On the Biodiversity and Biodeteriogenic Activity of Microbial Communities Present in the Hypogenic Environment of the Escoural Cave, Alentejo, Portugal
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Microbial Contamination: Assessment and Diversity
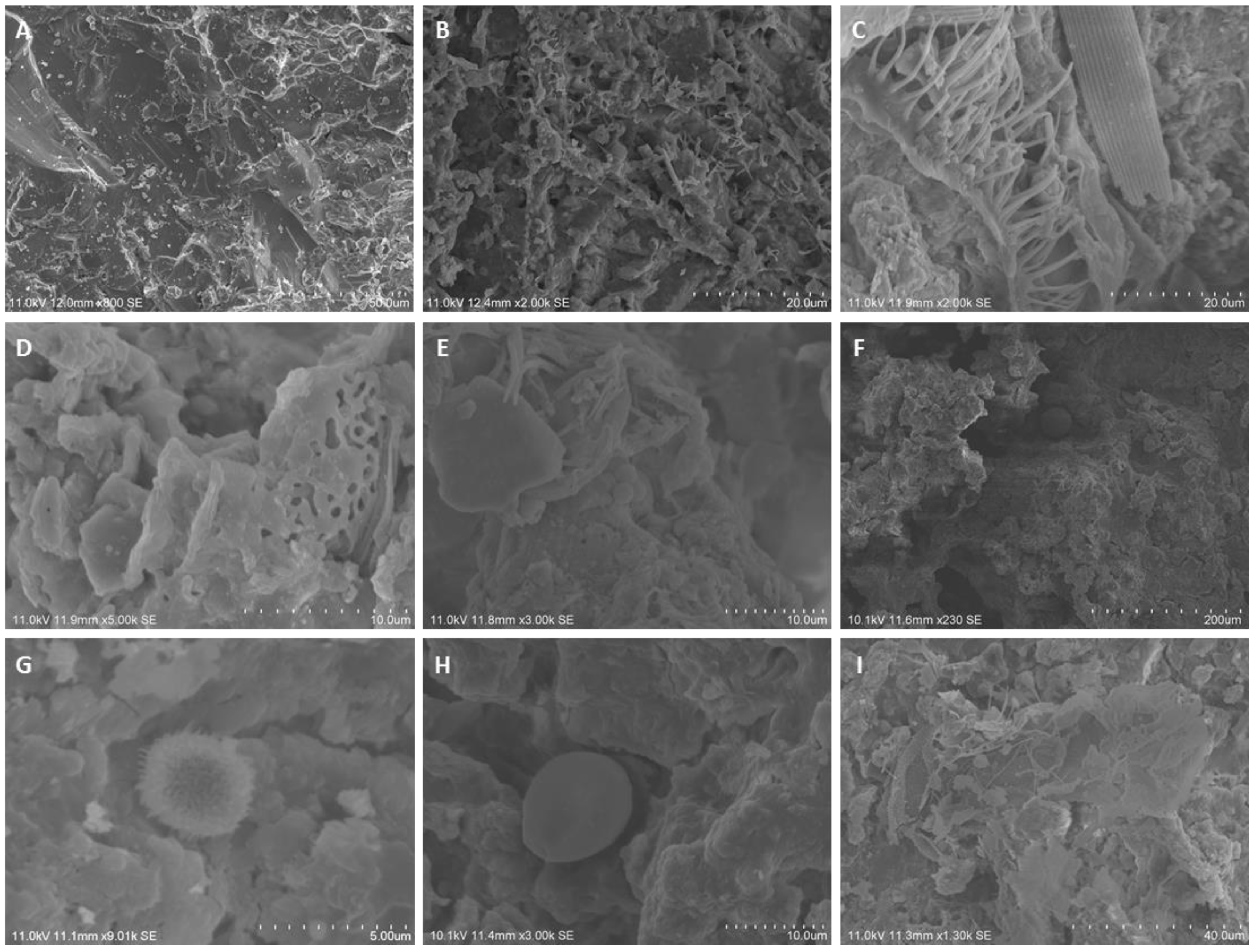
2.2.1. SEM
2.2.2. Isolation Procedure
2.2.3. High-Throughput Sequencing
2.3. Microbial Activity and Viability Assessment
2.4. Biodeteriogenic Activity
2.4.1. Raman Spectroscopy
2.4.2. Simulation Assays
3. Results
3.1. Assessment and Diversity of Microbial Contamination
3.2. Microbial Activity and Viability
3.3. Assessment of the Biodeteriogenic Ability
4. Discussion
4.1. Biological and Microbial Contamination
4.2. Microbial Diversity
4.3. Microbial Viability and Activity
4.4. Microbial Biodeteriogenic Activity
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berrouet, F. Les Altérations D’origine Biologique dans L’art Pariétal: Exemple des Relations Structurales et Conceptuelles entre le Mondmilch et les Représentations Paléolithiques: Cas Particulier de la Grotte de Lascaux et Enjeux Conservatoires. Ph.D. Thesis, Université de Bordeaux, Bordeaux, France, 2009. [Google Scholar]
- Cuezva, S.; Fernandez-Cortes, A.; Porca, E.; Pašić, L.; Jurado, V.; Hernández, M.; Serrano-Ortiz, P.; Hermosin, B.; Cañaveras, J.C.; Sanchez-Moral, S.; et al. The biogeochemical role of Actinobacteria in Altamira Cave, Spain. FEMS Microbiol. Ecol. 2012, 81, 281–290. [Google Scholar] [CrossRef] [Green Version]
- Dupont, J.; Jacquet, C.; Dennetière, B.; Lacoste, S.; Bousta, F.; Orial, G.; Cruaud, C.; Couloux, A.; Roquebert, M.-F. Inva-sion of the French Paleolithic painted cave of Lascaux by members of the Fusarium solani species complex. Mycologia 2007, 99, 526–533. [Google Scholar] [CrossRef]
- Saiz-Jimenez, C. Microbiological and environmental issues in show caves. World J. Microbiol. Biotechnol. 2012, 28, 2453–2464. [Google Scholar] [CrossRef]
- Jurado, V.; Porca, E.; Cuezva, S.; Fernandez-Cortes, A.; Moral, S.S.; Sáiz-Jiménez, C. Fungal outbreak in a show cave. Sci. Total Environ. 2010, 408, 3632–3638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nováková, A. Microscopic fungi isolated from the Domica Cave system (Slovak Karst National Park, Slovakia). A review. Int. J. Speleol. 2009, 38, 71–82. [Google Scholar] [CrossRef] [Green Version]
- Sugita, T.; Kikuchi, K.; Makimura, K.; Urata, K.; Someya, T.; Kamei, K.; Niimi, M.; Uehara, Y. Trichosporon Species Isolated from Guano Samples Obtained from Bat-Inhabited Caves in Japan. Appl. Environ. Microbiol. 2005, 71, 7626–7629. [Google Scholar] [CrossRef] [Green Version]
- Urzì, C.; Albertano, P. Studying phototrophic and heterotrophic microbial communities on stone monuments. Methods Enzymol. 2001, 336, 340–355. [Google Scholar] [CrossRef] [PubMed]
- Cañveras, J.C.; Sanchez-Moral, S.; Saiz-Jimenez, C. Microorganisms and Microbially Induced Fabrics in Cave Walls. Geomicrobiol. J. 2001, 18, 223–240. [Google Scholar] [CrossRef]
- Cuezva, S.; Sánchez-Moral, S.; Sáiz-Jiménez, C.; Cañaveras, J.C. Microbial communities and associated mineral fabrics in Altamira Cave, Spain. Int. J. Speleol. 2009, 38, 9. [Google Scholar]
- Northup, D.E.; Lavoie, K. Geomicrobiology of Caves: A Review. Geomicrobiol. J. 2001, 18, 199–222. [Google Scholar] [CrossRef]
- Garcia-Vallès, M.; Vendrell-Saz, M.; Krumbein, W.E.; Urzì, C. Coloured mineral coatings on monument surfaces as a result of biomineralization: The case of the Tarragona cathedral (Catalonia). Appl. Geochem. 1997, 12, 255–266. [Google Scholar] [CrossRef]
- Griffin, P.S.; Indictor, N.; Koestler, R.J. The biodeterioration of stone: A review of deterioration mechanisms, conservation case histories, and treatment. Int. Biodeterior. 1991, 28, 187–207. [Google Scholar] [CrossRef]
- Herrera, L.K.; Videla, H.A. Surface analysis and materials characterization for the study of biodeterioration and weathering effects on cultural property. Int. Biodeterior. Biodegrad. 2009, 63, 813–822. [Google Scholar] [CrossRef]
- Kusumi, A.; Li, X.; Osuga, Y.; Kawashima, A.; Gu, J.-D.; Nasu, M.; Katayama, Y. Bacterial Communities in Pigmented Biofilms Formed on the Sandstone Bas-Relief Walls of the Bayon Temple, Angkor Thom, Cambodia. Microbes Environ. 2013, 28, 422–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagai, K.; Suzuki, K.; Okada, G. Studies on the distribution of alkalophilic and alkali-tolerant soil fungi II: Fungal flora in two limestone caves in Japan. Mycoscience 1998, 39, 293–298. [Google Scholar] [CrossRef]
- Schiavon, N. Biodeterioration of calcareous and granitic building stones in urban environments. In Natural Stone, Weathering Phenomena, Conservation Strategies and Case Studies; Siegesmund, S., Weiss, T., Vollbrecht, V.A., Eds.; Geological Society of London Special Publication: London, UK, 2002; pp. 195–206. [Google Scholar] [CrossRef]
- Schiavon, N.; De Caro, T.; Kiros, A.; Caldeira, A.T.; Parisi, I.E.; Riccucci, C.; Gigante, G.E. A multi-analytical approach to investigate stone biodeterioration at a UNESCO world heritage site: The volcanic rock-hewn churches of Lalibela, Northern Ethiopia. Appl. Phys. Part A 2013, 113, 843–854. [Google Scholar] [CrossRef] [Green Version]
- Rosado, T.; Reis, A.; Mirão, J.; Candeias, A.; Vandenabeele, P.; Caldeira, A.T. Pink! Why not? On the unusual colour of Évora Cathedral. Int. Biodeterior. Biodegrad. 2014, 94, 121–127. [Google Scholar] [CrossRef] [Green Version]
- Rosado, T.; Dias, L.; Lança, M.; Nogueira, C.; Santos, R.; Martins, M.R.; Candeias, A.; Mirão, J.; Caldeira, A.T. Assessment of microbiota present on a Portuguese historical stone convent using high-throughput sequencing approaches. MicrobiologyOpen 2020, 9, 1067–1084. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Salvador, C.; Caldeira, A.T.; Angelini, E.; Schiavon, N. Biodegradation and microbial contamination of lime-stone surfaces: An experimental study from Batalha Monastery, Portugal. Corros. Mater. Degrad. 2021, 2, 31–45. [Google Scholar]
- Ariño, X.; Saiz-Jimenez, C. Colonization and deterioration processes in Roman mortars by cyanobacteria, algae and lichens. Aerobiologia 1996, 12, 9–18. [Google Scholar] [CrossRef]
- Tran, T.H.; Govin, A.; Guyonnet, R.; Grosseau, P.; Lors, C.; Garcia-Diaz, E.; Damidot, D.; Devès, O.; Ruot, B. Influence of the intrinsic characteristics of mortars on biofouling by Klebsormidium flaccidum. Int. Biodeterior. Biodegrad. 2012, 70, 31–39. [Google Scholar] [CrossRef] [Green Version]
- Altenburgera, P.; Kämpferb, P.; Makristathisc, A.; Lubitza, W.; Bussea, H.-J. Classification of bacteria isolated from a medieval wall painting. J. Biotechnol. 1996, 47, 39–52. [Google Scholar] [CrossRef]
- Cappitelli, F.; Abbruscato, P.; Foladori, P.; Zanardini, E.; Ranalli, G.; Principi, P.; Villa, F.; Polo, A.; Sorlini, C. Detection and Elimination of Cyanobacteria from Frescoes: The Case of the St. Brizio Chapel (Orvieto Cathedral, Italy). Microb. Ecol. 2009, 57, 633–639. [Google Scholar] [CrossRef]
- Gorbushina, A.A.; Heyrman, J.; Dornieden, T.; Gonzalez-Delvalle, M.; Krumbein, W.E.; Laiz, L.; Petersen, K.; Saiz-Jimenez, C.; Swings, J. Bacterial and fungal diversity and biodeterioration problems in mural painting environments of St. Martins church (Greene–Kreiensen, Germany). Int. Biodeterior. Biodegrad. 2004, 53, 13–24. [Google Scholar] [CrossRef]
- Gurtner, C.; Heyrman, J.; Piñar, G.; Lubitz, W.; Swings, J.; Rölleke, S. Comparative analyses of the bacterial diversity on two different biodeteriorated wall paintings by DGGE and 16S rDNA sequence analysis. Int. Biodeterior. Biodegrad. 2000, 46, 229–239. [Google Scholar] [CrossRef]
- Laiz, L.; Gonzalez, J.M.; Saiz-Jimenez, C. Microbial communities in caves: Ecology, physiology, and effects on paleo-lithic paintings. In Art, Biology, and Conservation: Biodeterioration of Works of Art; The Metropolitan Museum of Art: New York, NY, USA, 2003; pp. 210–225. [Google Scholar]
- Nugari, M.; Pietrini, A.; Caneva, G.; Imperi, F.; Visca, P. Biodeterioration of mural paintings in a rocky habitat: The Crypt of the Original Sin (Matera, Italy). Int. Biodeterior. Biodegrad. 2009, 63, 705–711. [Google Scholar] [CrossRef]
- Pepe, O.; Sannino, L.; Palomba, S.; Anastasio, M.; Blaiotta, G.; Villani, F.; Moschetti, G. Heterotrophic microorganisms in deteriorated medieval wall paintings in southern Italian churches. Microbiol. Res. 2010, 165, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Zucconi, L.; Gagliardi, M.; Isola, D.; Onofri, S.; Andaloro, M.C.; Pelosi, C.; Pogliani, P.; Selbmann, L. Biodeterioration agents dwelling in or on the wall paintings of the Holy Saviour’s cave (Vallerano, Italy). Int. Biodeterior. Biodegrad. 2012, 70, 40–46. [Google Scholar] [CrossRef]
- Bastian, F.; Alabouvette, C.; Saiz-Jimenez, C. Bacteria and free-living amoeba in the Lascaux Cave. Res. Microbiol. 2009, 160, 38–40. [Google Scholar] [CrossRef]
- Bastian, F.; Jurado, V.; Nováková, A.; Alabouvette, C.; Sáiz-Jiménez, C. The microbiology of Lascaux cave. Microbiology 2010, 156, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Cunningham, K.I.; Northup, D.E.; Pollastro, R.M.; Wright, W.G.; LaRock, E.J. Bacteria, fungi and biokarst in Lechu-guilla Cave, Carlsbad Caverns National Park, New Mexico. Environ. Geol. 1995, 25, 2–8. [Google Scholar] [CrossRef]
- González, I.; Laiz, L.; Hermosin, B.; Caballero, B.; Incerti, C.; Sáiz-Jiménez, C. Bacteria isolated from rock art paintings: The case of Atlanterra shelter (south Spain). J. Microbiol. Methods 1999, 36, 123–127. [Google Scholar] [CrossRef]
- Martin-Sanchez, P.M.; Nováková, A.; Bastian, F.; Alabouvette, C.; Saiz-Jimenez, C. Two new species of the genus Ochroconis, O. lascauxensis and O. anomala isolated from black stains in Lascaux Cave, France. Fungal Biol. 2012, 116, 574–589. [Google Scholar] [CrossRef] [PubMed]
- Martin-Sanchez, P.; Jurado, V.; Porca, E.; Bastian, F.; Lacanette, D.; Alabouvette, C.; Saiz-Jimenez, C. Airborne microorganisms in Lascaux Cave (France). Int. J. Speleol. 2014, 43, 295–303. [Google Scholar] [CrossRef]
- De Leo, F.; Iero, A.; Zammit, G.; Urzì, C. Chemoorganotrophic bacteria isolated from biodeteriorated surfaces in cave and catacombs. Int. J. Speleol. 2012, 41, 125–136. [Google Scholar] [CrossRef] [Green Version]
- Saarela, M.; Alakomi, H.-L.; Suihko, M.-L.; Maunuksela, L.; Raaska, L.; Mattila-Sandholm, T. Heterotrophic microorganisms in air and biofilm samples from Roman catacombs, with special emphasis on actinobacteria and fungi. Int. Biodeterior. Biodegrad. 2004, 54, 27–37. [Google Scholar] [CrossRef]
- Sanchez-Moral, S.; Canaveras, J.C.; Laiz, L.; Saiz-Jimenez, C.; Bedoya, J.; Luque, L. Biomediated Precipitation of Calcium Carbonate Metastable Phases in Hypogean Environments: A Short Review. Geomicrobiol. J. 2003, 20, 491–500. [Google Scholar] [CrossRef] [Green Version]
- Zammit, G.; Sánchez-Moral, S.; Albertano, P. Bacterially mediated mineralisation processes lead to biodeterioration of artworks in Maltese catacombs. Sci. Total Environ. 2011, 409, 2773–2782. [Google Scholar] [CrossRef] [PubMed]
- Cennamo, P.; Montuori, N.; Trojsi, G.; Fatigati, G.; Moretti, A. Biofilms in churches built in grottoes. Sci. Total Environ. 2016, 543, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Cañaveras, J.C.; Hoyos, M.; Sanchez-Moral, S.; Sanz-Rubio, E.; Bedoya, J.; Soler, V.; Groth, I.; Schumann, P.; Laiz, L.; Gonza-lez, I.; et al. Microbial Communities Associated with Hydromagnesite and Needle-Fiber Aragonite Deposits in a Karstic Cave (Altamira, Northern Spain). Geomicrobiol. J. 1999, 16, 9–25. [Google Scholar] [CrossRef] [Green Version]
- Groth, I.; Vettermann, R.; Schuetze, B.; Schumann, P.; Sáiz-Jiménez, C. Actinomycetes in Karstic caves of northern Spain (Altamira and Tito Bustillo). J. Microbiol. Methods 1999, 36, 115–122. [Google Scholar] [CrossRef]
- Laiz, L.; Groth, I.; Gonzalez, I.; Sáiz-Jiménez, C. Microbiological study of the dripping waters in Altamira cave (Santil-lana del Mar, Spain). J. Microbiol. Methods 1999, 36, 129–138. [Google Scholar] [CrossRef] [Green Version]
- Portillo, M.C.; Gonzalez, J.M. Comparing bacterial community fingerprints from white colonizations in Altamira Cave (Spain). World J. Microbiol. Biotechnol. 2009, 25, 1347–1352. [Google Scholar] [CrossRef]
- Portillo, M.C.; Saiz-Jimenez, C.; Gonzalez, J.M. Molecular characterization of total and metabolically active bacterial communities of “white colonizations” in the Altamira Cave, Spain. Res. Microbiol. 2009, 160, 41–47. [Google Scholar] [CrossRef]
- Portillo, M.C.; Gonzalez, J.M. Moonmilk deposits originate from specific bacterial communities in Altamira Cave (Spain). Microb. Ecol. 2011, 61, 182–189. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Moral, S.; Soler, V.; Cañaveras, J.; Sanz-Rubio, E.; Van Grieken, R.; Gysels, K. Inorganic deterioration affecting the Altamira Cave, N Spain: Quantitative approach to wall-corrosion (solutional etching) processes induced by visitors. Sci. Total Environ. 1999, 243–244, 67–84. [Google Scholar] [CrossRef]
- Sanchez-Moral, S.; Portillo, M.C.; Janices, I.; Cuezva, S.; Fernández-Cortés, A.; Cañaveras, J.C.; Gonzalez, J.M. The role of microorganisms in the formation of calcitic moonmilk deposits and speleothems in Altamira Cave. Geomorphology 2012, 139–140, 285–292. [Google Scholar] [CrossRef] [Green Version]
- Schabereiter-Gurtner, C.; Saiz-Jimenez, C.; Piñar, G.; Lubitz, W.; Rölleke, S. Altamira cave Paleolithic paintings harbor partly unknown bacterial communities. FEMS Microbiol. Lett. 2002, 211, 7–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beni, A.; Soki, E.; Lajtha, K.; Fekete, I. An optimized HPLC method for soil fungal biomass determination and its application to a detritus manipulation study. J. Microbiol. Methods 2014, 103, 124–130. [Google Scholar] [CrossRef]
- Saad, D.; Kinsey, G.; Paterson, R.R.M.; Gaylarde, C.C. Ergosterol analysis for the quantification of fungal growth on paint films. Proposal for a standard method. Surf. Coat. Int. Part B Coat. Trans. 2003, 86, 131–134. [Google Scholar] [CrossRef]
- Hippelein, M.; Rügamer, M. Ergosterol as an indicator of mould growth on building materials. Int. J. Hyg. Environ. Health 2004, 207, 379–385. [Google Scholar] [CrossRef]
- Mille-Lindblom, C.; Von Wachenfeldt, E.; Tranvik, L.J. Ergosterol as a measure of living fungal biomass: Persistence in environmental samples after fungal death. J. Microbiol. Methods 2004, 59, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Joblin, Y.; Moularat, S.; Anton, R.; Bousta, F.; Orial, G.; Robine, E.; Picon, O.; Bourouina, T. Detection of moulds by volatile organic compounds: Application to heritage conservation. Int. Biodeterior. Biodegrad. 2010, 64, 210–217. [Google Scholar] [CrossRef]
- Porca, E.; Jurado, V.; Martin-Sanchez, P.M.; Hermosin, B.; Bastian, F.; Alabouvette, C.; Saiz-Jimenez, C. Aerobiology: An ecological indicator for early detection and control of fungal outbreaks in caves. Ecol. Indic. 2011, 11, 1594–1598. [Google Scholar] [CrossRef]
- Araujo, A.C.; Lejeune, M.; Santos, A.I. Gruta do Escoural: Necropole Neolitica e Arte Rupestre Paleolitica; Instituto Portugues do Patrimonio Arquitectonico e Arqueologico: Lisbon, Portugal, 1995. [Google Scholar]
- Herlemann, D.P.R.; Labrenz, M.; Jürgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011, 5, 1571–1579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2012, 41, e1. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.L.C.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schubert, M.; Lindgreen, S.; Orlando, L. AdapterRemoval v2: Rapid adapter trimming, identification, and read merging. BMC Res. Notes 2016, 9, 88. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME Improves Sensitivity and Speed of Chimera Detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [Green Version]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [Green Version]
- Abarenkov, K.; Nilsson, R.H.; Larsson, K.-H.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Høiland, K.; Kjøller, R.; Larsson, E.; Pennanen, T.; et al. The UNITE database for molecular identification of fungi—Recent updates and future perspectives. New Phytol. 2010, 186, 281–285. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Boncler, M.; Różalski, M.; Krajewska, U.; Podsędek, A.; Watala, C. Comparison of PrestoBlue and MTT assays of cellular viability in the assessment of anti-proliferative effects of plant extracts on human endothelial cells. J. Pharmacol. Toxicol. Methods 2014, 69, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Rosado, T.; Martins, M.R.; Pires, M.; Mirão, J.; Candeias, A.; Caldeira, A.T. Enzymatic monitorization of mural paintings biodegradation and biodeterioration. Int. J. Conserv. Sci. 2013, 4, 603–612. [Google Scholar]
- Xu, M.; McCanna, D.J.; Sivak, J. Use of the viability reagent PrestoBlue in comparison with alamarBlue and MTT to assess the viability of human corneal epithelial cells. J. Pharmacol. Toxicol. Methods 2015, 71, 1–7. [Google Scholar] [CrossRef]
- Mariscal, A.; Lopez-Gigosos, R.M.; Carnero-Varo, M.; Fernández-Crehuet, J. Fluorescent assay based on resazurin for detection of activity of disinfectants against bacterial biofilm. Appl. Microbiol. Biotechnol. 2009, 82, 773–783. [Google Scholar] [CrossRef]
- Martín-Navarro, C.M.; López-Arencibia, A.; Sifaoui, I.; Reyes-Batlle, M.; Cabello-Vílchez, A.M.; Maciver, S.; Valladares, B.; Piñero, J.E.; Lorenzo-Morales, J. PrestoBlue® and AlamarBlue® are equally useful as agents to determine the viability of Acanthamoeba trophozoites. Exp. Parasitol. 2014, 145, S69–S72. [Google Scholar] [CrossRef]
- Malaurent, P.; Huneau, F.; Lastennet, R.; Fabre, R. Etudes Pour la Conservation des Parois de la Grotte d’Escoural-Portugal (Conservation Grottes Ornées No. 2004–24); Centre de Développement des Géosciences Appliquées—CDGA: Bordeaux, France, 2004. [Google Scholar]
- Arroyo, G.; Arroyo, I.; Arroyo, E. Microbiological analysis of Maltravieso Cave (Caceres), Spain. Int. Biodeterior. Biodegrad. 1997, 40, 131–139. [Google Scholar] [CrossRef]
- Barton, H.A.; Northup, D.E. Geomicrobiology in cave environments: Past, current and future perspectives. J. Cave Karst Stud. 2007, 69, 163–178. [Google Scholar]
- Bourges, F.; Genthon, P.; Genty, D.; Lorblanchet, M.; Mauduit, E.; D’Hulst, D. Conservation of prehistoric caves and stability of their inner climate: Lessons from Chauvet and other French caves. Sci. Total Environ. 2014, 493, 79–91. [Google Scholar] [CrossRef]
- Barton, H. Microbial life in the underworld: Biogenicity in secondary mineral formations. Geomicrobiol. J. 2001, 18, 359–368. [Google Scholar]
- Barton, H.A.; Jurado, V. What’s up down there? Microbial diversity in caves. Microbe 2007, 2, 132–138. [Google Scholar]
- Jurado, V.; Sanchez-Moral, S.; Saiz-Jimenez, C. Entomogenous fungi and the conservation of the cultural heritage: A review. Int. Biodeterior. Biodegrad. 2008, 62, 325–330. [Google Scholar] [CrossRef] [Green Version]
- Sterflinger, K. Fungi: Their role in deterioration of cultural heritage. Fungal Biol. Rev. 2010, 24, 47–55. [Google Scholar] [CrossRef]
- Wollenzien, U.; De Hoog, G.; Krumbein, W.; Urzí, C. On the isolation of microcolonial fungi occurring on and in marble and other calcareous rocks. Sci. Total Environ. 1995, 167, 287–294. [Google Scholar] [CrossRef]
- Lefevre, M. La “maladie verte” de Lascaux. Stud. Conserv. 1974, 19, 126–156. [Google Scholar] [CrossRef]




| Location | PB (au) | MTT (au) |
|---|---|---|
| GdE 3 | 1.80 ± 0.02 | 0.40 ± 0.04 |
| GdE 8 | 6.0 ± 0.5 | 0.50 ± 0.07 |
| GdE 23 | 0.49 ± 0.05 | 0.02 ± 0.005 |
| Fungi Genus | % |
|---|---|
| Penicillium | 29.22 |
| Mucor | 28.84 |
| Aspergillus | 18.22 |
| Cladosporium | 10.82 |
| Unknown | 7.78 |
| Fusarium | 2.85 |
| Trichoderma | 1.52 |
| Absidia | 0.57 |
| Acremonium | 0.38 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caldeira, A.T.; Schiavon, N.; Mauran, G.; Salvador, C.; Rosado, T.; Mirão, J.; Candeias, A. On the Biodiversity and Biodeteriogenic Activity of Microbial Communities Present in the Hypogenic Environment of the Escoural Cave, Alentejo, Portugal. Coatings 2021, 11, 209. https://doi.org/10.3390/coatings11020209
Caldeira AT, Schiavon N, Mauran G, Salvador C, Rosado T, Mirão J, Candeias A. On the Biodiversity and Biodeteriogenic Activity of Microbial Communities Present in the Hypogenic Environment of the Escoural Cave, Alentejo, Portugal. Coatings. 2021; 11(2):209. https://doi.org/10.3390/coatings11020209
Chicago/Turabian StyleCaldeira, Ana Teresa, Nick Schiavon, Guilhem Mauran, Cátia Salvador, Tânia Rosado, José Mirão, and António Candeias. 2021. "On the Biodiversity and Biodeteriogenic Activity of Microbial Communities Present in the Hypogenic Environment of the Escoural Cave, Alentejo, Portugal" Coatings 11, no. 2: 209. https://doi.org/10.3390/coatings11020209
APA StyleCaldeira, A. T., Schiavon, N., Mauran, G., Salvador, C., Rosado, T., Mirão, J., & Candeias, A. (2021). On the Biodiversity and Biodeteriogenic Activity of Microbial Communities Present in the Hypogenic Environment of the Escoural Cave, Alentejo, Portugal. Coatings, 11(2), 209. https://doi.org/10.3390/coatings11020209







