Synthesis and Biological Evaluation of New Pyridothienopyrimidine Derivatives as Antibacterial Agents and Escherichia coli Topoisomerase II Inhibitors
Abstract
:1. Introduction
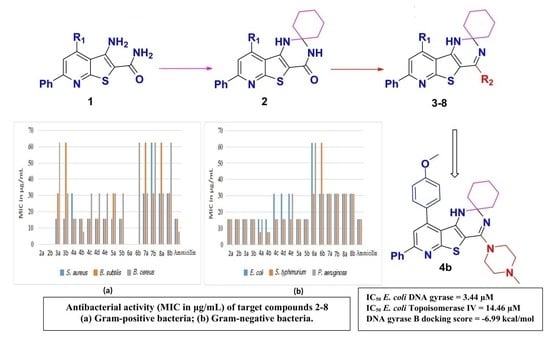
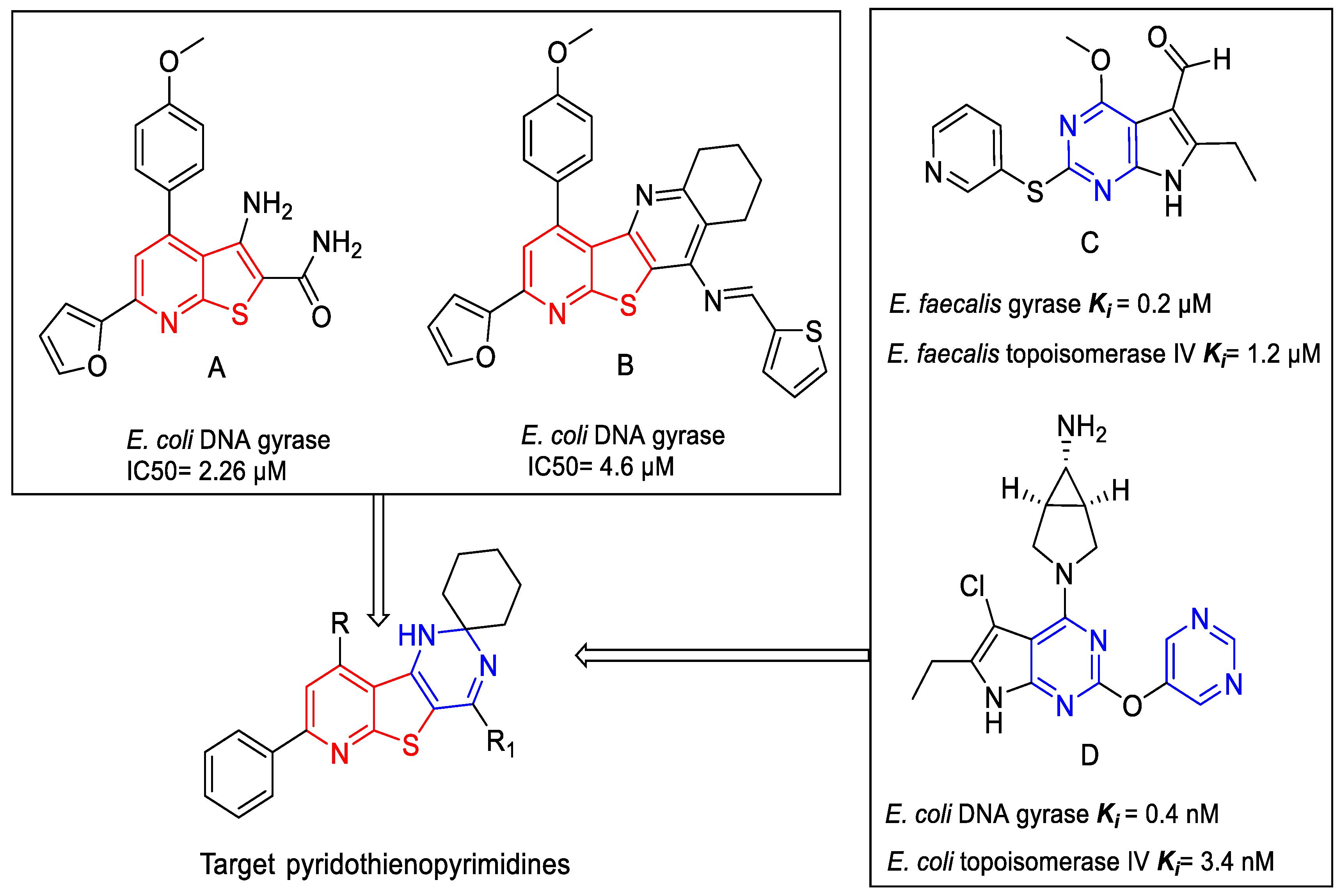
2. Results and Discussion
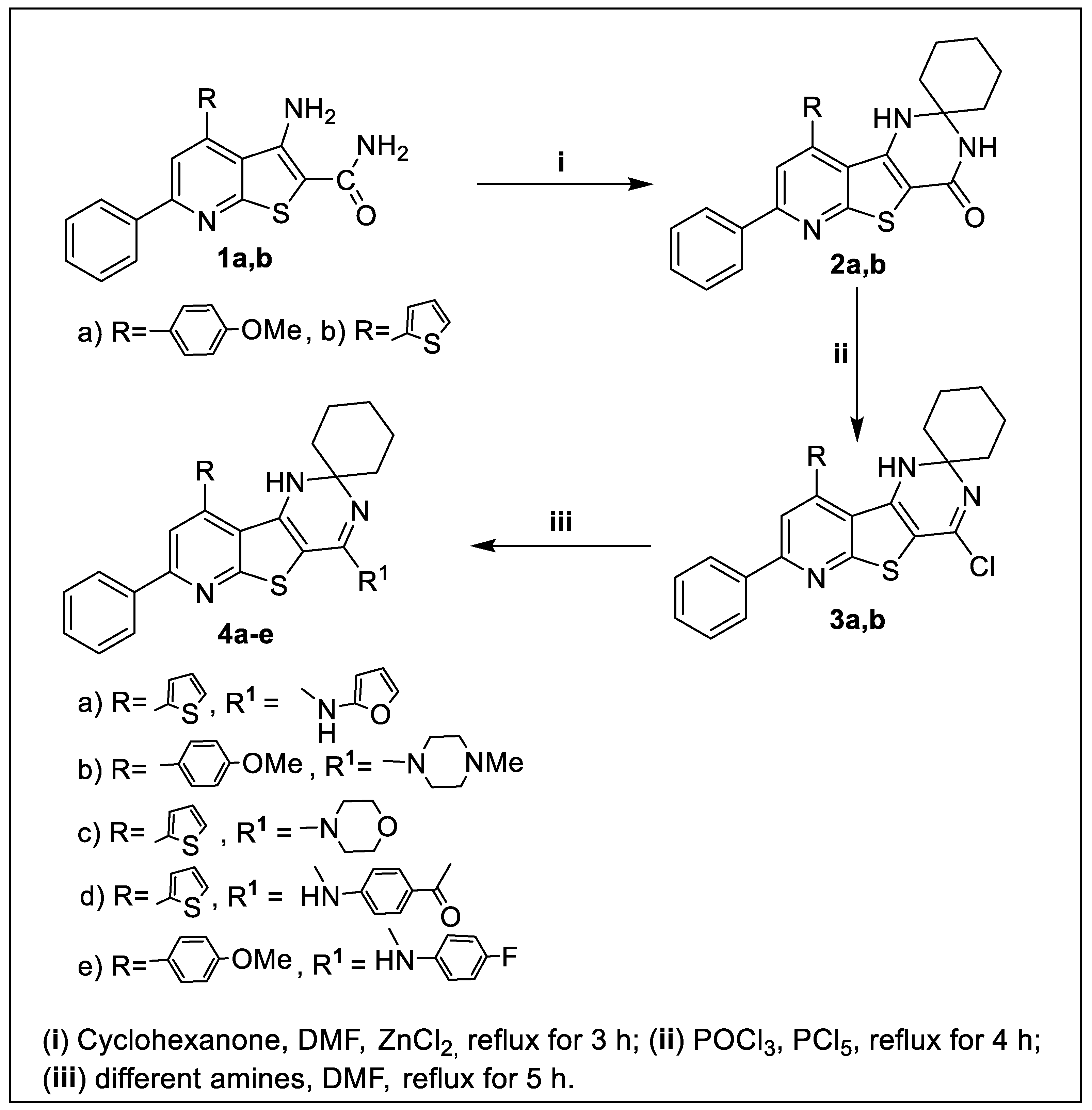
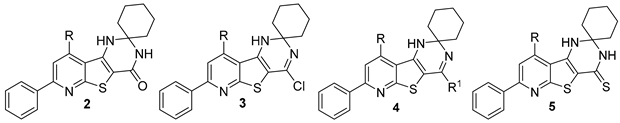
2.1. Chemistry
2.2. Antibacterial Activity
2.3. DNA Gyrase and Topoisomerase IV Inhibitory Activity
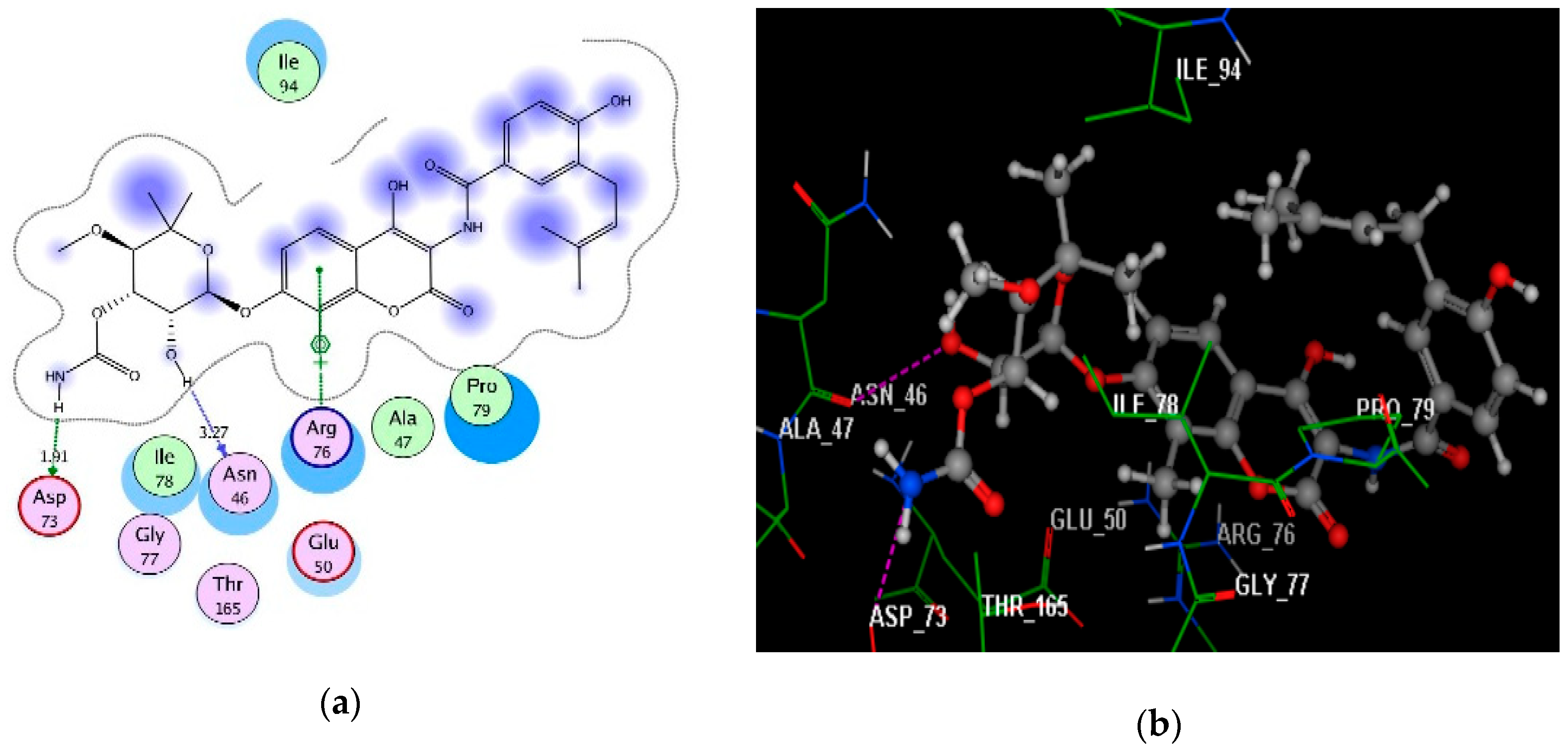
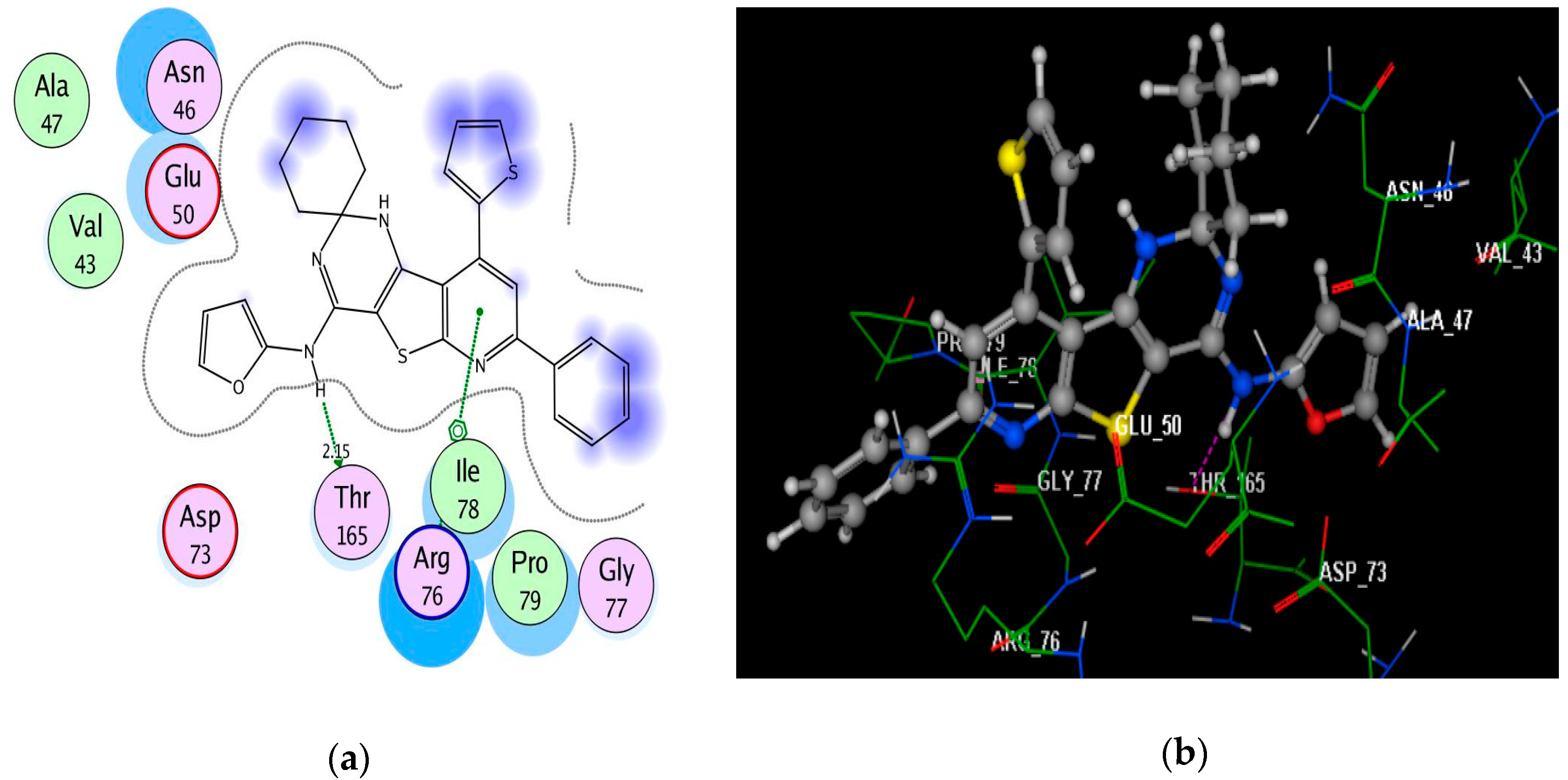
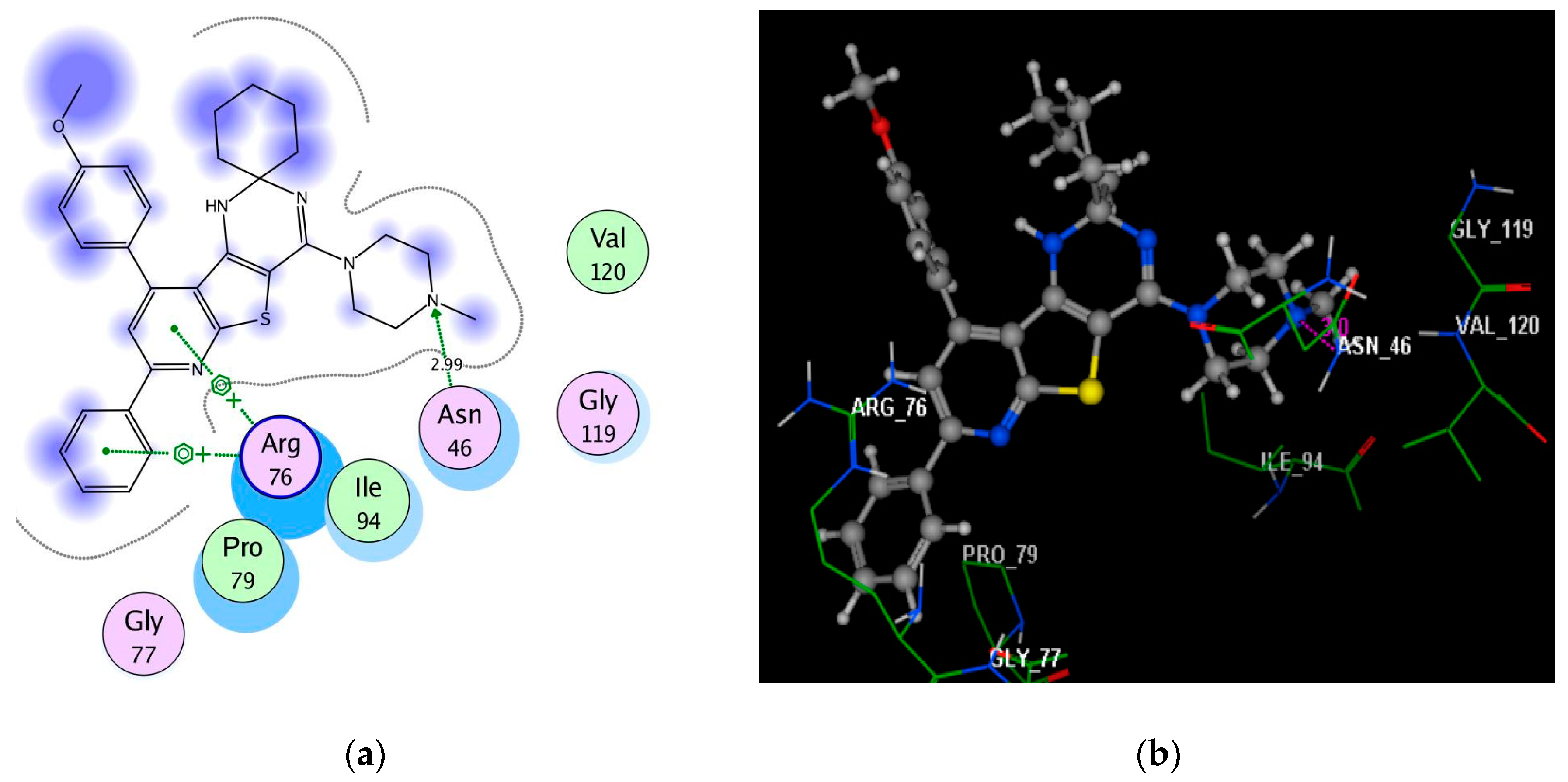
2.4. Molecular Docking Studies
3. Materials and Methods
3.1. Chemistry
3.1.1. General Consideration
3.1.2. Synthesis of Pyrido[3’,2’:4,5]Thieno[3,2-d]Pyrimidin]-4’(3’H)-ones 2a,b
3.1.3. Synthesis of 4’-Chloropyrido[3’,2’:4,5]Thieno[3,2-d]Pyrimidine Derivatives 3a,b
3.1.4. Synthesis of Pyrido[3’,2’:4,5]Thieno[3,2-d]Pyrimidin]-4’-Amines 4a–e
3.1.5. Synthesis of Pyrido[3’,2’:4,5]Thieno[3,2-d]Pyrimidine]-4’(3’H)-Thiones 5a,b
3.1.6. Synthesis of Ethyl 2-(Pyrido[3’,2’:4,5]Thieno[3,2-d]Pyrimidin-4-yl-Thio)Acetates 6a,b
3.1.7. Synthesis of Pyrido[3’,2’:4,5] Thieno[3,2-d]Pyrimidin]-4’-yl)thio)Acetohydrazides 7a,b
3.1.8. Synthesis of N’-(Arylidene)-2-(Pyrido[3’,2’:4,5]Thieno[3,2-d] Pyrimidin]-4’-yl)Thio) Acetohydrazides 8a,b
3.2. In Vitro Antibacterial Screening
3.3. DNA Gyrase Supercoiling and Topoisomerase IV Decatenation Inhibition Assays
3.4. Molecular Docking Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sifri, Z.C.; Chokshi, A.; Cennimo, D.; Horng, H. Global contributors to antibiotic resistance. J. Glob. Infect. Dis. 2019, 11, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Lomazzi, M.; Moore, M.; Johnson, A.; Balasegaram, M.; Borisch, B. Antimicrobial resistance–moving forward? BMC Public Health 2019, 19, 858. [Google Scholar] [CrossRef] [PubMed]
- De Kraker, M.E.A.; Stewardson, A.J.; Harbarth, S. Will 10 Million People Die a Year due to Antimicrobial Resistance by 2050? PLoS Med. 2016, 13, e1002184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gwynn, M.N.; Portnoy, A.; Rittenhouse, S.F.; Payne, D.J. Challenges of antibacterial discovery revisited. Ann. N. Y. Acad. Sci. 2010, 1213, 5–19. [Google Scholar] [CrossRef]
- Yi, L.; Lu, X. New Strategy on antimicrobial-resistance: Inhibitors of DNA replication enzymes. Curr. Med. Chem. 2019, 26, 1761–1787. [Google Scholar] [CrossRef]
- Klostermeier, D. Why two? On the role of (A-)symmetry in negative supercoiling of DNA by gyrase. Int. J. Mol. Sci. 2018, 19, 1489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berger, J.; Schoeffler, A. Recent advances in understanding structure–function relationships in the type II topoisomerase mechanism. Biochem. Soc. Trans. 2005, 33, 1465–1470. [Google Scholar] [CrossRef] [PubMed]
- Van Eijk, E.; Wittekoek, B.; Kuijper, E.J.; Smits, W.K. DNA replication proteins as potential targets for antimicrobials in drug-resistant bacterial pathogens. J. Antimicrob. Chemother. 2017, 72, 1275–1284. [Google Scholar] [CrossRef]
- Singh, S.B. Confronting the challenges of discovery of novel antibacterial agents. Bioorganic Med. Chem. Lett. 2014, 24, 3683–3689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collin, F.; Karkare, S.; Maxwell, A. Exploiting bacterial DNA gyrase as a drug target: Current state and perspectives. Appl. Microbiol. Biotechnol. 2011, 92, 479–497. [Google Scholar] [CrossRef] [Green Version]
- Aldred, K.J.; Kerns, R.J.; Osheroff, N. Mechanism of quinolone action and resistance. Biochemistry 2014, 53, 1565–1574. [Google Scholar] [CrossRef] [PubMed]
- Mustaev, A.; Malik, M.; Zhao, X.; Kurepina, N.; Luan, G.; Oppegard, L.M.; Hiasa, H.; Marks, K.R.; Kerns, R.J.; Berger, J.M.; et al. Fluoroquinolone-Gyrase-DNA Complexes. J. Biol. Chem. 2014, 289, 12300–12312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, E.S.; Hooper, D. Clinical importance and epidemiology of quinolone resistance. Infect. Chemother. 2014, 46, 226–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Correia, S.; Poeta, P.; Hébraud, M.; Capelo, J.L.; Igrejas, G. Mechanisms of quinolone action and resistance: Where do we stand? J. Med. Microbiol. 2017, 66, 551–559. [Google Scholar] [CrossRef]
- Badshah, S.L.; Ullah, A. New developments in non-quinolone-based antibiotics for the inhibiton of bacterial gyrase and topoisomerase IV. Eur. J. Med. Chem. 2018, 152, 393–400. [Google Scholar] [CrossRef]
- Jakopin, Ž.; Ilaš, J.; Barančoková, M.; Brvar, M.; Tammela, P.; Dolenc, M.S.; Tomašič, T.; Kikelj, D. Discovery of substituted oxadiazoles as a novel scaffold for DNA gyrase inhibitors. Eur. J. Med. Chem. 2017, 130, 171–184. [Google Scholar] [CrossRef] [Green Version]
- Zidar, N.; Macut, H.; Tomašič, T.; Mašič, L.P.; Ilaš, J.; Zega, A.; Tammela, P.; Kikelj, D. New N-phenyl-4,5-dibromopyrrolamides as DNA gyrase B inhibitors. MedChemComm 2019, 10, 1007–1017. [Google Scholar] [CrossRef]
- Kumar, S.; Narasimhan, B. Therapeutic potential of heterocyclic pyrimidine scaffolds. Chem. Central J. 2018, 12, 38. [Google Scholar] [CrossRef]
- Hung, J.M.; Arabshahi, H.J.; Leung, E.; Reynisson, J.; Barker, D. Synthesis and cytotoxicity of thieno[2,3-b]pyridine and furo[2,3-b]pyridine derivatives. Eur. J. Med. Chem. 2014, 86, 420–437. [Google Scholar] [CrossRef]
- Arabshahi, H.J.; Pilkington, L.I.; Jeon, C.Y.; Song, M.; Gridel, L.-M.; Zakharenko, A.L.; Lavrik, O.I.; Van Rensburg, M.; Leung, E.; Barker, D.; et al. A synthesis, in silico, in vitro and in vivo study of thieno[2,3-b]pyridine anticancer analogues. MedChemComm 2015, 6, 1987–1997. [Google Scholar] [CrossRef]
- Naguib, B.H.; El-Nassan, H.B. Synthesis of new thieno[2,3-b]pyridine derivatives as pim-1 inhibitors. J. Enzym. Inhib. Med. Chem. 2016, 31, 1718–1725. [Google Scholar] [CrossRef] [Green Version]
- Al-Trawneh, S.A.; El-Abadelah, M.M.; Zahra, J.A.; Al-Taweel, S.A.; Zani, F.; Incerti, M.; Cavazzoni, A.; Vicini, P. Synthesis and biological evaluation of tetracyclic thienopyridones as antibacterial and antitumor agents. Bioorganic Med. Chem. 2011, 19, 2541–2548. [Google Scholar] [CrossRef] [PubMed]
- El-Deen, E.M.M.; El-Hameed, E.K.A. Synthesis and in vitro biological evaluation of new tetracyclic pyridothienoquinolines as potential antimicrobial agents. Acta Pol. Pharm. Drug Res. 2017, 74, 837–847. [Google Scholar]
- Amorim, R.; De Meneses, M.D.F.; Borges, J.C.; Pinheiro, L.C.S.; Caldas, L.A.; Cirne-Santos, C.C.; De Mello, M.V.; De Souza, A.M.T.; Castro, H.C.; Paixão, I.C.N.D.P.; et al. Thieno[2,3-b]pyridine derivatives: A new class of antiviral drugs against Mayaro virus. Arch. Virol. 2017, 162, 1577–1587. [Google Scholar] [CrossRef] [PubMed]
- Schnute, M.E.; Anderson, D.J.; Brideau, R.J.; Ciske, F.L.; Collier, S.A.; Cudahy, M.M.; Eggen, M.; Genin, M.J.; Hopkins, T.A.; Judge, T.M.; et al. 2-Aryl-2-hydroxyethylamine substituted 4-oxo-4,7-dihydrothieno[2,3-b]pyridines as broad-spectrum inhibitors of human herpesvirus polymerases. Bioorganic Med. Chem. Lett. 2007, 17, 3349–3353. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, Y.; Wang, X.-Y.; Wang, B.; He, H.-Y.; Liu, J.-Y.; Xiang, M.-L.; He, J.; Wu, X.-H.; Yang, L. Synthesis, preliminary structure–activity relationships, and in vitro biological evaluation of 6-aryl-3-amino-thieno[2,3-b]pyridine derivatives as potential anti-inflammatory agents. Bioorganic Med. Chem. Lett. 2013, 23, 2349–2352. [Google Scholar] [CrossRef]
- Saito, K.; Nakao, A.; Shinozuka, T.; Shimada, K.; Matsui, S.; Oizumi, K.; Yano, K.; Ohata, K.; Nakai, D.; Nagai, Y.; et al. Discovery and structure–activity relationship of thienopyridine derivatives as bone anabolic agents. Bioorganic Med. Chem. 2013, 21, 1628–1642. [Google Scholar] [CrossRef]
- Gad-Elkareem, M.A.; Abdel-Fattah, A.M.; Elneairy, M.A.A. Pyridine-2(1 H)-thione in heterocyclic synthesis: Synthesis and antimicrobial activity of some new thio-substituted ethyl nicotinate, thieno[2,3- b ]pyridine and pyridothienopyrimidine derivatives. J. Sulfur Chem. 2011, 32, 273–286. [Google Scholar] [CrossRef]
- El-Aleam, R.H.A.; George, R.F.; Hassan, G.S.; Abdel-Rahman, H.M. Synthesis of 1,2,4-triazolo[1,5-a]pyrimidine derivatives: Antimicrobial activity, DNA Gyrase inhibition and molecular docking. Bioorganic Chem. 2020, 94, 103411. [Google Scholar] [CrossRef]
- Patil, S.B. Biological and medicinal significance of pyrimidines: A review. Int. J. Pharm Sci. Res. 2018, 9, 44–52. [Google Scholar] [CrossRef]
- Fayed, A.A.; Amr, A.E.-G.E.; Al-Omar, M.A.; Mostafa, E.E. Synthesis and antimicrobial activity of some new substituted pyrido[3′,2′:4,5]thieno[3,2-d]-pyrimidinone derivatives. Russ. J. Bioorganic Chem. 2014, 40, 308–313. [Google Scholar] [CrossRef]
- El-Essawy, F.A.; Boshta, N.M.; El-Sawaf, A.K.; Nassar, A.A.; Khalafallah, M.S. Synthesis of novel 3-substituted pyridothienopyrimidine derivatives with biological evaluation as antimicrobial agents. Chem. Res. Chin. Univ. 2016, 32, 967–972. [Google Scholar] [CrossRef]
- Kadah, M.S. Synthesis of some new thienopyridine and pyridothienopyrimidine derivatives with expected antitumor activity. Int. J. Med Sci. 2016, 3, 5–10. [Google Scholar] [CrossRef]
- Naguib, B.H.; El-Nassan, H.B.; Abdelghany, T.M. Synthesis of new pyridothienopyrimidinone derivatives as Pim-1 inhibitors. J. Enzym. Inhib. Med. Chem. 2017, 32, 457–467. [Google Scholar] [CrossRef]
- El-Nassan, H.B.; Naguib, B.H.; Beshay, E.A. Synthesis of new pyridothienopyrimidinone and pyridothienotriazolopyrimidine derivatives as pim-1 inhibitors. J. Enzym. Inhib. Med. Chem. 2017, 33, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Aziz, Y.M.A.; Said, M.M.; El Shihawy, H.A.; Abouzid, K.A. Discovery of novel tricyclic pyrido[3′,2′:4,5]thieno[3,2-d]pyrimidin-4-amine derivatives as VEGFR-2 inhibitors. Bioorganic Chem. 2015, 60, 1–12. [Google Scholar] [CrossRef] [PubMed]
- El-Deen, E.M.; El-Meguid, E.A.A.; Hasabelnaby, S.; Karam, E.A.; Nossier, E.S.; El-Deen, E.M.; El-Meguid, E.A.A. Synthesis, Docking Studies, and In Vitro Evaluation of Some Novel Thienopyridines and Fused Thienopyridine-Quinolines as Antibacterial Agents and DNA Gyrase Inhibitors. Molecules 2019, 24, 3650. [Google Scholar] [CrossRef] [Green Version]
- Tari, L.W.; Trzoss, M.; Bensen, D.C.; Li, X.; Chen, Z.; Lam, T.; Zhang, J.; Creighton, C.J.; Cunningham, M.L.; Kwan, B.; et al. Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. Bioorganic Med. Chem. Lett. 2013, 23, 1529–1536. [Google Scholar] [CrossRef] [PubMed]
- Trzoss, M.; Bensen, D.C.; Li, X.; Chen, Z.; Lam, T.; Zhang, J.; Creighton, C.J.; Cunningham, M.L.; Kwan, B.; Stidham, M.; et al. Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE), Part II: Development of inhibitors with broad spectrum, Gram-negative antibacterial activity. Bioorganic Med. Chem. Lett. 2013, 23, 1537–1543. [Google Scholar] [CrossRef]
- Patel, R.; Park, S. An evolving role of piperazine moieties in drug design and discovery. Mini-Rev. Med. Chem. 2013, 13, 1579–1601. [Google Scholar] [CrossRef]
- Patil, P.; Madhavachary, R.; Kurpiewska, K.; Kalinowska-Tłuścik, J.; Dömling, A. De Novo assembly of highly substituted morpholines and piperazines. Org. Lett. 2017, 19, 642–645. [Google Scholar] [CrossRef] [PubMed]
- Popiołek, Ł. Hydrazide–hydrazones as potential antimicrobial agents: Overview of the literature since 2010. Med. Chem. Res. 2016, 26, 287–301. [Google Scholar] [CrossRef] [Green Version]
- Rollas, S.; Küçükgüzel, S.G. Biological activities of hydrazone derivatives. Molecules 2007, 12, 1910–1939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michael, J.M.; El-Zahar, M.I.; El-Masry, A.M.; Mohi El-Deen, E.M. New pyrido [3,2:4,5] thieno [3,2-D] pyrimidines of possible antimicrobial activity. Al-Azhar Bull. Sci. 1992, 3, 767. [Google Scholar]
- Sharanin, Y.A.; Motrosova, S.V. Cyclization Reaction of Nitriles Part 56. Synthesis and transformations of substituted 6-aryl-4-(2-thienyl)-3-cyano-pyridine-2(1h)-thiones. Zh. Org. Khim. 1996, 32, 1251–1255. [Google Scholar]
- Elzahabi, H.S.A.; Nossier, E.S.; Khalifa, N.M.; AlAsfoury, R.A.; El-Manawaty, M.A. Anticancer evaluation and molecular modeling of multi-targeted kinase inhibitors based pyrido[2,3-d]pyrimidine scaffold. J. Enzym. Inhib. Med. Chem. 2018, 33, 546–557. [Google Scholar] [CrossRef] [Green Version]
- Holdgate, G.A.; Tunnicliffe, A.; Ward, W.H.J.; Weston, S.A.; Rosenbrock, G.; Barth, P.T.; Taylor, I.W.F.; Pauptit, R.A.; Timms, D. The entropic penalty of ordered water accounts for weaker binding of the antibiotic novobiocin to a resistant mutant of DNA gyrase: A thermodynamic and crystallographic study. Biochem. 1997, 36, 9663–9673. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- Phillips, J.W.; Goetz, M.A.; Smith, S.K.; Zink, D.L.; Polishook, J.; Onishi, R.; Salowe, S.; Wiltsie, J.; Allocco, J.; Sigmund, J.; et al. Discovery of Kibdelomycin, A Potent new class of bacterial type II topoisomerase inhibitor by chemical-genetic profiling in staphylococcus aureus. Chem. Biol. 2011, 18, 955–965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maxwell, A.; Burton, N.P.; O’Hagan, N. High-throughput assays for DNA gyrase and other topoisomerases. Nucleic Acids Res. 2006, 34, e104. [Google Scholar] [CrossRef] [Green Version]
- Bellon, S.; Parsons, J.D.; Wei, Y.; Hayakawa, K.; Swenson, L.L.; Charifson, P.S.; Lippke, J.A.; Aldape, R.; Gross, C.H. Crystal structures of escherichia coli topoisomerase IV pare subunit (24 and 43 Kilodaltons): A single residue dictates differences in novobiocin potency against topoisomerase IV and DNA gyrase. Antimicrob. Agents Chemother. 2004, 48, 1856–1864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |

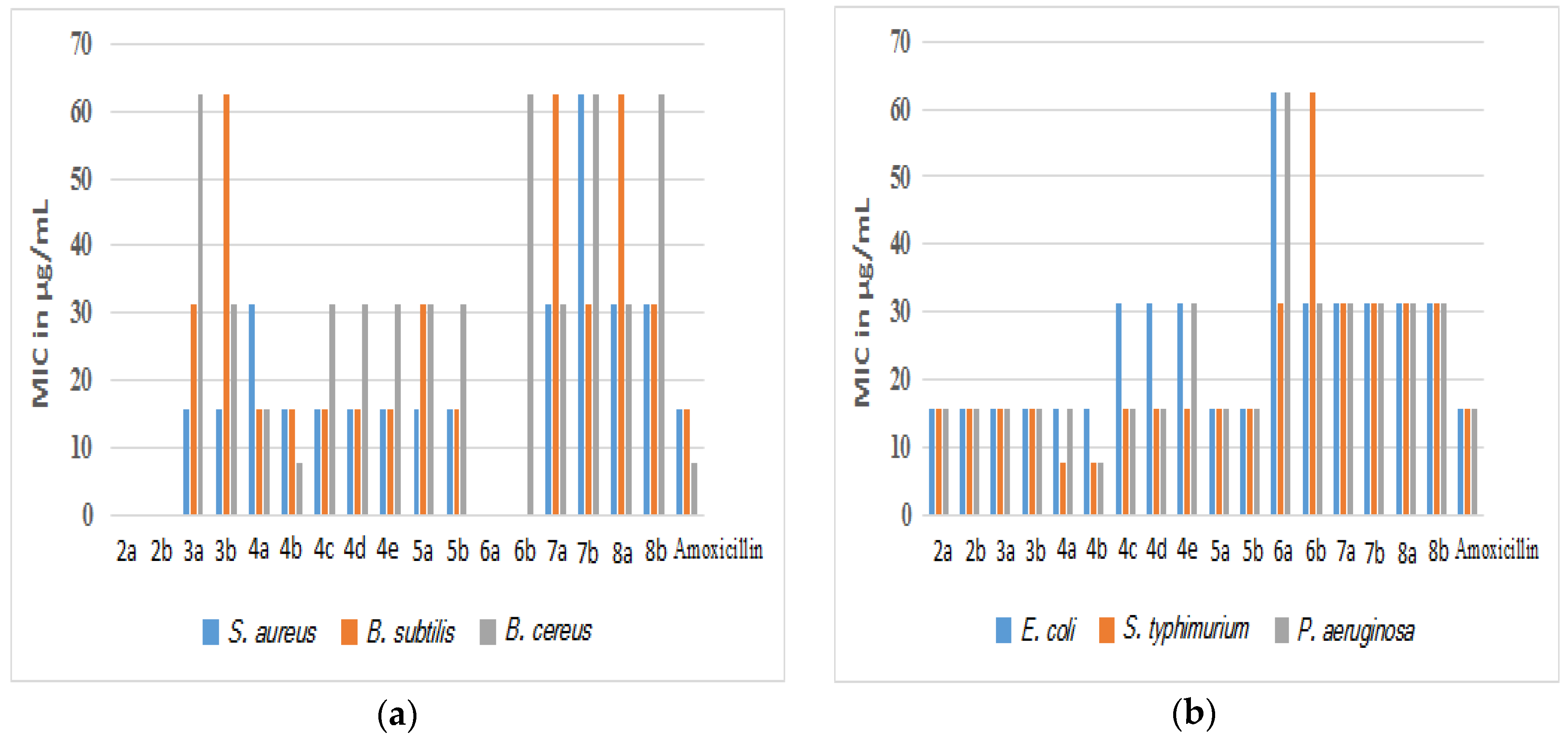
| Compound | Gram-Positive Bacteria | Gram-Negative Bacteria | ||||
|---|---|---|---|---|---|---|
| S. aureus | B. subtilis | B. cereus | E. coli | S. typhimurium | P. aeruginosa | |
| 2a | >125 | >125 | >125 | 15.63 | 15.63 | 15.63 |
| 2b | >125 | >125 | >125 | 15.63 | 15.63 | 15.63 |
| 3a | 15.63 | 31.25 | 62.5 | 15.63 | 15.63 | 15.63 |
| 3b | 15.63 | 62.5 | 31.25 | 15.63 | 15.63 | 15.63 |
| 4a | 31.25 | 15.63 | 15.63 | 15.63 | 7.81 | 15.63 |
| 4b | 15.63 | 15.63 | 7.81 | 15.63 | 7.81 | 7.81 |
| 4c | 15.63 | 15.63 | 31.25 | 31.25 | 15.63 | 15.63 |
| 4d | 15.63 | 15.63 | 31.25 | 31.25 | 15.63 | 15.63 |
| 4e | 15.63 | 15.63 | 31.25 | 31.25 | 15.63 | 31.25 |
| 5a | 15.63 | 31.25 | 31.25 | 15.63 | 15.63 | 15.63 |
| 5b | 15.63 | 15.63 | 31.25 | 15.63 | 15.63 | 15.63 |
| 6a | >125 | >125 | >125 | 62.5 | 31.25 | 62.5 |
| 6b | >125 | >125 | 62.5 | 31.25 | 62.5 | 31.25 |
| 7a | 31.25 | 62.5 | 31.25 | 31.25 | 31.25 | 31.25 |
| 7b | 62.5 | 31.25 | 62.5 | 31.25 | 31.25 | 31.25 |
| 8a | 31.25 | 62.5 | 31.25 | 31.25 | 31.25 | 31.25 |
| 8b | 31.25 | 31.25 | 62.5 | 31.25 | 31.25 | 31.25 |
| Amoxicillin | 15.63 | 15.63 | 7.81 | 15.63 | 15.63 | 15.63 |
| IC50(μM) | ||||
|---|---|---|---|---|
| Compound | R | R1 | DNA Gyrase Supercoiling | Topoisomerase IV Decatenation |
| 2a | 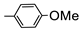 | 8.30 | 21.99 | |
| 2b | 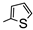 | 10.42 | 22.03 | |
| 3a |  | 8.99 | 21.78 | |
| 3b | 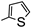 | 6.96 | 17.50 | |
| 4a | 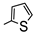 |  | 5.77 | 14.89 |
| 4b |  |  | 3.44 | 14.46 |
| 5a |  | 12.99 | 23.25 | |
| 5b- | 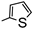 | 14.23 | 17.24 | |
| Ciprofloxacin | 3.52 | 17.57 | ||
| Novobiocin | 4.19 | 14.59 | ||
| Compound No. | Docking Score (Kcal/mol) | Amino Acid Residues (Bond Length Ao) | Atoms of Compound | Type of Bond |
|---|---|---|---|---|
| Novobiocin | −6.80 | Asn46(3.27); Asp73(1.91); Arg76 | H(OH)(oxan-4-yl); H(OCONH2); C6H2(coumarin) | H-don H-don Arene–cation |
| 2a | −5.25 | Arg76; Gly77(2.79) | pyridine; O(pyrimidone) | Arene–cation H-acc |
| 2b | −5.74 | Arg76; Gly77(2.25) | pyridine; O(pyrimidone) | Arene–cation H-acc |
| 3a | −5.65 | Arg76; Thr165(2.50) | pyridine; N-3(pyrimidine) | Arene–cation H-acc |
| 3b | −5.90 | Arg76; Thr165(2.27) | pyridine; N-3(pyrimidine) | Arene–cation H-acc |
| 4a | −6.64 | Arg76; Thr165(2.15) | pyridine; H(NH-furan-2-yl) | Arene–cation H-don |
| 4b | −6.99 | Asn46(2.99); Arg76; Arg76 | N(piperazine); pyridine; phenyl | H-acc Arene–cation Arene–cation |
| 5a | −5.30 | Arg76 | pyridine | Arene–cation |
| 5b | −5.55 | Arg76 | pyridine | Arene–cation |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohi El-Deen, E.M.; Abd El-Meguid, E.A.; Karam, E.A.; Nossier, E.S.; Ahmed, M.F. Synthesis and Biological Evaluation of New Pyridothienopyrimidine Derivatives as Antibacterial Agents and Escherichia coli Topoisomerase II Inhibitors. Antibiotics 2020, 9, 695. https://doi.org/10.3390/antibiotics9100695
Mohi El-Deen EM, Abd El-Meguid EA, Karam EA, Nossier ES, Ahmed MF. Synthesis and Biological Evaluation of New Pyridothienopyrimidine Derivatives as Antibacterial Agents and Escherichia coli Topoisomerase II Inhibitors. Antibiotics. 2020; 9(10):695. https://doi.org/10.3390/antibiotics9100695
Chicago/Turabian StyleMohi El-Deen, Eman M., Eman A. Abd El-Meguid, Eman A. Karam, Eman S. Nossier, and Marwa F. Ahmed. 2020. "Synthesis and Biological Evaluation of New Pyridothienopyrimidine Derivatives as Antibacterial Agents and Escherichia coli Topoisomerase II Inhibitors" Antibiotics 9, no. 10: 695. https://doi.org/10.3390/antibiotics9100695
APA StyleMohi El-Deen, E. M., Abd El-Meguid, E. A., Karam, E. A., Nossier, E. S., & Ahmed, M. F. (2020). Synthesis and Biological Evaluation of New Pyridothienopyrimidine Derivatives as Antibacterial Agents and Escherichia coli Topoisomerase II Inhibitors. Antibiotics, 9(10), 695. https://doi.org/10.3390/antibiotics9100695