Clin. Microbiol. Infect. 2022, 28, 1499.e7–1499.e14. [Google Scholar] [CrossRef]
- Li, C.; Tate, H.; Huang, X.; Hsu, C.H.; Harrison, L.B.; Zhao, S.; Fortenberry, G.Z.; Dessai, U.; McDermott, P.F.; Strain, E.A. The spread of pESI-mediated extended-spectrum cephalosporin resistance in Salmonella serovars-Infantis, Senftenberg, and Alachua isolated from food animal sources in the United States. PLoS ONE 2024, 19, e0299354. [Google Scholar] [CrossRef]
- Dos Santos, A.M.P.; Panzenhagen, P.; Ferrari, R.G.; Conte-Junior, C.A. Large-scale genomic analysis reveals the pESI-like megaplasmid presence in Salmonella Agona, Muenchen, Schwarzengrund, and Senftenberg. Food Microbiol. 2022, 108, 104112. [Google Scholar] [CrossRef] [PubMed]
- Kasturi, K.N.; Drgon, T. Real-Time PCR Method for Detection of Salmonella spp. in Environmental Samples. Appl. Environ. Microbiol. 2017, 83, e00644-17. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Frye, J.G.; Hu, J.; Fedorka-Cray, P.J.; Gautom, R.; Boyle, D.S. Multiplex PCR-based method for identification of common clinical serotypes of Salmonella enterica subsp. enterica. J. Clin. Microbiol. 2006, 44, 3608–3615. [Google Scholar] [CrossRef]
- Kang, H.; Kim, H.; Lee, J.; Jeon, J.H.; Kim, S.; Park, Y.; Joo, I.; Kim, H. Genetic Characteristics of Multidrug-Resistant Salmonella Isolated from Poultry Meat in South Korea. Microorganisms 2024, 12, 1646. [Google Scholar] [CrossRef]
- Animal and Plant Quarantine Agency, Ministry of Agriculture Food and Rural Affairs, Ministry of Food and Drug Safety. National Antimicrobial Usage and Resistance Monitoring in Animals and Animal Products, 2023; Animal and Plant Quarantine Agency, Ministry of Agriculture Food and Rural Affairs, Ministry of Food and Drug Safety: Seoul, Republic of Korea, 2023. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 31st ed.; CLSI supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2021. [Google Scholar]
- National Antimicrobial Resistance Monitoring System. The National Antimicrobial Resistance Monitoring System: Enteric Bacteria; National Antimicrobial Resistance Monitoring System: Atlanta, GA, USA, 2021. [Google Scholar]
- Kim, D.; Yoon, E.J.; Hong, J.S.; Choi, M.H.; Kim, H.S.; Kim, Y.R.; Kim, Y.A.; Uh, Y.; Shin, K.S.; Shin, J.H.; et al. Major Bloodstream Infection-Causing Bacterial Pathogens and Their Antimicrobial Resistance in South Korea, 2017-2019: Phase I Report From Kor-GLASS. Front. Microbiol. 2021, 12, 799084. [Google Scholar] [CrossRef]
- Kang, J.H.; Bae, I.K.; Kwon, S.B.; Jeong, S.H.; Lee, J.; Lee, W.G.; Kang, J.O.; Ahn, J.Y.; Hong, S.G.; Shin, J.H.; et al. Prevalence of Ambler Class A Extended-Spectrum beta-Lactamase-Producing Escherichia coli and Klebsiella pneumoniae Isolates in Korea. Korean J. Clin. Microbiol. 2005, 8, 17–25. [Google Scholar]
- Kim, Y.; Cho, H.; Jang, B.; Lee, M.; Park, K.T. Molecular characterization of emerging multi-drug resistant Clostridium perfringens isolated from pork production chains in Korea. Food Microbiol. 2025, 128, 104729. [Google Scholar] [CrossRef]
- Zhang, S.; den Bakker, H.C.; Li, S.; Chen, J.; Dinsmore, B.A.; Lane, C.; Lauer, A.C.; Fields, P.I.; Deng, X. SeqSero2: Rapid and Improved Salmonella Serotype Determination Using Whole-Genome Sequencing Data. Appl. Environ. Microbiol. 2019, 85, e01746-19. [Google Scholar] [CrossRef]
- Davis, S.; Pettengill, J.B.; Luo, Y.; Payne, J.; Shpuntoff, A.; Rand, H.; Strain, E. CFSAN SNP Pipeline: An automated method for constructing SNP matrices from next-generation sequence data. PeerJ Comput. Sci. 2015, 1, e20. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Pightling, A.W.; Pettengill, J.B.; Luo, Y.; Baugher, J.D.; Rand, H.; Strain, E. Interpreting Whole-Genome Sequence Analyses of Foodborne Bacteria for Regulatory Applications and Outbreak Investigations. Front. Microbiol. 2018, 9, 1482. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Robertson, J.; Nash, J.H.E. MOB-suite: Software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb. Genom. 2018, 4, e000206. [Google Scholar] [CrossRef]
- Cho, H.; Kim, Y.; Hassan, A.; Park, K.T. Whole-genome sequence-based comparison of antimicrobial resistant diarrheagenic Escherichia coli in pork and chicken production chains in Korea. Int. J. Food Microbiol. 2025, 431, 111085. [Google Scholar] [CrossRef]
- Lee, I.; Ouk Kim, Y.; Park, S.C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).