Recent Updates on Mechanisms of Resistance to 5-Fluorouracil and Reversal Strategies in Colon Cancer Treatment
Simple Summary
Abstract
1. Introduction
2. Mechanisms of Action of 5-Fluorouracil
3. Classical Mechanisms of Resistance
3.1. Alterations in Drug Transports
3.2. Evasion of Apoptosis
3.3. Changes in Cell Cycle and DNA-Damage Repair Kinetics
3.4. Involvement of Autophagy
3.5. Epithelial-to-Mesenchymal Transition (EMT)
3.6. Involvement of Cancer Stem Cells
3.7. Interactions within the Tumor Microenvironment
3.8. Epigenetic Alterations
3.9. Dysregulations of miRNAs
3.10. Redox Imbalances
4. Mechanism of Resistance by Key 5-Fluorouracil Enzymes
4.1. Amplification of Thymidylate Synthase
4.2. Suppressed Expression of Thymidine Phosphorylase
4.3. Overexpression of Dihydropyrimidine Dehydrogenase
4.4. Overexpression of Methylenetetrahydrofolate Reductase
5. Reversal Strategies
5.1. Small Molecule Inhibitors
5.2. Plant-Derived Small Molecules
5.3. Non-Coding RNAs Regulators
5.4. Targeted Immunotherapy
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Keum, N.; Giovannucci, E. Global burden of colorectal cancer: Emerging trends, risk factors and prevention strategies. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 713–732. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.C.; Ding, H.; Wang, J.; Chan, P.S.; Huang, J. Prevalence and risk factors of colorectal cancer in Asia. Intest. Res. 2019, 17, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Idrees, M.; Tejani, M. Current Treatment Strategies for Elderly Patients with Metastatic Colon Cancer. Cureus 2019, 11, e4713. [Google Scholar] [CrossRef] [PubMed]
- Heidelberger, C.; Chaudhuri, N.K.; Danneberg, P.; Mooren, D.; Griesbach, L.; Duschinsky, R.; Schnitzer, R.J.; Pleven, E.; Scheiner, J. Fluorinated pyrimidines, a new class of tumour-inhibitory compounds. Nature 1957, 179, 663–666. [Google Scholar] [CrossRef] [PubMed]
- Vodenkova, S.; Buchler, T.; Cervena, K.; Veskrnova, V.; Vodicka, P.; Vymetalkova, V. 5-fluorouracil and other fluoropyrimidines in colorectal cancer: Past, present and future. Pharmacol. Ther. 2020, 206, 107447. [Google Scholar] [CrossRef]
- Xie, Y.-H.; Chen, Y.-X.; Fang, J.-Y. Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct. Target. Ther. 2020, 5, 22. [Google Scholar] [CrossRef]
- Christensen, S.; Van der Roest, B.; Besselink, N.; Janssen, R.; Boymans, S.; Martens, J.W.M.; Yaspo, M.-L.; Priestley, P.; Kuijk, E.; Cuppen, E.; et al. 5-Fluorouracil treatment induces characteristic T>G mutations in human cancer. Nat. Commun. 2019, 10, 4571. [Google Scholar] [CrossRef]
- Gu, J.; Li, Z.; Zhou, J.; Sun, Z.; Bai, C. Response prediction to oxaliplatin plus 5-fluorouracil chemotherapy in patients with colorectal cancer using a four-protein immunohistochemical model. Oncol. Lett. 2019, 18, 2091–2101. [Google Scholar] [CrossRef]
- Yao, W.; Zhu, S.; Li, P.; Zhang, S. Large tumor suppressor kinase 2 overexpression attenuates 5-FU-resistance in colorectal cancer via activating the JNK-MIEF1-mitochondrial division pathway. Cancer Cell Int. 2019, 19, 97. [Google Scholar] [CrossRef]
- Salem, M.E.; Yin, J.; Goldberg, R.M.; Pederson, L.D.; Wolmark, N.; Alberts, S.R.; Taieb, J.; Marshall, J.L.; Lonardi, S.; Yoshino, T.; et al. Evaluation of the change of outcomes over a 10-year period in patients with stage III colon cancer: Pooled analysis of 6501 patients treated with fluorouracil, leucovorin, and oxaliplatin in the ACCENT database. Ann. Oncol. 2020, 31, 480–486. [Google Scholar] [CrossRef]
- Ishikawa, K.; Kawano, Y.; Arihara, Y.; Kubo, T.; Takada, K.; Murase, K.; Miyanishi, K.; Kobune, M.; Kato, J. BH3 profiling discriminates the anti-apoptotic status of 5-fluorouracil-resistant colon cancer cells. Oncol. Rep. 2019, 42, 2416–2425. [Google Scholar] [CrossRef] [PubMed]
- Rutman, R.J.; Cantarow, A.; Paschkis, K.E. Studies in 2-Acetylaminofluorene Carcinogenesis: III. The Utilization of Uracil-2-C14 by Preneoplastic Rat Liver and Rat Hepatoma. Cancer Res. 1954, 14, 119–123. [Google Scholar] [PubMed]
- Jubeen, F.; Liaqat, A.; Sultan, M.; Zafar Iqbal, S.; Sajid, I.; Sher, F. Green synthesis and biological evaluation of novel 5-fluorouracil derivatives as potent anticancer agents. Saudi Pharm. J. 2019, 27, 1164–1173. [Google Scholar] [CrossRef] [PubMed]
- Merloni, F.; Ranallo, N.; Scortichini, L.; Giampieri, R.; Berardi, R. Tailored therapy in patients treated with fluoropyrimidines: Focus on the role of dihydropyrimidine dehydrogenase. Cancer Drug Resist. 2019. [Google Scholar] [CrossRef]
- Wigle, T.J.; Tsvetkova, E.V.; Welch, S.A.; Kim, R.B. DPYD and Fluorouracil-Based Chemotherapy: Mini Review and Case Report. Pharmaceutics 2019, 11, 199. [Google Scholar] [CrossRef]
- Lin, S.; Yue, J.; Guan, X.; Yuan, P.; Wang, J.; Luo, Y.; Fan, Y.; Cai, R.; Li, Q.; Chen, S.; et al. Polymorphisms of MTHFR and TYMS predict capecitabine-induced hand-foot syndrome in patients with metastatic breast cancer. Cancer Commun. 2019, 39, 57. [Google Scholar] [CrossRef]
- Yao, C.W.; Kang, K.A.; Piao, M.J.; Ryu, Y.S.; Fernando, P.M.D.J.; Oh, M.C.; Park, J.E.; Shilnikova, K.; Na, S.-Y.; Jeong, S.U.; et al. Reduced Autophagy in 5-Fluorouracil Resistant Colon Cancer Cells. Biomol. Ther. 2017, 25, 315–320. [Google Scholar] [CrossRef]
- Ratti, M.; Hahne, J.C.; Toppo, L.; Castelli, E.; Petrelli, F.; Passalacqua, R.; Barni, S.; Tomasello, G.; Ghidini, M. Major innovations and clinical applications of disodium-levofolinate: A review of available preclinical and clinical data. Ther. Adv. Med. Oncol. 2019, 11, 1758835919853954. [Google Scholar] [CrossRef]
- Robey, R.W.; Pluchino, K.M.; Hall, M.D.; Fojo, A.T.; Bates, S.E.; Gottesman, M.M. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer 2018, 18, 452–464. [Google Scholar] [CrossRef]
- Han, S.-H.; Kim, J.W.; Kim, M.; Kim, J.H.; Lee, K.-W.; Kim, B.-H.; Oh, H.-K.; Kim, D.-W.; Kang, S.-B.; Kim, H.; et al. Prognostic implication of ABC transporters and cancer stem cell markers in patients with stage III colon cancer receiving adjuvant FOLFOX-4 chemotherapy. Oncol. Lett. 2019, 17, 5572–5580. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; Wang, J.; Yang, J.; Yang, G. Abnormal expression of ABCD3 is an independent prognostic factor for colorectal cancer. Oncol. Lett. 2020, 19, 3567–3577. [Google Scholar] [CrossRef] [PubMed]
- Rehman, Z.; Fahim, A.; Bhatti, A.; Sadia, H.; John, P. Co-expression of HIF-1alpha, MDR1 and LAPTM4B in peripheral blood of solid tumors. PeerJ 2019, 7, e6309. [Google Scholar] [CrossRef] [PubMed]
- Dvorak, P.; Hlavac, V.; Mohelnikova-Duchonova, B.; Liska, V.; Pesta, M.; Soucek, P. Downregulation of ABC Transporters in Non-neoplastic Tissues Confers Better Prognosis for Pancreatic and Colorectal Cancer Patients. J. Cancer 2017, 8, 1959–1971. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, J.-M.; Wei, W.; Yang, R.; Chen, D.; Ma, X.-D.; Jiang, G.-M.; Wang, B.-L. Regulation of ATP-binding cassette subfamily B member 1 by Snail contributes to chemoresistance in colorectal cancer. Cancer Sci. 2020, 111, 84–97. [Google Scholar] [CrossRef]
- Gao, Q.; Li, X.-X.; Xu, Y.-M.; Zhang, J.-Z.; Rong, S.; Qin, Y.-Q.; Fang, J. IRE1α-targeting downregulates ABC transporters and overcomes drug resistance of colon cancer cells. Cancer Lett. 2020, 476, 67–74. [Google Scholar] [CrossRef]
- Yin, W.; Zhong, G.; Fan, H.; Xia, H. The Effect of Compound Sophora on Fluorouracil and Oxaliplatin Resistance in Colorectal Cancer Cells. Evid. Based Complement. Altern. Med. 2019, 2019, 7564232. [Google Scholar] [CrossRef]
- Riahi-Chebbi, I.; Souid, S.; Othman, H.; Haoues, M.; Karoui, H.; Morel, A.; Srairi-Abid, N.; Essafi, M.; Essafi-Benkhadir, K. The Phenolic compound Kaempferol overcomes 5-fluorouracil resistance in human resistant LS174 colon cancer cells. Sci. Rep. 2019, 9, 195. [Google Scholar] [CrossRef]
- Chen, N.; Kong, Y.; Wu, Y.; Gao, Q.; Fu, J.; Sun, X.; Geng, Q. CAC1 knockdown reverses drug resistance through the downregulation of P-gp and MRP-1 expression in colorectal cancer. PLoS ONE 2019, 14, e0222035. [Google Scholar] [CrossRef]
- Zhang, S.; Chatterjee, T.; Godoy, C.; Wu, L.; Liu, Q.J.; Carmon, K.S. GPR56 Drives Colorectal Tumor Growth and Promotes Drug Resistance through Upregulation of MDR1 Expression via a RhoA-Mediated Mechanism. Mol. Cancer Res. 2019, 17, 2196–2207. [Google Scholar] [CrossRef]
- Sun, L.; Ke, J.; He, Z.; Chen, Z.; Huang, Q.; Ai, W.; Wang, G.; Wei, Y.; Zou, X.; Zhang, S.; et al. HES1 Promotes Colorectal Cancer Cell Resistance To 5-Fu by Inducing Of EMT and ABC Transporter Proteins. J. Cancer 2017, 8, 2802–2808. [Google Scholar] [CrossRef]
- Hu, H.; Wang, M.; Guan, X.; Yuan, Z.; Liu, Z.; Zou, C.; Wang, G.; Gao, X.; Wang, X. Loss of ABCB4 attenuates the caspase-dependent apoptosis regulating resistance to 5-Fu in colorectal cancer. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef] [PubMed]
- Pasello, M.; Giudice, A.M.; Scotlandi, K. The ABC subfamily A transporters: Multifaceted players with incipient potentialities in cancer. Semin. Cancer Biol. 2020, 60, 57–71. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Feng, L.; Liu, P.; Duan, W. ANRIL promotes chemoresistance via disturbing expression of ABCC1 by regulating the expression of Let-7a in colorectal cancer. Biosci. Rep. 2018, 38, BSR20180620. [Google Scholar] [CrossRef] [PubMed]
- Schaller, L.; Lauschke, V.M. The genetic landscape of the human solute carrier (SLC) transporter superfamily. Hum. Genet. 2019, 138, 1359–1377. [Google Scholar] [CrossRef] [PubMed]
- Young, J. The SLC28 (CNT) and SLC29 (ENT) nucleoside transporter families: A 30-year collaborative odyssey. Biochem. Soc. Trans. 2016, 44, 869–876. [Google Scholar] [CrossRef]
- Mohelnikova-Duchonova, B.; Melichar, B.; Soucek, P. FOLFOX/FOLFIRI pharmacogenetics: The call for a personalized approach in colorectal cancer therapy. World J. Gastroenterol. 2014, 20, 10316–10330. [Google Scholar] [CrossRef]
- Phua, L.C.; Mal, M.; Koh, P.K.; Cheah, P.Y.; Chan, E.C.Y.; Ho, H.K. Investigating the role of nucleoside transporters in the resistance of colorectal cancer to 5-fluorouracil therapy. Cancer Chemother. Pharmacol. 2013, 71, 817–823. [Google Scholar] [CrossRef]
- Wang, W.-B.; Yang, Y.; Zhao, Y.-P.; Zhang, T.-P.; Liao, Q.; Shu, H. Recent studies of 5-fluorouracil resistance in pancreatic cancer. World J. Gastroenterol. 2014, 20, 15682–15690. [Google Scholar] [CrossRef]
- Ren, A.; Sun, S.; Li, S.; Chen, T.; Shu, Y.; Du, M.; Zhu, L. Genetic variants in SLC22A3 contribute to the susceptibility to colorectal cancer. Int. J. Cancer 2019, 145, 154–163. [Google Scholar] [CrossRef]
- Kikuchi, D.; Saito, M.; Saito, K.; Watanabe, Y.; Matsumoto, Y.; Kanke, Y.; Onozawa, H.; Hayase, S.; Sakamoto, W.; Ishigame, T.; et al. Upregulated solute carrier family 37 member 1 in colorectal cancer is associated with poor patient outcome and metastasis. Oncol. Lett. 2018, 15, 2065–2072. [Google Scholar] [CrossRef]
- Sheng, N.; Yan, L.; You, W.; Tan, G.; Gong, J.; Chen, H.; Yang, Y.; Hu, L.; Wang, Z. Knockdown of SLC39A7 inhibits cell growth and induces apoptosis in human colorectal cancer cells. Acta Biochim. Biophys. Sin. 2017, 49, 926–934. [Google Scholar] [CrossRef] [PubMed]
- Or, C.-H.R.; Huang, C.-W.; Chang, C.-C.; Lai, Y.-C.; Chen, Y.-J.; Chang, C.-C. Obatoclax, a Pan-BCL-2 Inhibitor, Downregulates Survivin to Induce Apoptosis in Human Colorectal Carcinoma Cells Via Suppressing WNT/β-catenin Signaling. Int. J. Mol. Sci. 2020, 21, 1773. [Google Scholar] [CrossRef] [PubMed]
- Knoll, G.; Riffelsberger, P.; Raats, D.; Kranenburg, O.; Ehrenschwender, M. NOXA-dependent contextual synthetic lethality of BCL-XL inhibition and “osmotic reprogramming” in colorectal cancer. Cell Death Dis. 2020, 11, 257. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Ghosh, B.; Chakraborty, T.; Roy, S. Bavachinin mitigates DMH induced colon cancer in rats by altering p53/Bcl2/BAX signaling associated with apoptosis. Biotech. Histochem. 2020. [Google Scholar] [CrossRef]
- Al-Obeed, O.; El-Obeid, A.S.; Matou-Nasri, S.; Vaali-Mohammed, M.-A.; AlHaidan, Y.; Elwatidy, M.; Al Dosary, H.; Alehaideb, Z.; Alkhayal, K.; Haseeb, A.; et al. Herbal melanin inhibits colorectal cancer cell proliferation by altering redox balance, inducing apoptosis, and modulating MAPK signaling. Cancer Cell Int. 2020, 20, 126. [Google Scholar] [CrossRef]
- Scherr, A.-L.; Gdynia, G.; Salou, M.; Radhakrishnan, P.; Duglova, K.; Heller, A.; Keim, S.; Kautz, N.; Jassowicz, A.; Elssner, C.; et al. Bcl-xL is an oncogenic driver in colorectal cancer. Cell Death Dis. 2016, 7, e2342. [Google Scholar] [CrossRef]
- Fu, J.; Xu, Y.; Yang, Y.; Liu, Y.; Ma, L.; Zhang, Y. Aspirin suppresses chemoresistance and enhances antitumor activity of 5-Fu in 5-Fu-resistant colorectal cancer by abolishing 5-Fu-induced NF-κB activation. Sci. Rep. 2019, 9, 16937. [Google Scholar] [CrossRef]
- Yang, Y.; Ma, L.; Xu, Y.; Liu, Y.; Li, W.; Cai, J.; Zhang, Y. Enalapril overcomes chemoresistance and potentiates antitumor efficacy of 5-FU in colorectal cancer by suppressing proliferation, angiogenesis, and NF-κB/STAT3-regulated proteins. Cell Death Dis. 2020, 11, 477. [Google Scholar] [CrossRef]
- Koosha, S.; Mohamed, Z.; Sinniah, A.; Alshawsh, M.A. Investigation into the Molecular Mechanisms underlying the Anti-proliferative and Anti-tumorigenesis activities of Diosmetin against HCT-116 Human Colorectal Cancer. Sci. Rep. 2019, 9, 5148. [Google Scholar] [CrossRef]
- Dai, X.; Zhou, B.; Xie, Y.; Lou, J.; Li, K. Bufalin and 5-fluorouracil synergistically induce apoptosis in colorectal cancer cells. Oncol. Lett. 2018, 15, 8019–8026. [Google Scholar] [CrossRef]
- Ramesh, P.; Medema, J.P. BCL-2 family deregulation in colorectal cancer: Potential for BH3 mimetics in therapy. Apoptosis 2020, 25, 305–320. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, X.; Liu, J.; Zhang, T. Knockdown of REG Iα Enhances the Sensitivity to 5-Fluorouracil of Colorectal Cancer Cells via Cyclin D1/CDK4 Pathway and BAX/BCL-2 Pathways. Cancer Biother. Radiopharm. 2019, 34, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Lan, W.; Zhao, J.; Shang, H.; Peng, J.; Chen, W.; Lin, J. Anlotinib overcomes multiple drug resistant of the colorectal cancer cells via inactivating PI3K/AKT pathway. bioRxiv 2019, 821801. [Google Scholar] [CrossRef]
- Srivastava, R.; Cao, Z.; Nedeva, C.; Naim, S.; Bachmann, D.; Rabachini, T.; Gangoda, L.; Shahi, S.; Glab, J.; Menassa, J.; et al. BCL-2 family protein BOK is a positive regulator of uridine metabolism in mammals. Proc. Natl. Acad. Sci. USA 2019, 116, 15469–15474. [Google Scholar] [CrossRef]
- Benhalilou, N.; Alsamri, H.; Alneyadi, A.; Athamneh, K.; Alrashedi, A.; Altamimi, N.; Al Dhaheri, Y.; Eid, A.H.; Iratni, R. Origanum majorana Ethanolic Extract Promotes Colorectal Cancer Cell Death by Triggering Abortive Autophagy and Activation of the Extrinsic Apoptotic Pathway. Front. Oncol. 2019, 9, 795. [Google Scholar] [CrossRef]
- Țigu, A.B.; Toma, V.-A.; Moț, A.C.; Jurj, A.; Moldovan, C.S.; Fischer-Fodor, E.; Berindan-Neagoe, I.; Pârvu, M. The Synergistic Antitumor Effect of 5-Fluorouracil Combined with Allicin against Lung and Colorectal Carcinoma Cells. Molecules 2020, 25, 1947. [Google Scholar] [CrossRef]
- Koosha, S.; Mohamed, Z.; Sinniah, A.; Ibrahim, Z.A.; Seyedan, A.; Alshawsh, M.A. Antiproliferative and apoptotic activities of 8-prenylnaringenin against human colon cancer cells. Life Sci. 2019, 232, 116633. [Google Scholar] [CrossRef]
- Rani, I.; Sharma, B.; Kumar, S.; Kaur, S.; Agnihotri, N. Apoptosis mediated chemosensitization of tumor cells to 5-fluorouracil on supplementation of fish oil in experimental colon carcinoma. Tumor Biol. 2017, 39, 1010428317695019. [Google Scholar] [CrossRef]
- Lee, J.; Kang, J.-S.; Choi, B.-Y.; Keum, Y.-S. Sensitization of 5-Fluorouracil-Resistant SNUC5 Colon Cancer Cells to Apoptosis by α-Mangostin. Biomol. Ther. 2016, 24, 604–609. [Google Scholar] [CrossRef][Green Version]
- Lee, J.H.; Lee, S.-W. Enhancement of chemosensitivity in 5-fluorouracil-resistant colon cancer cells with carcinoembryonic antigen-specific RNA aptamer. Mol. Biol. Rep. 2019, 46, 3835–3842. [Google Scholar] [CrossRef]
- Cervantes-Villagrana, R.D.; Albores-García, D.; Cervantes-Villagrana, A.R.; García-Acevez, S.J. Tumor-induced neurogenesis and immune evasion as targets of innovative anti-cancer therapies. Signal Transduct. Target. Ther. 2020, 5, 99. [Google Scholar] [CrossRef]
- Xiao, W.; Ibrahim, M.L.; Redd, P.S.; Klement, J.D.; Lu, C.; Yang, D.; Savage, N.M.; Liu, K. Loss of Fas Expression and Function Is Coupled with Colon Cancer Resistance to Immune Checkpoint Inhibitor Immunotherapy. Mol. Cancer Res. 2019, 17, 420–430. [Google Scholar] [CrossRef] [PubMed]
- Bower, J.J.; Vance, L.D.; Psioda, M.; Smith-Roe, S.L.; Simpson, D.A.; Ibrahim, J.G.; Hoadley, K.A.; Perou, C.M.; Kaufmann, W.K. Patterns of cell cycle checkpoint deregulation associated with intrinsic molecular subtypes of human breast cancer cells. NPJ Breast Cancer 2017, 3, 9. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-J.; Chung, Y.-C.; Chang, H.-L.; Chang, H.-P.; Chou, J.-L.; Lin, C.-C.; Chen, C.-H.; Hsu, C.-P. Synergistic Effect of Combined Treatment with Longan Flower Extract and 5-Fluorouracil on Colorectal Cancer Cells. Nutr. Cancer 2020, 72, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Francipane, M.G.; Bulanin, D.; Lagasse, E. Establishment and Characterization of 5-Fluorouracil-Resistant Human Colorectal Cancer Stem-Like Cells: Tumor Dynamics under Selection Pressure. Int. J. Mol. Sci. 2019, 20, 1817. [Google Scholar] [CrossRef]
- Liu, T.; Wang, X.; Hu, W.; Fang, Z.; Jin, Y.; Fang, X.; Miao, Q.R. Epigenetically Down-Regulated Acetyltransferase PCAF Increases the Resistance of Colorectal Cancer to 5-Fluorouracil. Neoplasia 2019, 21, 557–570. [Google Scholar] [CrossRef]
- Nakagawa, Y.; Kajihara, A.; Takahashi, A.; Kondo, N.; Mori, E.; Kirita, T.; Ohnishi, T. The BRCA2 gene is a potential molecular target during 5-fluorouracil therapy in human oral cancer cells. Oncol. Rep. 2014, 31, 2001–2006. [Google Scholar] [CrossRef]
- Liu, Z.; Yu, M.; Fei, B.; Sun, J.; Wang, D. Identification Of Natural Compound Derivative For Inhibition Of XLF And Overcoming Chemoresistance In Colorectal Cancer Cells. Drug Des. Dev. Ther. 2019, 13, 3823–3834. [Google Scholar] [CrossRef]
- Srinivas, U.S.; Dyczkowski, J.; Beißbarth, T.; Gaedcke, J.; Mansour, W.Y.; Borgmann, K.; Dobbelstein, M. 5-Fluorouracil sensitizes colorectal tumor cells towards double stranded DNA breaks by interfering with homologous recombination repair. Oncotarget 2015, 6, 12574–12586. [Google Scholar] [CrossRef][Green Version]
- Liu, Z.; Yu, M.; Fei, B.; Sun, J.; Wang, D. Nonhomologous end joining key factor XLF enhances both 5-florouracil and oxaliplatin resistance in colorectal cancer. Onco Targets Ther. 2019, 12, 2095–2104. [Google Scholar] [CrossRef]
- Chen, X.; Li, W.-F.; Wu, X.; Zhang, H.-C.; Chen, L.; Zhang, P.-Y.; Liu, L.-Y.; Ma, D.; Chen, T.; Zhou, L.; et al. Dicer regulates non-homologous end joining and is associated with chemosensitivity in colon cancer patients. Carcinogenesis 2017, 38, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Mei, L.; Zhang, J.; He, K.; Zhang, J. Ataxia telangiectasia and Rad3-related inhibitors and cancer therapy: Where we stand. J. Hematol. Oncol. 2019, 12, 43. [Google Scholar] [CrossRef] [PubMed]
- Akasaka, T.; Tsujii, M.; Kondo, J.; Hayashi, Y.; Ying, J.; Lu, Y.; Kato, M.; Yamada, T.; Yamamoto, S.; Inoue, T.; et al. 5FU resistance abrogates the amplified cytotoxic effects induced by inhibiting checkpoint kinase 1 in p53mutated colon cancer cells. Int. J. Oncol. 2015, 46, 63–70. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Narayan, S.; Jaiswal, A.S.; Sharma, R.; Nawab, A.; Duckworth, L.V.; Law, B.K.; Zajac-Kaye, M.; George, T.J.; Sharma, J.; Sharma, A.K.; et al. NSC30049 inhibits Chk1 pathway in 5-FU-resistant CRC bulk and stem cell populations. Oncotarget 2017, 8, 57246–57264. [Google Scholar] [CrossRef]
- Yao, J.; Huang, A.; Zheng, X.; Liu, T.; Lin, Z.; Zhang, S.; Yang, Q.; Zhang, T.; Ma, H. 53BP1 loss induces chemoresistance of colorectal cancer cells to 5-fluorouracil by inhibiting the ATM-CHK2-P53 pathway. J. Cancer Res. Clin. Oncol. 2017, 143, 419–431. [Google Scholar] [CrossRef]
- Maiuthed, A.; Ninsontia, C.; Erlenbach-Wuensch, K.; Ndreshkjana, B.; Muenzner, J.K.; Caliskan, A.; Husayn, A.P.; Chaotham, C.; Hartmann, A.; Vial Roehe, A.; et al. Cytoplasmic p21 Mediates 5-Fluorouracil Resistance by Inhibiting Pro-Apoptotic Chk2. Cancers 2018, 10, 373. [Google Scholar] [CrossRef]
- Yun, C.W.; Lee, S.H. The Roles of Autophagy in Cancer. Int. J. Mol. Sci. 2018, 19, 3466. [Google Scholar] [CrossRef]
- Gu, X.-Y.; Jiang, Y.; Li, M.-Q.; Han, P.; Liu, Y.-L.; Cui, B.-B. Over-expression of EGFR regulated by RARA contributes to 5-FU resistance in colon cancer. Aging 2020, 12, 156–177. [Google Scholar] [CrossRef]
- Zhang, P.; Lai, Z.-L.; Chen, H.-F.; Zhang, M.; Wang, A.; Jia, T.; Sun, W.-Q.; Zhu, X.-M.; Chen, X.-F.; Zhao, Z.; et al. Curcumin synergizes with 5-fluorouracil by impairing AMPK/ULK1-dependent autophagy, AKT activity and enhancing apoptosis in colon cancer cells with tumor growth inhibition in xenograft mice. J. Exp. Clin. Cancer Res. 2017, 36, 190. [Google Scholar] [CrossRef]
- Yang, J.-W.; Zhang, Q.-H.; Liu, T. Autophagy facilitates anticancer effect of 5-fluorouracil in HCT-116 cells. J. Cancer Res. Ther. 2018, 14, S1141–S1147. [Google Scholar] [CrossRef]
- Zhang, R.; Pan, T.; Xiang, Y.; Zhang, M.; Feng, J.; Liu, S.; Duan, T.; Chen, P.; Zhai, B.; Chen, X.; et al. β-Elemene Reverses the Resistance of p53-Deficient Colorectal Cancer Cells to 5-Fluorouracil by Inducing Pro-death Autophagy and Cyclin D3-Dependent Cycle Arrest. Front. Bioeng. Biotechnol. 2020, 8, 378. [Google Scholar] [CrossRef] [PubMed]
- Alnuqaydan, A.M.; Rah, B.; Almutary, A.G.; Chauhan, S.S. Synergistic antitumor effect of 5-fluorouracil and withaferin-A induces endoplasmic reticulum stress-mediated autophagy and apoptosis in colorectal cancer cells. Am. J. Cancer Res. 2020, 10, 799–815. [Google Scholar] [PubMed]
- Liu, S.; Lin, H.; Wang, D.; Li, Q.; Luo, H.; Li, G.; Chen, X.; Li, Y.; Chen, P.; Zhai, B.; et al. PCDH17 increases the sensitivity of colorectal cancer to 5-fluorouracil treatment by inducing apoptosis and autophagic cell death. Signal Transduct. Target. Ther. 2019, 4, 53. [Google Scholar] [CrossRef] [PubMed]
- Woźniak, M.; Makuch, S.; Winograd, K.; Wiśniewski, J.; Ziółkowski, P.; Agrawal, S. 6-Shogaol enhances the anticancer effect of 5-fluorouracil, oxaliplatin, and irinotecan via increase of apoptosis and autophagy in colon cancer cells in hypoxic/aglycemic conditions. BMC Complement. Med. Ther. 2020, 20, 141. [Google Scholar] [CrossRef]
- Li, X.; Zhou, Y.; Li, Y.; Yang, L.; Ma, Y.; Peng, X.; Yang, S.; Liu, J.; Li, H. Autophagy: A novel mechanism of chemoresistance in cancers. Biomed. Pharmacother. 2019, 119, 109415. [Google Scholar] [CrossRef]
- Vu, T.; Datta, P.K. Regulation of EMT in Colorectal Cancer: A Culprit in Metastasis. Cancers 2017, 9, 171. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Gao, N.; Bian, D.; Zhai, Q.; Yang, P.; Li, M.; Wang, X. Correlation between FAK and EGF-Induced EMT in Colorectal Cancer Cells. J. Oncol. 2020, 2020, 5428920. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Lai, Q.; He, C.; Fang, Y.; Yan, Q.; Zhang, Y.; Wang, X.; Gu, C.; Wang, Y.; Ye, L.; et al. RUNX1 promotes tumour metastasis by activating the Wnt/β-catenin signalling pathway and EMT in colorectal cancer. J. Exp. Clin. Cancer Res. 2019, 38, 334. [Google Scholar] [CrossRef]
- Ji, M.; Li, W.; He, G.; Zhu, D.; Lv, S.; Tang, W.; Jian, M.; Zheng, P.; Yang, L.; Qi, Z.; et al. Zinc-α2-glycoprotein 1 promotes EMT in colorectal cancer by filamin A mediated focal adhesion pathway. J. Cancer 2019, 10, 5557–5566. [Google Scholar] [CrossRef]
- Wang, K.; Song, K.; Ma, Z.; Yao, Y.; Liu, C.; Yang, J.; Xiao, H.; Zhang, J.; Zhang, Y.; Zhao, W. Identification of EMT-related high-risk stage II colorectal cancer and characterisation of metastasis-related genes. Br. J. Cancer 2020. [Google Scholar] [CrossRef]
- Lu, Y.; Zhang, R.; Zhang, X.; Zhang, B.; Yao, Q. Curcumin may reverse 5-fluorouracil resistance on colonic cancer cells by regulating TET1-NKD-Wnt signal pathway to inhibit the EMT progress. Biomed. Pharmacother. 2020, 129, 110381. [Google Scholar] [CrossRef]
- Liang, L.; Wu, J.; Luo, J.; Wang, L.; Chen, Z.X.; Han, C.L.; Gan, T.Q.; Huang, J.A.; Cai, Z.W. Oxymatrine reverses 5-fluorouracil resistance by inhibition of colon cancer cell epithelial-mesenchymal transition and NF-κB signaling in vitro. Oncol. Lett. 2020, 19, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Babaei-Jadidi, R.; Lorenzi, F.; Spencer-Dene, B.; Clarke, P.; Domingo, E.; Tulchinsky, E.; Vries, R.G.J.; Kerr, D.; Pan, Y.; et al. An FBXW7-ZEB2 axis links EMT and tumour microenvironment to promote colorectal cancer stem cells and chemoresistance. Oncogenesis 2019, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Gasiulė, S.; Dreize, N.; Kaupinis, A.; Ražanskas, R.; Čiupas, L.; Stankevičius, V.; Kapustina, Ž.; Laurinavičius, A.; Valius, M.; Vilkaitis, G. Molecular Insights into miRNA-Driven Resistance to 5-Fluorouracil and Oxaliplatin Chemotherapy: miR-23b Modulates the Epithelial-Mesenchymal Transition of Colorectal Cancer Cells. J. Clin. Med. 2019, 8, 2115. [Google Scholar] [CrossRef] [PubMed]
- Galle, E.; Thienpont, B.; Cappuyns, S.; Venken, T.; Busschaert, P.; Van Haele, M.; Van Cutsem, E.; Roskams, T.; van Pelt, J.; Verslype, C.; et al. DNA methylation-driven EMT is a common mechanism of resistance to various therapeutic agents in cancer. Clin. Epigenet. 2020, 12, 27. [Google Scholar] [CrossRef]
- Li, Y.; Huang, S.; Li, Y.; Zhang, W.; He, K.; Zhao, M.; Lin, H.; Li, D.; Zhang, H.; Zheng, Z.; et al. Decreased expression of LncRNA SLC25A25-AS1 promotes proliferation, chemoresistance, and EMT in colorectal cancer cells. Tumor Biol. 2016, 1–11. [Google Scholar] [CrossRef]
- Chen, D.-L.; Xu, R.-H. The emerging role of long non-coding RNAs in the drug resistance of colorectal cancer. Int. J. Clin. Exp. Pathol. 2018, 11, 4735–4743. [Google Scholar]
- Deng, J.-J.; Zhang, W.; Xu, X.-M.; Zhang, F.; Tao, W.-P.; Ye, J.-J.; Ge, W. Twist mediates an aggressive phenotype in human colorectal cancer cells. Int. J. Oncol. 2016, 48, 1117–1124. [Google Scholar] [CrossRef]
- Francescangeli, F.; Contavalli, P.; De Angelis, M.L.; Careccia, S.; Signore, M.; Haas, T.L.; Salaris, F.; Baiocchi, M.; Boe, A.; Giuliani, A.; et al. A pre-existing population of ZEB2+ quiescent cells with stemness and mesenchymal features dictate chemoresistance in colorectal cancer. J. Exp. Clin. Cancer Res. 2020, 39, 2. [Google Scholar] [CrossRef]
- Mohd-Zahid, M.H.; Mohamud, R.; Che Abdullah, C.A.; Lim, J.; Alem, H.; Wan Hanaffi, W.N.; Iskandar, Z.A. Colorectal cancer stem cells: A review of targeted drug delivery by gold nanoparticles. RSC Adv. 2020, 10, 973–985. [Google Scholar] [CrossRef]
- Iyer, D.N.; Sin, W.-Y.; Ng, L. Linking stemness with colorectal cancer initiation, progression, and therapy. World J. Stem Cells 2019, 11, 519–534. [Google Scholar] [CrossRef]
- Quarni, W.; Dutta, R.; Green, R.; Katiri, S.; Patel, B.; Mohapatra, S.S.; Mohapatra, S. Mithramycin A Inhibits Colorectal Cancer Growth by Targeting Cancer Stem Cells. Sci. Rep. 2019, 9, 15202. [Google Scholar] [CrossRef] [PubMed]
- Jahanafrooz, Z.; Mosafer, J.; Akbari, M.; Hashemzaei, M.; Mokhtarzadeh, A.; Baradaran, B. Colon cancer therapy by focusing on colon cancer stem cells and their tumor microenvironment. J. Cell. Physiol. 2020, 235, 4153–4166. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhao, J.; Ding, J.; Wang, Z.; Du, J.; Wu, C. Knocking down LSD1 inhibits the stemness features of colorectal cancer stem cells. Braz. J. Med. Biol. Res. 2020, 53, e9230. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Dong, J.; Haiech, J.; Kilhoffer, M.-C.; Zeniou, M. Cancer Stem Cell Quiescence and Plasticity as Major Challenges in Cancer Therapy. Stem Cells Int. 2016, 2016, 1740936. [Google Scholar] [CrossRef] [PubMed]
- Trisciuoglio, D.; Tupone, M.G.; Desideri, M.; Di Martile, M.; Gabellini, C.; Buglioni, S.; Pallocca, M.; Alessandrini, G.; D’Aguanno, S.; Del Bufalo, D. BCL-XL overexpression promotes tumor progression-associated properties. Cell Death Dis. 2017, 8, 3216. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Xia, L.; Wang, H.; Oyang, L.; Su, M.; Liu, Q.; Lin, J.; Tan, S.; Tian, Y.; Liao, Q.; et al. Cancer stem cells in progression of colorectal cancer. Oncotarget 2017, 9, 33403–33415. [Google Scholar] [CrossRef]
- Begicevic, R.-R.; Falasca, M. ABC Transporters in Cancer Stem Cells: Beyond Chemoresistance. Int. J. Mol. Sci. 2017, 18, 2362. [Google Scholar] [CrossRef]
- Kim, E.; Davidson, L.A.; Zoh, R.S.; Hensel, M.E.; Patil, B.S.; Jayaprakasha, G.K.; Callaway, E.S.; Allred, C.D.; Turner, N.D.; Weeks, B.R.; et al. Homeostatic responses of colonic LGR5+ stem cells following acute in vivo exposure to a genotoxic carcinogen. Carcinogenesis 2016, 37, 206–214. [Google Scholar] [CrossRef]
- Das, P.; Islam, M.; Lam, A. The Roles of Cancer Stem Cells and Therapy Resistance in Colorectal Carcinoma. Cells 2020, 9, 1392. [Google Scholar] [CrossRef]
- Steinbichler, T.B.; Dudás, J.; Skvortsov, S.; Ganswindt, U.; Riechelmann, H.; Skvortsova, I.-I. Therapy resistance mediated by cancer stem cells. Semin. Cancer Biol. 2018, 53, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, W.; Cai, H.; Du, B.; Zhang, L.; Ma, W.; Hu, Y.; Feng, S.; Miao, G. MACC1 facilitates chemoresistance and cancer stem cell-like properties of colon cancer cells through the PI3K/AKT signaling pathway. Mol. Med. Rep. 2017, 16, 8747–8754. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.-J.; Kang, G.-J.; Kang, J.-I.; Boo, H.-J.; Hyun, J.W.; Koh, Y.S.; Chang, W.-Y.; Kim, Y.R.; Kwon, J.-M.; Maeng, Y.H.; et al. Over-activation of AKT signaling leading to 5-Fluorouracil resistance in SNU-C5/5-FU cells. Oncotarget 2018, 9, 19911–19928. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Xu, X.; Chen, D.; Zhao, F.; Wang, W. Therapeutic potential of targeting the Wnt/β-catenin signaling pathway in colorectal cancer. Biomed. Pharmacother. 2019, 110, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Urushibara, S.; Tsubota, T.; Asai, R.; Azumi, J.; Ashida, K.; Fujiwara, Y.; Shiota, G. WNT/β-Catenin Signaling Inhibitor IC-2 Suppresses Sphere Formation and Sensitizes Colorectal Cancer Cells to 5-Fluorouracil. Anticancer Res. 2017, 37, 4085–4091. [Google Scholar]
- Dermani, F.K.; Amini, R.; Saidijam, M.; Pourjafar, M.; Saki, S.; Najafi, R. Zerumbone inhibits epithelial-mesenchymal transition and cancer stem cells properties by inhibiting the β-catenin pathway through miR-200c. J. Cell. Physiol. 2018, 233, 9538–9547. [Google Scholar] [CrossRef]
- Todaro, M.; Alea, M.P.; Di Stefano, A.B.; Cammareri, P.; Vermeulen, L.; Iovino, F.; Tripodo, C.; Russo, A.; Gulotta, G.; Medema, J.P.; et al. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell 2007, 1, 389–402. [Google Scholar] [CrossRef]
- Tsunekuni, K.; Konno, M.; Haraguchi, N.; Koseki, J.; Asai, A.; Matsuoka, K.; Kobunai, T.; Takechi, T.; Doki, Y.; Mori, M.; et al. CD44/CD133-Positive Colorectal Cancer Stem Cells are Sensitive to Trifluridine Exposure. Sci. Rep. 2019, 9, 14861. [Google Scholar] [CrossRef]
- Vincent, A.; Ouelkdite-Oumouchal, A.; Souidi, M.; Leclerc, J.; Neve, B.; Van Seuningen, I. Colon cancer stemness as a reversible epigenetic state: Implications for anticancer therapies. World J. Stem Cells 2019, 11, 920–936. [Google Scholar] [CrossRef]
- Paschall, A.V.; Yang, D.; Lu, C.; Redd, P.S.; Choi, J.-H.; Heaton, C.M.; Lee, J.R.; Nayak-Kapoor, A.; Liu, K. CD133+CD24lo defines a 5-Fluorouracil-resistant colon cancer stem cell-like phenotype. Oncotarget 2016, 7, 78698–78712. [Google Scholar] [CrossRef]
- Park, Y.Y.; An, C.H.; Oh, S.T.; Chang, E.D.; Lee, J. Expression of CD133 is associated with poor prognosis in stage II colorectal carcinoma. Medicine 2019, 98, e16709. [Google Scholar] [CrossRef]
- Miller, T.J.; McCoy, M.J.; Hemmings, C.; Bulsara, M.K.; Iacopetta, B.; Platell, C.F. The prognostic value of cancer stem-like cell markers SOX2 and CD133 in stage III colon cancer is modified by expression of the immune-related markers FoxP3, PD-L1 and CD3. Pathology 2017, 49, 721–730. [Google Scholar] [CrossRef]
- Márquez-González, R.M.; Saucedo-Sariñana, A.M.; Barros-Núñez, P.; Gallegos-Arreola, M.P.; Pineda-Razo, T.D.; Marin-Contreras, M.E.; Flores-Martínez, S.E.; Sánchez-Corona, J.; Rosales-Reynoso, M.A. CD44 Genotypes Are Associated with Susceptibility and Tumor Characteristics in Colorectal Cancer Patients. Tohoku J. Exp. Med. 2020, 250, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Tang, Y.; Xie, L.; Huang, A.; Xue, C.; Gu, Z.; Wang, K.; Zong, S. The Prognostic and Clinical Value of CD44 in Colorectal Cancer: A Meta-Analysis. Front. Oncol. 2019, 9, 309. [Google Scholar] [CrossRef] [PubMed]
- Eom, D.-W.; Hong, S.-M.; Kim, G.; Bae, Y.K.; Jang, K.-T.; Yu, E. Prognostic Significance of CD44v6, CD133, CD166, and ALDH1 Expression in Small Intestinal Adenocarcinoma. Appl. Immunohistochem. Mol. Morphol. AIMM 2015, 23, 682–688. [Google Scholar] [CrossRef]
- Kozovska, Z.; Patsalias, A.; Bajzik, V.; Durinikova, E.; Demkova, L.; Jargasova, S.; Smolkova, B.; Plava, J.; Kucerova, L.; Matuskova, M. ALDH1A inhibition sensitizes colon cancer cells to chemotherapy. BMC Cancer 2018, 18, 656. [Google Scholar] [CrossRef] [PubMed]
- Sivamaruthi, B.S.; Kesika, P.; Chaiyasut, C. The Role of Probiotics in Colorectal Cancer Management. Evid. Based Complement. Altern. Med. 2020, 2020, 3535982. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Ha, E.-M. Combination Therapy of Lactobacillus plantarum Supernatant and 5-Fluouracil Increases Chemosensitivity in Colorectal Cancer Cells. J. Microbiol. Biotechnol. 2016, 26, 1490–1503. [Google Scholar] [CrossRef] [PubMed]
- Olmeda, F.; Ben Amar, M. Clonal pattern dynamics in tumor: The concept of cancer stem cells. Sci. Rep. 2019, 9, 15607. [Google Scholar] [CrossRef]
- Dominijanni, A.; Gmeiner, W.H. Improved potency of F10 relative to 5-fluorouracil in colorectal cancer cells with p53 mutations. Cancer Drug Resist. 2018, 1, 48–58. [Google Scholar] [CrossRef]
- Huang, Y.-C.; Li, Z.; Li, J.; Lyu, F.-J. Interaction between Stem Cells and the Microenvironment for Musculoskeletal Repair. Stem Cells Int. 2020, 2020, 7587428. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.-L.; Wang, Z.-J.; Wei, G.-H.; Yang, Y.; Wang, X.-W. Changes in extracellular matrix in different stages of colorectal cancer and their effects on proliferation of cancer cells. World J. Gastrointest. Oncol. 2020, 12, 267–275. [Google Scholar] [CrossRef]
- Manmuan, S.; Manmuan, P. Fucoxanthin enhances 5-FU chemotherapeutic efficacy in colorectal cancer cells by affecting MMP-9 invasive proteins. J. Appl. Pharm. Sci. 2019, 9, 7–14. [Google Scholar]
- Bauer, J.; Emon, M.A.B.; Staudacher, J.J.; Thomas, A.L.; Zessner-Spitzenberg, J.; Mancinelli, G.; Krett, N.; Saif, M.T.; Jung, B. Increased stiffness of the tumor microenvironment in colon cancer stimulates cancer associated fibroblast-mediated prometastatic activin A signaling. Sci. Rep. 2020, 10, 50. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Miao, Z.; Yang, Q.; Wang, Y.; Zhang, J. The Dynamic Roles of Mesenchymal Stem Cells in Colon Cancer. Can. J. Gastroenterol. Hepatol. 2018, 2018, 7628763. [Google Scholar] [CrossRef]
- Herrera, M.; Galindo-Pumariño, C.; García-Barberán, V.; Peña, C. A Snapshot of The Tumor Microenvironment in Colorectal Cancer: The Liquid Biopsy. Int. J. Mol. Sci. 2019, 20, 6016. [Google Scholar] [CrossRef]
- Wei, R.; Liu, S.; Zhang, S.; Min, L.; Zhu, S. Cellular and Extracellular Components in Tumor Microenvironment and Their Application in Early Diagnosis of Cancers. Anal. Cell. Pathol. 2020, 2020, 6283796. [Google Scholar] [CrossRef]
- Yahaya, M.A.F.; Lila, M.A.M.; Ismail, S.; Zainol, M.; Afizan, N.A.R.N.M. Tumour-Associated Macrophages (TAMs) in Colon Cancer and How to Reeducate Them. J. Immunol. Res. 2019, 2019, 2368249. [Google Scholar] [CrossRef]
- Zhong, X.; Chen, B.; Yang, Z. The Role of Tumor-Associated Macrophages in Colorectal Carcinoma Progression. Cell. Physiol. Biochem. 2018, 45, 356–365. [Google Scholar] [CrossRef]
- Ben-Shmuel, A.; Biber, G.; Barda-Saad, M. Unleashing Natural Killer Cells in the Tumor Microenvironment—The Next Generation of Immunotherapy? Front. Immunol. 2020, 11, 275. [Google Scholar] [CrossRef]
- Bassani, B.; Baci, D.; Gallazzi, M.; Poggi, A.; Bruno, A.; Mortara, L. Natural Killer Cells as Key Players of Tumor Progression and Angiogenesis: Old and Novel Tools to Divert Their Pro-Tumor Activities into Potent Anti-Tumor Effects. Cancers 2019, 11, 461. [Google Scholar] [CrossRef]
- Guo, G.; Wang, Y.; Zhou, Y.; Quan, Q.; Zhang, Y.; Wang, H.; Zhang, B.; Xia, L. Immune cell concentrations among the primary tumor microenvironment in colorectal cancer patients predicted by clinicopathologic characteristics and blood indexes. J. Immunother. Cancer 2019, 7, 179. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Wang, Y.; Tiruthani, K. Tumor immune microenvironment and nano-immunotherapeutics in colorectal cancer. Nanomed. Nanotechnol. Biol. Med. 2019, 21, 102034. [Google Scholar] [CrossRef]
- Guo, F.F.; Cui, J.W. The Role of Tumor-Infiltrating B Cells in Tumor Immunity. J. Oncol. 2019, 2019, 2592419. [Google Scholar] [CrossRef]
- Zhang, G.; Luo, X.; Zhang, W.; Chen, E.; Xu, J.; Wang, F.; Cao, G.; Ju, Z.; Jin, D.; Huang, X.; et al. CXCL-13 Regulates Resistance to 5-Fluorouracil in Colorectal Cancer. Cancer Res. Treat. 2020, 52, 622–633. [Google Scholar] [CrossRef]
- Kazanietz, M.G.; Durando, M.; Cooke, M. CXCL13 and Its Receptor CXCR5 in Cancer: Inflammation, Immune Response, and Beyond. Front. Endocrinol. 2019, 10, 471. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhang, X.; Guo, H.; Fu, L.; Pan, G.; Sun, Y. CXCL13-CXCR5 axis promotes the growth and invasion of colon cancer cells via PI3K/AKT pathway. Mol. Cell. Biochem. 2015, 400, 287–295. [Google Scholar] [CrossRef]
- Yu, X.; Shi, W.; Zhang, Y.; Wang, X.; Sun, S.; Song, Z.; Liu, M.; Zeng, Q.; Cui, S.; Qu, X. CXCL12/CXCR4 axis induced miR-125b promotes invasion and confers 5-fluorouracil resistance through enhancing autophagy in colorectal cancer. Sci. Rep. 2017, 7, 42226. [Google Scholar] [CrossRef]
- Huang, W.-S.; Hsieh, M.-C.; Huang, C.-Y.; Kuo, Y.-H.; Tung, S.-Y.; Shen, C.-H.; Hsieh, Y.-Y.; Teng, C.-C.; Lee, K.-F.; Chen, T.-C.; et al. The Association of CXC Receptor 4 Mediated Signaling Pathway with Oxaliplatin-Resistant Human Colorectal Cancer Cells. PLoS ONE 2016, 11, e0159927. [Google Scholar] [CrossRef]
- Ma, J.; Sun, X.; Wang, Y.; Chen, B.; Qian, L.; Wang, Y. Fibroblast-derived CXCL12 regulates PTEN expression and is associated with the proliferation and invasion of colon cancer cells via PI3k/Akt signaling. Cell Commun. Signal. 2019, 17, 119. [Google Scholar] [CrossRef]
- Wang, D.; Yang, L.; Yu, W.; Wu, Q.; Lian, J.; Li, F.; Liu, S.; Li, A.; He, Z.; Liu, J.; et al. Colorectal cancer cell-derived CCL20 recruits regulatory T cells to promote chemoresistance via FOXO1/CEBPB/NF-κB signaling. J. Immunother. Cancer 2019, 7, 215. [Google Scholar] [CrossRef] [PubMed]
- Romano, G.; Santi, L.; Bianco, M.R.; Giuffrè, M.R.; Pettinato, M.; Bugarin, C.; Garanzini, C.; Savarese, L.; Leoni, S.; Cerrito, M.G.; et al. The TGF-β pathway is activated by 5-fluorouracil treatment in drug resistant colorectal carcinoma cells. Oncotarget 2016, 7, 22077–22091. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Yang, C.; Wang, S.; Shi, D.; Zhang, C.; Lin, X.; Xiong, B. M2 macrophages confer resistance to 5-fluorouracil in colorectal cancer through the activation of CCL22/PI3K/AKT signaling. Onco Targets Ther. 2019, 12, 3051–3063. [Google Scholar] [CrossRef]
- Jayasingam, S.D.; Citartan, M.; Thang, T.H.; Mat Zin, A.A.; Ang, K.C.; Ch’ng, E.S. Evaluating the Polarization of Tumor-Associated Macrophages Into M1 and M2 Phenotypes in Human Cancer Tissue: Technicalities and Challenges in Routine Clinical Practice. Front. Oncol. 2020, 9, 1512. [Google Scholar] [CrossRef]
- Malesci, A.; Bianchi, P.; Celesti, G.; Basso, G.; Marchesi, F.; Grizzi, F.; Di Caro, G.; Cavalleri, T.; Rimassa, L.; Palmqvist, R.; et al. Tumor-associated macrophages and response to 5-fluorouracil adjuvant therapy in stage III colorectal cancer. Oncoimmunology 2017, 6, e1342918. [Google Scholar] [CrossRef]
- Liu, T.; Han, C.; Wang, S.; Fang, P.; Ma, Z.; Xu, L.; Yin, R. Cancer-associated fibroblasts: An emerging target of anti-cancer immunotherapy. J. Hematol. Oncol. 2019, 12, 86. [Google Scholar] [CrossRef]
- Gonçalves-Ribeiro, S.; Díaz-Maroto, N.G.; Berdiel-Acer, M.; Soriano, A.; Guardiola, J.; Martínez-Villacampa, M.; Salazar, R.; Capellà, G.; Villanueva, A.; Martínez-Balibrea, E.; et al. Carcinoma-associated fibroblasts affect sensitivity to oxaliplatin and 5FU in colorectal cancer cells. Oncotarget 2016, 7, 59766–59780. [Google Scholar] [CrossRef]
- Yadav, V.K.; Huang, Y.-J.; George, T.A.; Wei, P.-L.; Sumitra, M.R.; Ho, C.-L.; Chang, T.-H.; Wu, A.T.H.; Huang, H.-S. Preclinical Evaluation of the Novel Small-Molecule MSI-N1014 for Treating Drug-Resistant Colon Cancer via the LGR5/β-catenin/miR-142-3p Network and Reducing Cancer-Associated Fibroblast Transformation. Cancers 2020, 12, 1590. [Google Scholar] [CrossRef]
- Zhang, R.; Qi, F.; Zhao, F.; Li, G.; Shao, S.; Zhang, X.; Yuan, L.; Feng, Y. Cancer-associated fibroblasts enhance tumor-associated macrophages enrichment and suppress NK cells function in colorectal cancer. Cell Death Dis. 2019, 10, 273. [Google Scholar] [CrossRef]
- Liu, M.-L.; Zang, F.; Zhang, S.-J. RBCK1 contributes to chemoresistance and stemness in colorectal cancer (CRC). Biomed. Pharmacother. 2019, 118, 109250. [Google Scholar] [CrossRef]
- Koh, B.; Jeon, H.; Kim, D.; Kang, D.; Kim, K.R. Effect of fibroblast co-culture on the proliferation, viability and drug response of colon cancer cells. Oncol. Lett. 2019, 17, 2409–2417. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, G.J. Regulation of heterogeneous cancer-associated fibroblasts: The molecular pathology of activated signaling pathways. J. Exp. Clin. Cancer Res. 2020, 39, 112. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Yan, C.; Mu, L.; Huang, K.; Li, X.; Tao, D.; Wu, Y.; Qin, J. Fibroblast-Derived Exosomes Contribute to Chemoresistance through Priming Cancer Stem Cells in Colorectal Cancer. PLoS ONE 2015, 10, e0125625. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, R.-X.; Chan, K.-W.; Hu, J.; Zhang, J.; Wei, L.; Tan, H.; Yang, X.; Liu, H. Exosomal transfer of p-STAT3 promotes acquired 5-FU resistance in colorectal cancer cells. J. Exp. Clin. Cancer Res. 2019, 38, 320. [Google Scholar] [CrossRef]
- Hu, Y.-B.; Yan, C.; Mu, L.; Mi, Y.-L.; Zhao, H.; Hu, H.; Li, X.-L.; Tao, D.-D.; Wu, Y.-Q.; Gong, J.-P.; et al. Exosomal Wnt-induced dedifferentiation of colorectal cancer cells contributes to chemotherapy resistance. Oncogene 2019, 38, 1951–1965. [Google Scholar] [CrossRef]
- Hon, K.W.; Ab-Mutalib, N.S.; Abdullah, N.M.A.; Jamal, R.; Abu, N. Extracellular Vesicle-derived circular RNAs confers chemoresistance in Colorectal cancer. Sci. Rep. 2019, 9, 16497. [Google Scholar] [CrossRef] [PubMed]
- Bigagli, E.; Luceri, C.; Guasti, D.; Cinci, L. Exosomes secreted from human colon cancer cells influence the adhesion of neighboring metastatic cells: Role of microRNA-210. Cancer Biol. Ther. 2016, 17, 1062–1069. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.-H.; Tian, D.; Yang, Z.-C.; Li, J.-L. Exosomal miR-21 promotes proliferation, invasion and therapy resistance of colon adenocarcinoma cells through its target PDCD4. Sci. Rep. 2020, 10, 8271. [Google Scholar] [CrossRef]
- Shinohara, H.; Kuranaga, Y.; Kumazaki, M.; Sugito, N.; Yoshikawa, Y.; Takai, T.; Taniguchi, K.; Ito, Y.; Akao, Y. Regulated Polarization of Tumor-Associated Macrophages by miR-145 via Colorectal Cancer-Derived Extracellular Vesicles. J. Immunol. 2017, 199, 1505–1515. [Google Scholar] [CrossRef]
- Xiao, Y.; Li, Y.; Yuan, Y.; Liu, B.; Pan, S.; Liu, Q.; Qi, X.; Zhou, H.; Dong, W.; Jia, L. The potential of exosomes derived from colorectal cancer as a biomarker. Clin. Chim. Acta 2019, 490, 186–193. [Google Scholar] [CrossRef]
- Handy, D.E.; Castro, R.; Loscalzo, J. Epigenetic modifications: Basic mechanisms and role in cardiovascular disease. Circulation 2011, 123, 2145–2156. [Google Scholar] [CrossRef] [PubMed]
- Babaei, K.; Khaksar, R.; Zeinali, T.; Hemmati, H.; Bandegi, A.; Samidoust, P.; Ashoobi, M.T.; Hashemian, H.; Delpasand, K.; Talebinasab, F.; et al. Epigenetic profiling of MUTYH, KLF6, WNT1 and KLF4 genes in carcinogenesis and tumorigenesis of colorectal cancer. BioMedicine 2019, 9, 22. [Google Scholar] [CrossRef]
- Cheng, Y.; He, C.; Wang, M.; Ma, X.; Mo, F.; Yang, S.; Han, J.; Wei, X. Targeting epigenetic regulators for cancer therapy: Mechanisms and advances in clinical trials. Signal Transduct. Target. Ther. 2019, 4, 62. [Google Scholar] [CrossRef]
- Zhang, H.; Lu, Y.; Wu, J.; Feng, J. LINC00460 Hypomethylation Promotes Metastasis in Colorectal Carcinoma. Front. Genet. 2019, 10, 880. [Google Scholar] [CrossRef]
- Zhang, H.; Sun, X.; Lu, Y.; Wu, J.; Feng, J. DNA-methylated gene markers for colorectal cancer in TCGA database. Exp. Ther. Med. 2020, 19, 3042–3050. [Google Scholar] [CrossRef]
- Shen, Y.; Tong, M.; Liang, Q.; Guo, Y.; Sun, H.Q.; Zheng, W.; Ao, L.; Guo, Z.; She, F. Epigenomics alternations and dynamic transcriptional changes in responses to 5-fluorouracil stimulation reveal mechanisms of acquired drug resistance of colorectal cancer cells. Pharm. J. 2018, 18, 23–28. [Google Scholar] [CrossRef]
- Shi, L.; Li, X.; Wu, Z.; Li, X.; Nie, J.; Guo, M.; Mei, Q.; Han, W. DNA methylation-mediated repression of miR-181a/135a/302c expression promotes the microsatellite-unstable colorectal cancer development and 5-FU resistance via targeting PLAG1. J. Genet. Genom. 2018, 45, 205–214. [Google Scholar] [CrossRef]
- He, J.; Pei, L.; Jiang, H.; Yang, W.; Chen, J.; Liang, H. Chemoresistance of colorectal cancer to 5-fluorouracil is associated with silencing of the BNIP3 gene through aberrant methylation. J. Cancer 2017, 8, 1187–1196. [Google Scholar] [CrossRef]
- Wang, B.; Lu, F.-Y.; Shi, R.-H.; Feng, Y.-D.; Zhao, X.-D.; Lu, Z.-P.; Xiao, L.; Zhou, G.-Q.; Qiu, J.-M.; Cheng, C.-E. MiR-26b regulates 5-FU-resistance in human colorectal cancer via down-regulation of Pgp. Am. J. Cancer Res. 2018, 8, 2518–2527. [Google Scholar]
- Fouad, M.A.; Salem, S.E.; Hussein, M.M.; Zekri, A.R.N.; Hafez, H.F.; El Desouky, E.D.; Shouman, S.A. Impact of Global DNA Methylation in Treatment Outcome of Colorectal Cancer Patients. Front. Pharmacol. 2018, 9, 1173. [Google Scholar] [CrossRef]
- Baharudin, R.; Ab Mutalib, N.-S.; Othman, S.N.; Sagap, I.; Rose, I.M.; Mohd Mokhtar, N.; Jamal, R. Identification of Predictive DNA Methylation Biomarkers for Chemotherapy Response in Colorectal Cancer. Front. Pharmacol. 2017, 8, 47. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.; Huang, J.; Niu, H.; Wang, J.; Si, Y.; Bai, Z.; Ding, W. Epigenetic regulation of osteopontin splicing isoform c defines its role as a microenvironmental factor to promote the survival of colon cancer cells from 5-FU treatment. Res. Sq 2020. [Google Scholar] [CrossRef]
- Jiang, G.; Lin, J.; Wang, W.; Sun, M.; Chen, K.; Wang, F. WNT5A Promoter Methylation Is Associated with Better Responses and Longer Progression-Free Survival in Colorectal Cancer Patients Treated with 5-Fluorouracil-Based Chemotherapy. Genet. Test. Mol. Biomark. 2017, 21, 74–79. [Google Scholar] [CrossRef]
- Wen, S.; Wang, X.; Wang, Y.; Shen, J.; Pu, J.; Liang, H.; Chen, C.; Liu, L.; Dai, P. Nucleoside diphosphate kinase 2 confers acquired 5-fluorouracil resistance in colorectal cancer cells. Artif. Cells Nanomed. Biotechnol. 2018, 46, 896–905. [Google Scholar] [CrossRef] [PubMed]
- Paschall, A.V.; Yang, D.; Lu, C.; Choi, J.-H.; Li, X.; Liu, F.; Figueroa, M.; Oberlies, N.H.; Pearce, C.; Bollag, W.B.; et al. H3K9 Trimethylation Silences Fas Expression To Confer Colon Carcinoma Immune Escape and 5-Fluorouracil Chemoresistance. J. Immunol. 2015, 195, 1868–1882. [Google Scholar] [CrossRef]
- Zhan, W.; Liao, X.; Liu, J.; Tian, T.; Yu, L.; Li, R. USP38 regulates the stemness and chemoresistance of human colorectal cancer via regulation of HDAC3. Oncogenesis 2020, 9, 48. [Google Scholar] [CrossRef]
- Moody, L.; Dvoretskiy, S.; An, R.; Mantha, S.; Pan, Y.-X. The Efficacy of miR-20a as a Diagnostic and Prognostic Biomarker for Colorectal Cancer: A Systematic Review and Meta-Analysis. Cancers 2019, 11, 1111. [Google Scholar] [CrossRef]
- Perilli, L.; Tessarollo, S.; Albertoni, L.; Curtarello, M.; Pastò, A.; Brunetti, E.; Fassan, M.; Rugge, M.; Indraccolo, S.; Amadori, A.; et al. Silencing of miR-182 is associated with modulation of tumorigenesis through apoptosis induction in an experimental model of colorectal cancer. BMC Cancer 2019, 19, 821. [Google Scholar] [CrossRef]
- Li, H.; Zhang, X.; Jin, Z.; Yin, T.; Duan, C.; Sun, J.; Xiong, R.; Li, Z. MiR-122 Promotes the Development of Colon Cancer by Targeting ALDOA in Vitro. Technol. Cancer Res. Treat. 2019, 18, 1533033819871300. [Google Scholar] [CrossRef]
- Angius, A.; Pira, G.; Scanu, A.M.; Uva, P.; Sotgiu, G.; Saderi, L.; Manca, A.; Serra, C.; Uleri, E.; Piu, C.; et al. MicroRNA-425-5p Expression Affects BRAF/RAS/MAPK Pathways In Colorectal Cancers. Int. J. Med. Sci. 2019, 16, 1480–1491. [Google Scholar] [CrossRef]
- Mukohyama, J.; Isobe, T.; Hu, Q.; Hayashi, T.; Watanabe, T.; Maeda, M.; Yanagi, H.; Qian, X.; Yamashita, K.; Minami, H.; et al. miR-221 targets QKI to enhance the tumorigenic capacity of human colorectal cancer stem cells. Cancer Res. 2019, 79, 5151–5158. [Google Scholar] [CrossRef] [PubMed]
- Carter, V.J.; O’brien, J.S.; Burton, F.J.; Oxford, G.B.; Stephen, V.; Hallion, J.; Bishop, C.; Galbraith, J.N.; Eichenberger, R.M.; Sarojini, H.; et al. The microRNA-200 family acts as an oncogene in colorectal cancer by inhibiting the tumor suppressor RASSF2. Oncol. Lett. 2019, 18, 3994–4007. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Shi, X.; Wang, N.; Peng, W.; Cheng, Z. MicroRNA-215-3p Suppresses the Growth, Migration, and Invasion of Colorectal Cancer by Targeting FOXM1. Technol. Cancer Res. Treat. 2019, 18, 1533033819874776. [Google Scholar] [CrossRef]
- Gu, C.; Cai, J.; Xu, Z.; Zhou, S.; Ye, L.; Yan, Q.; Zhang, Y.; Fang, Y.; Liu, Y.; Tu, C.; et al. MiR-532-3p suppresses colorectal cancer progression by disrupting the ETS1/TGM2 axis-mediated Wnt/β-catenin signaling. Cell Death Dis. 2019, 10, 739. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Yang, C.; Zhang, L.; Wei, L.; Lin, J.; Zhao, J.; Peng, J. Spica Prunellae Extract Enhances Fluorouracil Sensitivity of 5-Fluorouracil-Resistant Human Colon Carcinoma HCT-8/5-FU Cells via TOP2α and miR-494. Biomed. Res. Int. 2019, 2019, 5953619. [Google Scholar] [CrossRef]
- Nowak-Sliwinska, P.; Scapozza, L.; Ruiz i Altaba, A. Drug repurposing in oncology: Compounds, pathways, phenotypes and computational approaches for colorectal cancer. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 434–454. [Google Scholar] [CrossRef]
- Zhang, L.; Li, B.; Zhang, B.; Zhang, H.; Suo, J. miR-361 enhances sensitivity to 5-fluorouracil by targeting the FOXM1-ABCC5/10 signaling pathway in colorectal cancer. Oncol. Lett. 2019, 18, 4064–4073. [Google Scholar] [CrossRef]
- Barisciano, G.; Colangelo, T.; Rosato, V.; Muccillo, L.; Taddei, M.L.; Ippolito, L.; Chiarugi, P.; Galgani, M.; Bruzzaniti, S.; Matarese, G.; et al. miR-27a is a master regulator of metabolic reprogramming and chemoresistance in colorectal cancer. Br. J. Cancer 2020, 122, 1354–1366. [Google Scholar] [CrossRef]
- Karimi Dermani, F.; Najafi, R. miR-200c as a Predictive Biomarker for 5-Fluorouracil Chemosensitivity in Colorectal Cancer. J. Gastrointest. Cancer 2018, 49, 102–103. [Google Scholar] [CrossRef]
- Zhang, Y.; Talmon, G.; Wang, J. MicroRNA-587 antagonizes 5-FU-induced apoptosis and confers drug resistance by regulating PPP2R1B expression in colorectal cancer. Cell Death Dis. 2015, 6, e1845. [Google Scholar] [CrossRef]
- Zhang, H.; Tang, J.; Li, C.; Kong, J.; Wang, J.; Wu, Y.; Xu, E.; Lai, M. MiR-22 regulates 5-FU sensitivity by inhibiting autophagy and promoting apoptosis in colorectal cancer cells. Cancer Lett. 2014, 356. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Ye, M.-L.; Zhang, Y.-P.; Li, W.-J.; Li, M.-T.; Wang, H.-Z.; Qiu, X.; Xu, Y.; Yin, J.-W.; Hu, Q.; et al. MicroRNA-375-3p enhances chemosensitivity to 5-fluorouracil by targeting thymidylate synthase in colorectal cancer. Cancer Sci. 2020, 111, 1528–1541. [Google Scholar] [CrossRef] [PubMed]
- Ren, T.-J.; Liu, C.; Hou, J.-F.; Shan, F.-X. CircDDX17 reduces 5-fluorouracil resistance and hinders tumorigenesis in colorectal cancer by regulating miR-31-5p/KANK1 axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1743–1754. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Liang, Y.; Shen, L.; Shen, L. MicroRNA-204 modulates colorectal cancer cell sensitivity in response to 5-fluorouracil-based treatment by targeting high mobility group protein A2. Biol. Open 2016, 5, 563–570. [Google Scholar] [CrossRef]
- Yang, Y.; Bao, Y.; Yang, G.-K.; Wan, J.; Du, L.-J.; Ma, Z.-H. MiR-214 sensitizes human colon cancer cells to 5-FU by targeting Hsp27. Cell. Mol. Biol. Lett. 2019, 24, 22. [Google Scholar] [CrossRef]
- Liu, G.; Zhou, J.; Dong, M. Down-regulation of miR-543 expression increases the sensitivity of colorectal cancer cells to 5-Fluorouracil through the PTEN/PI3K/AKT pathway. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Zhao, C.; Zhao, Q.; Zhang, C.; Wang, G.; Yao, Y.; Huang, X.; Zhan, F.; Zhu, Y.; Shi, J.; Chen, J.; et al. miR-15b-5p resensitizes colon cancer cells to 5-fluorouracil by promoting apoptosis via the NF-κB/XIAP axis. Sci. Rep. 2017, 7, 4194. [Google Scholar] [CrossRef]
- Liu, J.; Huang, Y.; Wang, H.; Wu, D. MiR-106a-5p promotes 5-FU resistance and the metastasis of colorectal cancer by targeting TGFβR2. Int. J. Clin. Exp. Pathol. 2018, 11, 5622–5634. [Google Scholar]
- Meng, X.; Fu, R. miR-206 regulates 5-FU resistance by targeting Bcl-2 in colon cancer cells. Onco Targets Ther. 2018, 11, 1757–1765. [Google Scholar] [CrossRef]
- Park, G.-B.; Jeong, J.-Y.; Kim, D. Modified TLR-mediated downregulation of miR-125b-5p enhances CD248 (endosialin)-induced metastasis and drug resistance in colorectal cancer cells. Mol. Carcinog. 2020, 59, 154–167. [Google Scholar] [CrossRef]
- Xu, G.; Zhu, H.; Xu, J.; Wang, Y.; Zhang, Y.; Zhang, M.; Zhu, D. Long non-coding RNA POU6F2-AS2 promotes cell proliferation and drug resistance in colon cancer by regulating miR-377/BRD4. J. Cell. Mol. Med. 2020, 24, 4136–4149. [Google Scholar] [CrossRef] [PubMed]
- Bian, Z.; Zhang, J.; Li, M.; Feng, Y.; Yao, S.; Song, M.; Qi, X.; Fei, B.; Yin, Y.; Hua, D.; et al. Long non-coding RNA LINC00152 promotes cell proliferation, metastasis, and confers 5-FU resistance in colorectal cancer by inhibiting miR-139-5p. Oncogenesis 2017, 6, 395. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Li, J.; Zhou, L.; Han, J.; Liu, R.; Zhang, H.; Ning, T.; Gao, Z.; Liu, B.; Chen, X.; et al. The c-Myc/miR-27b-3p/ATG10 regulatory axis regulates chemoresistance in colorectal cancer. Theranostics 2020, 10, 1981–1996. [Google Scholar] [CrossRef] [PubMed]
- PARK, G.-B.; Kim, D. TLR-mediated miR-125b-5p downregulation enhances CD248-induced metastasis and drug resistance in colorectal cancer cells. Cancer Biol. 2019. [Google Scholar] [CrossRef]
- Aggarwal, V.; Tuli, H.S.; Varol, A.; Thakral, F.; Yerer, M.B.; Sak, K.; Varol, M.; Jain, A.; Khan, M.A.; Sethi, G. Role of Reactive Oxygen Species in Cancer Progression: Molecular Mechanisms and Recent Advancements. Biomolecules 2019, 9, 735. [Google Scholar] [CrossRef]
- Moreira, H.; Szyjka, A.; Paliszkiewicz, K.; Barg, E. Prooxidative Activity of Celastrol Induces Apoptosis, DNA Damage, and Cell Cycle Arrest in Drug-Resistant Human Colon Cancer Cells. Oxid. Med. Cell. Longev. 2019, 2019, 6793957. [Google Scholar] [CrossRef]
- Yan, J.; Dou, X.; Zhou, J.; Xiong, Y.; Mo, L.; Li, L.; Lei, Y. Tubeimoside-I sensitizes colorectal cancer cells to chemotherapy by inducing ROS-mediated impaired autophagolysosomes accumulation. J. Exp. Clin. Cancer Res. 2019, 38, 353. [Google Scholar] [CrossRef]
- Liu, C.; Zhao, Y.; Wang, J.; Yang, Y.; Zhang, Y.; Qu, X.; Peng, S.; Yao, Z.; Zhao, S.; He, B.; et al. FoxO3 reverses 5-fluorouracil resistance in human colorectal cancer cells by inhibiting the Nrf2/TR1 signaling pathway. Cancer Lett. 2020, 470, 29–42. [Google Scholar] [CrossRef]
- Liu, T.; Han, Y.; Yu, C.; Ji, Y.; Wang, C.; Chen, X.; Wang, X.; Shen, J.; Zhang, Y.; Lang, J.-Y. MYC predetermines the sensitivity of gastrointestinal cancer to antifolate drugs through regulating TYMS transcription. EBioMedicine 2019, 48, 289–300. [Google Scholar] [CrossRef]
- Turgeon, M.-O.; Perry, N.J.S.; Poulogiannis, G. DNA Damage, Repair, and Cancer Metabolism. Front. Oncol. 2018, 8, 15. [Google Scholar] [CrossRef]
- Jiang, H.; Li, B.; Wang, F.; Ma, C.; Hao, T. Expression of ERCC1 and TYMS in colorectal cancer patients and the predictive value of chemotherapy efficacy. Oncol. Lett. 2019, 18, 1157–1162. [Google Scholar] [CrossRef]
- Bai, W.; Wu, Y.; Zhang, P.; Xi, Y. Correlations between expression levels of thymidylate synthase, thymidine phosphorylase and dihydropyrimidine dehydrogenase, and efficacy of 5-fluorouracil-based chemotherapy for advanced colorectal cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 12333–12345. [Google Scholar] [PubMed]
- Wakasa, K.; Kawabata, R.; Nakao, S.; Hattori, H.; Taguchi, K.; Uchida, J.; Yamanaka, T.; Maehara, Y.; Fukushima, M.; Oda, S. Dynamic Modulation of Thymidylate Synthase Gene Expression and Fluorouracil Sensitivity in Human Colorectal Cancer Cells. PLoS ONE 2015, 10, e0123076. [Google Scholar] [CrossRef]
- Showalter, S.L.; Showalter, T.N.; Witkiewicz, A.; Havens, R.; Kennedy, E.P.; Hucl, T.; Kern, S.E.; Yeo, C.J.; Brody, J.R. Evaluating the drug-target relationship between thymidylate synthase expression and tumor response to 5-fluorouracil. Is it time to move forward? Cancer Biol. Ther. 2008, 7, 986–994. [Google Scholar] [CrossRef] [PubMed]
- Hammond, W.A.; Swaika, A.; Mody, K. Pharmacologic resistance in colorectal cancer: A review. Ther. Adv. Med. Oncol. 2015, 8, 57–84. [Google Scholar] [CrossRef] [PubMed]
- Nie, Q.; Guo, X.; Liu, H.; Zeng, L.; Wang, X.; Wen, S.; Wang, W.; Lu, Y.; Wang, Q.; Peng, W. Effects of DPYD and TS gene polymorphisms on chemosensitivity of 5-FU in advanced colorectal cancer. Int. J. Clin. Exp. Med. 2019, 12, 9380–9386. [Google Scholar]
- Ntavatzikos, A.; Spathis, A.; Patapis, P.; Machairas, N.; Vourli, G.; Peros, G.; Papadopoulos, I.; Panayiotides, I.; Koumarianou, A. TYMS/KRAS/BRAF molecular profiling predicts survival following adjuvant chemotherapy in colorectal cancer. World J. Gastrointest. Oncol. 2019, 11, 551–566. [Google Scholar] [CrossRef]
- Wang, M.; Hu, H.; Wang, Y.; Huang, Q.; Huang, R.; Chen, Y.; Ma, T.; Qiao, T.; Zhang, Q.; Wu, H.; et al. Long non-coding RNA TUG1 mediates 5-fluorouracil resistance by acting as a ceRNA of miR-197-3p in colorectal cancer. J. Cancer 2019, 10, 4603–4613. [Google Scholar] [CrossRef]
- Che, J.; Pan, L.; Yang, X.; Liu, Z.; Huang, L.; Wen, C.; Lin, A.; Liu, H. Thymidine phosphorylase expression and prognosis in colorectal cancer treated with 5-fluorouracil-based chemotherapy: A meta-analysis. Mol. Clin. Oncol. 2017, 7, 943–952. [Google Scholar] [CrossRef]
- Kugimiya, N.; Harada, E.; Suehiro, Y.; Suga, A.; Takemoto, Y.; Hamano, K. Determination of thymidine phosphorylase expression level facilitates recurrence risk stratification in stage II/III colorectal cancer following adjuvant chemotherapy with oral fluoropyrimidines. Oncol. Lett. 2019, 17, 5267–5274. [Google Scholar] [CrossRef]
- Gajjar, K.K.; Vora, H.H.; Kobawala, T.P.; Trivedi, T.I.; Ghosh, N.R. Deciphering the potential value of 5-fluorouracil metabolic enzymes in predicting prognosis and treatment response of colorectal cancer patients. Int. J. Biol. Markers 2018, 33, 180–188. [Google Scholar] [CrossRef]
- Ogawa, M.; Watanabe, M.; Mitsuyama, Y.; Anan, T.; Ohkuma, M.; Kobayashi, T.; Eto, K.; Yanaga, K. Thymidine phosphorylase mRNA expression may be a predictor of response to post-operative adjuvant chemotherapy with S-1 in patients with stage III colorectal cancer. Oncol. Lett. 2014, 8, 2463–2468. [Google Scholar] [CrossRef][Green Version]
- Shigeta, K.; Ishii, Y.; Hasegawa, H.; Okabayashi, K.; Kitagawa, Y. Evaluation of 5-fluorouracil metabolic enzymes as predictors of response to adjuvant chemotherapy outcomes in patients with stage II/III colorectal cancer: A decision-curve analysis. World J. Surg. 2014, 38, 3248–3256. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, J.; Yao, X.; Jiang, C.; Ni, P.; Cheng, L.; Liu, J.; Ni, S.; Chen, Q.; Li, Q.; et al. Bevacizumab-enhanced antitumor effect of 5-fluorouracil via upregulation of thymidine phosphorylase through vascular endothelial growth factor A/vascular endothelial growth factor receptor 2-specificity protein 1 pathway. Cancer Sci. 2018, 109, 3294–3304. [Google Scholar] [CrossRef]
- Li, L.-H.; Dong, H.; Zhao, F.; Tang, J.; Chen, X.; Ding, J.; Men, H.-T.; Luo, W.-X.; Du, Y.; Ge, J.; et al. The upregulation of dihydropyrimidine dehydrogenase in liver is involved in acquired resistance to 5-fluorouracil. Eur. J. Cancer 2013, 49, 1752–1760. [Google Scholar] [CrossRef]
- Zhang, Y.-H.; Luo, D.-D.; Wan, S.-B.; Qu, X.-J. S1PR2 inhibitors potently reverse 5-FU resistance by downregulating DPD expression in colorectal cancer. Pharmacol. Res. 2020, 155, 104717. [Google Scholar] [CrossRef]
- Pathania, S.; Bhatia, R.; Baldi, A.; Singh, R.; Rawal, R.K. Drug metabolizing enzymes and their inhibitors’ role in cancer resistance. Biomed. Pharmacother. 2018, 105, 53–65. [Google Scholar] [CrossRef]
- Zhang, Y.-H.; Shi, W.-N.; Wu, S.-H.; Miao, R.-R.; Sun, S.-Y.; Luo, D.-D.; Wan, S.-B.; Guo, Z.-K.; Wang, W.-Y.; Yu, X.-F.; et al. SphK2 confers 5-fluorouracil resistance to colorectal cancer via upregulating H3K56ac-mediated DPD expression. Oncogene 2020. [Google Scholar] [CrossRef]
- Chen, X.-L.; Wang, Y.-M.; Zhao, F.; Chen, Z.; Yang, X.; Sun, C.; Gao, Y.; Yang, T.-G.; Tian, G.; Chen, Y.-M.; et al. Methylenetetrahydrofolate reductase polymorphisms and colorectal cancer prognosis: A meta-analysis. J. Gene Med. 2019, 21, e3114. [Google Scholar] [CrossRef]
- Naghibalhossaini, F.; Shefaghat, M.; Mansouri, A.; Jaberi, H.; Tatar, M.; Eftekhar, E. The Impact of Thymidylate Synthase and Methylenetetrahydrofolate Reductase Genotypes on Sensitivity to 5-Fluorouracil Treatment in Colorectal Cancer Cells. Acta Med. Iran. 2017, 55, 751–758. [Google Scholar]
- Pawłowski, P.H.; Szczęsny, P.; Rempoła, B.; Poznańska, A.; Poznański, J. Combined in silico and 19F NMR analysis of 5-fluorouracil metabolism in yeast at low ATP conditions. Biosci. Rep. 2019, 39, BSR20192847. [Google Scholar] [CrossRef] [PubMed]
- Tampellini, M.; Bironzo, P.; Di Maio, M.; Scagliotti, G.V. Thymidine phosphorylase: The unforeseen driver in colorectal cancer treatment? Future Oncol. 2018, 14, 1223–1231. [Google Scholar] [CrossRef] [PubMed]
- Kumamoto, K.; Kuwabara, K.; Tajima, Y.; Amano, K.; Hatano, S.; Ohsawa, T.; Okada, N.; Ishibashi, K.; Haga, N.; Ishida, H. Thymidylate synthase and thymidine phosphorylase mRNA expression in primary lesions using laser capture microdissection is useful for prediction of the efficacy of FOLFOX treatment in colorectal cancer patients with liver metastasis. Oncol. Lett. 2012, 3, 983–989. [Google Scholar] [CrossRef] [PubMed]
- Goto, T.; Shinmura, K.; Yokomizo, K.; Sakuraba, K.; Kitamura, Y.; Shirahata, A.; Saito, M.; Kigawa, G.; Nemoto, H.; Sanada, Y.; et al. Expression levels of thymidylate synthase, dihydropyrimidine dehydrogenase, and thymidine phosphorylase in patients with colorectal cancer. Anticancer Res. 2012, 32, 1757–1762. [Google Scholar] [PubMed]
- Lindskog, E.B.; Wettergren, Y.; Odin, E.; Gustavsson, B.; Derwinger, K. Thymidine Phosphorylase Gene Expression in Stage III Colorectal Cancer. Clin. Med. Insights Oncol. 2012, 6, 347–353. [Google Scholar] [CrossRef]
- Sakowicz-Burkiewicz, M.; Przybyla, T.; Wesserling, M.; Bielarczyk, H.; Maciejewska, I.; Pawelczyk, T. Suppression of TWIST1 enhances the sensitivity of colon cancer cells to 5-fluorouracil. Int. J. Biochem. Cell Biol. 2016, 78, 268–278. [Google Scholar] [CrossRef]
- Mori, R.; Yoshida, K.; Futamura, M.; Suetsugu, T.; Shizu, K.; Tanahashi, T.; Tanaka, Y.; Matsuhashi, N.; Yamaguchi, K. The inhibition of thymidine phosphorylase can reverse acquired 5FU-resistance in gastric cancer cells. Gastric Cancer 2019, 22, 497–505. [Google Scholar] [CrossRef]
- Zizzo, N.; Passantino, G.; D’alessio, R.M.; Tinelli, A.; Lopresti, G.; Patruno, R.; Tricarico, D.; Maqoud, F.; Scala, R.; Zito, F.A.; et al. Thymidine Phosphorylase Expression and Microvascular Density Correlation Analysis in Canine Mammary Tumor: Possible Prognostic Factor in Breast Cancer. Front. Vet. Sci. 2019, 6, 368. [Google Scholar] [CrossRef]
- Hasegawa, K.; Okamoto, H.; Kawamura, K.; Kato, R.; Kobayashi, Y.; Sekiya, T.; Udagawa, Y. The effect of chemotherapy or radiotherapy on thymidine phosphorylase and dihydropyrimidine dehydrogenase expression in cancer of the uterine cervix. Eur. J. Obstet. Gynecol. Reprod. Biol. 2012, 163, 67–70. [Google Scholar] [CrossRef]
- Farina-Sarasqueta, A.; van Lijnschoten, G.; Rutten, H.J.T.; van den Brule, A.J.C. Value of gene polymorphisms as markers of 5-FU therapy response in stage III colon carcinoma: A pilot study. Cancer Chemother. Pharmacol. 2010, 66, 1167–1171. [Google Scholar] [CrossRef][Green Version]
- Yadak, R.; Breur, M.; Bugiani, M. Gastrointestinal Dysmotility in MNGIE: From thymidine phosphorylase enzyme deficiency to altered interstitial cells of Cajal. Orphanet J. Rare Dis. 2019, 14, 33. [Google Scholar] [CrossRef] [PubMed]
- Falcão de Campos, C.; Oliveira Santos, M.; Roque, R.; Conceição, I.; de Carvalho, M. Mitochondrial Neurogastrointestinal Encephalomyopathy: Novel Pathogenic Mutation in Thymidine Phosphorylase Gene in a Patient from Cape Verde Islands. Case Rep. Neurol. Med. 2019, 2019, 5976410. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Yang, X.; Liu, L.; Xu, Y. Leukoencephalopathy in Mitochondrial Neurogastrointestinal Encephalomyopathy-Like Syndrome with Polymerase-Gamma Mutations. Ann. Indian Acad. Neurol. 2019, 22, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Filosto, M.; Cotti Piccinelli, S.; Caria, F.; Gallo Cassarino, S.; Baldelli, E.; Galvagni, A.; Volonghi, I.; Scarpelli, M.; Padovani, A. Mitochondrial Neurogastrointestinal Encephalomyopathy (MNGIE-MTDPS1). J. Clin. Med. 2018, 7, 389. [Google Scholar] [CrossRef] [PubMed]
- Jacob, J.; Gopalan, R.; Lakshmanaperumalsamy, P.; Illuri, R.; Bhosle, D.; Sangli, G.K.; Mundkinajeddu, D. Evaluation of Anti-psoriatic Potential of the Fruit Rind of Punica granatum L. Pharmacogn. J. 2019, 11, 466–468. [Google Scholar] [CrossRef]
- Javaid, S.; Shaikh, M.; Fatima, N.; Choudhary, M.I. Natural compounds as angiogenic enzyme thymidine phosphorylase inhibitors: In vitro biochemical inhibition, mechanistic, and in silico modeling studies. PLoS ONE 2019, 14, e0225056. [Google Scholar] [CrossRef]
- Li, W.; Yue, H. Thymidine phosphorylase: A potential new target for treating cardiovascular disease. Trends Cardiovasc. Med. 2018, 28, 157–171. [Google Scholar] [CrossRef]
- Warren, H.R.; Hejmadi, M. Effect of hypoxia on chemosensitivity to 5-fluorouracil in SH-SY5Y neuroblastoma cells. Biosci. Horiz. Int. J. Stud. Res. 2016, 9. [Google Scholar] [CrossRef]
- Jing, X.; Yang, F.; Shao, C.; Wei, K.; Xie, M.; Shen, H.; Shu, Y. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol. Cancer 2019, 18, 157. [Google Scholar] [CrossRef]
- Nijhuis, A.; Thompson, H.; Adam, J.; Parker, A.; Gammon, L.; Lewis, A.; Bundy, J.G.; Soga, T.; Jalaly, A.; Propper, D.; et al. Remodelling of microRNAs in colorectal cancer by hypoxia alters metabolism profiles and 5-fluorouracil resistance. Hum. Mol. Genet. 2017, 26, 1552–1564. [Google Scholar] [CrossRef]
- Palmirotta, R.; Lovero, D.; Delacour, H.; Le Roy, A.; Cremades, S.; Silvestris, F. Rare Dihydropyrimidine Dehydrogenase Variants and Toxicity by Floropyrimidines: A Case Report. Front. Oncol. 2019, 9, 139. [Google Scholar] [CrossRef]
- Latchman, J.; Guastella, A.; Tofthagen, C. 5-Fluorouracil toxicity and dihydropyrimidine dehydrogenase enzyme: Implications for practice. Clin. J. Oncol. Nurs. 2014, 18, 581–585. [Google Scholar] [CrossRef] [PubMed]
- Mukherji, D.; Massih, S.A.; Tfayli, A.; Kanso, M.; Faraj, W. Three different polymorphisms of the DPYD gene associated with severe toxicity following administration of 5-FU: A case report. J. Med. Case Rep. 2019, 13, 76. [Google Scholar] [CrossRef] [PubMed]
- Amstutz, U.; Henricks, L.M.; Offer, S.M.; Barbarino, J.; Schellens, J.H.M.; Swen, J.J.; Klein, T.E.; McLeod, H.L.; Caudle, K.E.; Diasio, R.B.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Dihydropyrimidine Dehydrogenase Genotype and Fluoropyrimidine Dosing: 2017 Update. Clin. Pharmacol. Ther. 2018, 103, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Lunenburg, C.A.T.C.; van der Wouden, C.H.; Nijenhuis, M.; Crommentuijn-van Rhenen, M.H.; de Boer-Veger, N.J.; Buunk, A.M.; Houwink, E.J.F.; Mulder, H.; Rongen, G.A.; van Schaik, R.H.N.; et al. Dutch Pharmacogenetics Working Group (DPWG) guideline for the gene–drug interaction of DPYD and fluoropyrimidines. Eur. J. Hum. Genet. 2020, 28, 508–517. [Google Scholar] [CrossRef]
- De Falco, V.; Natalicchio, M.I.; Napolitano, S.; Coppola, N.; Conzo, G.; Martinelli, E.; Zanaletti, N.; Vitale, P.; Giunta, E.F.; Vietri, M.T.; et al. A case report of a severe fluoropyrimidine-related toxicity due to an uncommon DPYD variant. Medicine 2019, 98, e15759. [Google Scholar] [CrossRef]
- Meulendijks, D.; Henricks, L.M.; Sonke, G.S.; Deenen, M.J.; Froehlich, T.K.; Amstutz, U.; Largiader, C.R.; Jennings, B.A.; Marinaki, A.M.; Sanderson, J.D.; et al. Clinical relevance of DPYD variants c.1679T>G, c.1236G>A/HapB3, and c.1601G>A as predictors of severe fluoropyrimidine-associated toxicity: A systematic review and meta-analysis of individual patient data. Lancet. Oncol. 2015, 16, 1639–1650. [Google Scholar] [CrossRef]
- Nie, Q.; Shrestha, S.; Tapper, E.E.; Trogstad-Isaacson, C.S.; Bouchonville, K.J.; Lee, A.M.; Wu, R.; Jerde, C.R.; Wang, Z.; Kubica, P.A.; et al. Quantitative Contribution of rs75017182 to Dihydropyrimidine Dehydrogenase mRNA Splicing and Enzyme Activity. Clin. Pharmacol. Ther. 2017, 102, 662–670. [Google Scholar] [CrossRef]
- Sun, M.; Zhong, J.; Zhang, L.; Shi, S. Genetic impact of methylenetetrahydrofolate reductase (MTHFR) polymorphism on the susceptibility to colorectal polyps: A meta-analysis. BMC Med. Genet. 2019, 20, 94. [Google Scholar] [CrossRef]
- Jiang, H.; Shen, Y. Methylene tetrahydrofolate reductase (MTHFR) gene rs1801133 C>T polymorphisms and response to 5-FU based chemotherapy in patients with colorectal cancer: A meta-analysis. Pteridines 2019, 30, 126–132. [Google Scholar] [CrossRef]
- Kim, K.-R.; Yoon, J.-H.; Shim, H.-J.; Hwang, J.-E.; Bae, W.-K.; Chung, I.-J.; Kim, H.-N.; Shin, M.-H.; Cho, S.-H. Role of depth of response and MTHFR genotype as predictors of fluorouracil rechallenge therapy for refractory metastatic colorectal cancer. Oncol. Lett. 2017, 14, 2491–2498. [Google Scholar] [CrossRef] [PubMed]
- Zawiah, M.; Yousef, A.-M.; Kadi, T.; Yousef, M.; Majdalawi, K.; Al-Yacoub, S.; Al-Hiary, R.; Tantawi, D.; Mukred, R.; Ajaj, A.R. Early disease relapse in a patient with colorectal cancer who harbors genetic variants of DPYD, TYMS, MTHFR and DHFR after treatment with 5-fluorouracil-based chemotherapy. Drug Metab. Pers. Ther. 2018, 33, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Botticelli, A.; Borro, M.; Onesti, C.E.; Strigari, L.; Gentile, G.; Cerbelli, B.; Romiti, A.; Occhipinti, M.; Sebastiani, C.; Lionetto, L.; et al. Degradation Rate of 5-Fluorouracil in Metastatic Colorectal Cancer: A New Predictive Outcome Biomarker? PLoS ONE 2016, 11, e0163105. [Google Scholar] [CrossRef] [PubMed]
- Tsukihara, H.; Tsunekuni, K.; Takechi, T. Folic Acid-Metabolizing Enzymes Regulate the Antitumor Effect of 5-Fluoro-2′-Deoxyuridine in Colorectal Cancer Cell Lines. PLoS ONE 2016, 11, e0163961. [Google Scholar] [CrossRef]
- Okada, K.; Hakata, S.; Terashima, J.; Gamou, T.; Habano, W.; Ozawa, S. Combination of the histone deacetylase inhibitor depsipeptide and 5-fluorouracil upregulates major histocompatibility complex class II and p21 genes and activates caspase-3/7 in human colon cancer HCT-116 cells. Oncol. Rep. 2016, 36, 1875–1885. [Google Scholar] [CrossRef]
- Terranova-Barberio, M.; Pecori, B.; Roca, M.S.; Imbimbo, S.; Bruzzese, F.; Leone, A.; Muto, P.; Delrio, P.; Avallone, A.; Budillon, A.; et al. Synergistic antitumor interaction between valproic acid, capecitabine and radiotherapy in colorectal cancer: Critical role of p53. J. Exp. Clin. Cancer Res. 2017, 36, 177. [Google Scholar] [CrossRef]
- Sveen, A.; Bruun, J.; Eide, P.W.; Eilertsen, I.A.; Ramirez, L.; Murumägi, A.; Arjama, M.; Danielsen, S.A.; Kryeziu, K.; Elez, E.; et al. Colorectal Cancer Consensus Molecular Subtypes Translated to Preclinical Models Uncover Potentially Targetable Cancer Cell Dependencies. Clin. Cancer Res. 2018, 24, 794–806. [Google Scholar] [CrossRef]
- Kryeziu, K.; Bruun, J.; Guren, T.K.; Sveen, A.; Lothe, R.A. Combination therapies with HSP90 inhibitors against colorectal cancer. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 240–247. [Google Scholar] [CrossRef]
- He, S.; Smith, D.L.; Sequeira, M.; Sang, J.; Bates, R.C.; Proia, D.A. The HSP90 inhibitor ganetespib has chemosensitizer and radiosensitizer activity in colorectal cancer. Investig. New Drugs 2014, 32, 577–586. [Google Scholar] [CrossRef]
- Gong, S.; Xu, D.; Zhu, J.; Zou, F.; Peng, R. Efficacy of the MEK Inhibitor Cobimetinib and its Potential Application to Colorectal Cancer Cells. Cell. Physiol. Biochem. 2018, 47, 680–693. [Google Scholar] [CrossRef]
- Varghese, V.; Magnani, L.; Harada-Shoji, N.; Mauri, F.; Szydlo, R.M.; Yao, S.; Lam, E.W.-F.; Kenny, L.M. FOXM1 modulates 5-FU resistance in colorectal cancer through regulating TYMS expression. Sci. Rep. 2019, 9, 1505. [Google Scholar] [CrossRef] [PubMed]
- Jing, C.; Li, H.; Du, Y.; Cao, H.; Liu, S.; Wang, Z.; Ma, R.; Feng, J.; Wu, J. MEK inhibitor enhanced the antitumor effect of oxaliplatin and 5-fluorouracil in MEK1 Q56P-mutant colorectal cancer cells. Mol. Med. Rep. 2019, 19, 1092–1100. [Google Scholar] [CrossRef] [PubMed]
- Długosz-Pokorska, A.; Pięta, M.; Janecki, T.; Janecka, A. New uracil analogs as downregulators of ABC transporters in 5-fluorouracil-resistant human leukemia HL-60 cell line. Mol. Biol. Rep. 2019, 46, 5831–5839. [Google Scholar] [CrossRef] [PubMed]
- Sunami, E.; Kusumoto, T.; Ota, M.; Sakamoto, Y.; Yoshida, K.; Tomita, N.; Maeda, A.; Teshima, J.; Okabe, M.; Tanaka, C.; et al. S-1 and Oxaliplatin Versus Tegafur-uracil and Leucovorin as Postoperative Adjuvant Chemotherapy in Patients With High-risk Stage III Colon Cancer (ACTS-CC 02): A Randomized, Open-label, Multicenter, Phase III Superiority Trial. Clin. Colorectal Cancer 2020, 19, 22–31. [Google Scholar] [CrossRef]
- Kim, H.G.; Kim, C.W.; Lee, D.H.; Lee, J.-S.; Oh, E.-T.; Park, H.J. Quinacrine-Mediated Inhibition of Nrf2 Reverses Hypoxia-Induced 5-Fluorouracil Resistance in Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 4366. [Google Scholar] [CrossRef]
- Nishizawa, Y.; Ikeda, R.; Yamamoto, M.; Kawahara, K.; Shinsato, Y.; Minami, K.; Nitta, M.; Terazono, H.; Akiyama, S.-I.; Furukawa, T.; et al. 5-Aza-2-deoxycytidine Enhances the Sensitivity of 5-Fluorouracil by Demethylation of the Thymidine Phosphorylase Promoter. Anticancer Res. 2019, 39, 4129–4136. [Google Scholar] [CrossRef]
- Falcone, A.; André, T.; Edeline, J.; François, E.; Taieb, J.; Phelip, J.; Portales, F.; Price, T.; Becquart, M.; Moreno Vera, S.; et al. Safety and efficacy of trifluridine/tipiracil in previously treated metastatic colorectal cancer (mCRC): Preliminary results from the phase IIIb, international, open-label, early-access PRECONNECT study. Ann. Oncol. 2018, 29, v104–v105. [Google Scholar] [CrossRef]
- Minegaki, T.; Suzuki, A.; Mori, M.; Tsuji, S.; Yamamoto, S.; Watanabe, A.; Tsuzuki, T.; Tsunoda, T.; Yamamoto, A.; Tsujimoto, M.; et al. Histone deacetylase inhibitors sensitize 5-fluorouracil-resistant MDA-MB-468 breast cancer cells to 5-fluorouracil. Oncol. Lett. 2018, 16, 6202–6208. [Google Scholar] [CrossRef]
- Jang, H.Y.; Kim, D.H.; Lee, H.J.; Kim, W.D.; Kim, S.-Y.; Hwang, J.J.; Lee, S.J.; Moon, D.H. Schedule-dependent synergistic effects of 5-fluorouracil and selumetinib in KRAS or BRAF mutant colon cancer models. Biochem. Pharmacol. 2019, 160, 110–120. [Google Scholar] [CrossRef]
- Shimada, T.; Tsuruta, M.; Hasegawa, H.; Okabayashi, K.; Shigeta, K.; Ishida, T.; Asada, Y.; Suzumura, H.; Koishikawa, K.; Akimoto, S.; et al. Heat shock protein 27 knockdown using nucleotide-based therapies enhances sensitivity to 5-FU chemotherapy in SW480 human colon cancer cells. Oncol. Rep. 2018, 39, 1119–1124. [Google Scholar] [CrossRef]
- Lavitrano, M.; Ianzano, L.; Bonomo, S.; Cialdella, A.; Cerrito, M.G.; Pisano, F.; Missaglia, C.; Giovannoni, R.; Romano, G.; McLean, C.M.; et al. BTK inhibitors synergise with 5-FU to treat drug-resistant TP53-null colon cancers. J. Pathol. 2020, 250, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Rodríguez, M.G.; Sánchez-Barrera, C.Á.; Callejas, B.E.; García-Castillo, V.; Beristain-Terrazas, D.L.; Delgado-Buenrostro, N.L.; Chirino, Y.I.; León-Cabrera, S.A.; Rodríguez-Sosa, M.; Gutierrez-Cirlos, E.B.; et al. Use of STAT6 Phosphorylation Inhibitor and Trimethylglycine as New Adjuvant Therapies for 5-Fluorouracil in Colitis-Associated Tumorigenesis. Int. J. Mol. Sci. 2020, 21, 2130. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Klement, J.D.; Yang, D.; Albers, T.; Lebedyeva, I.O.; Waller, J.L.; Liu, K. SUV39H1 regulates human colon carcinoma apoptosis and cell cycle to promote tumor growth. Cancer Lett. 2020, 476, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Yue, T.; Huang, Z.; Zhu, J.; Bu, D.; Wang, X.; Pan, Y.; Liu, Y.; Wang, P. Inhibition of hydrogen sulfide synthesis reverses acquired resistance to 5-FU through miR-215-5p-EREG/TYMS axis in colon cancer cells. Cancer Lett. 2019, 466, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Andreucci, E.; Ruzzolini, J.; Peppicelli, S.; Bianchini, F.; Laurenzana, A.; Carta, F.; Supuran, C.T.; Calorini, L. The carbonic anhydrase IX inhibitor SLC-0111 sensitises cancer cells to conventional chemotherapy. J. Enzyme Inhib. Med. Chem. 2019, 34, 117–123. [Google Scholar] [CrossRef]
- Xi, C.; Wang, L.; Yu, J.; Ye, H.; Cao, L.; Gong, Z. Inhibition of cyclin-dependent kinases by AT7519 is effective to overcome chemoresistance in colon and cervical cancer. Biochem. Biophys. Res. Commun. 2019, 513, 589–593. [Google Scholar] [CrossRef]
- Xi, C.; Wang, L.; Yu, J.; Ye, H.; Cao, L.; Gong, Z. Inhibition of eukaryotic translation initiation factor 4E is effective against chemo-resistance in colon and cervical cancer. Biochem. Biophys. Res. Commun. 2018, 503, 2286–2292. [Google Scholar] [CrossRef]
- Ye, Q.; Liu, K.; Shen, Q.; Li, Q.; Hao, J.; Han, F.; Jiang, R.-W. Reversal of Multidrug Resistance in Cancer by Multi-Functional Flavonoids. Front. Oncol. 2019, 9, 487. [Google Scholar] [CrossRef]
- Wang, Z.; Zhan, Y.; Xu, J.; Wang, Y.; Sun, M.; Chen, J.; Liang, T.; Wu, L.; Xu, K. β-Sitosterol Reverses Multidrug Resistance via BCRP Suppression by Inhibiting the p53-MDM2 Interaction in Colorectal Cancer. J. Agric. Food Chem. 2020, 68, 3850–3858. [Google Scholar] [CrossRef]
- Kong, D.; Zhang, D.; Chu, X.; Wang, J. Schizandrin A enhances chemosensitivity of colon carcinoma cells to 5-fluorouracil through up-regulation of miR-195. Biomed. Pharmacother. 2018, 99, 176–183. [Google Scholar] [CrossRef]
- Liu, H.; Liu, H.; Zhou, Z.; Parise, R.A.; Chu, E.; Schmitz, J.C. Herbal formula Huang Qin Ge Gen Tang enhances 5-fluorouracil antitumor activity through modulation of the E2F1/TS pathway. Cell Commun. Signal. 2018, 16, 7. [Google Scholar] [CrossRef]
- La, X.; Zhang, L.; Li, Z.; Li, H.; Yang, Y. (-)-Epigallocatechin Gallate (EGCG) Enhances the Sensitivity of Colorectal Cancer Cells to 5-FU by Inhibiting GRP78/NF-κB/miR-155-5p/MDR1 Pathway. J. Agric. Food Chem. 2019, 67, 2510–2518. [Google Scholar] [CrossRef]
- Li, T.; Chen, Y.; Zhang, J.; Liu, S. LncRNA TUG1 promotes cells proliferation and inhibits cells apoptosis through regulating AURKA in epithelial ovarian cancer cells. Medicine 2018, 97, e12131. [Google Scholar] [CrossRef]
- Yang, C.; Pan, Y.; Deng, S.P. Downregulation of lncRNA CCAT1 enhances 5-fluorouracil sensitivity in human colon cancer cells. BMC Mol. Cell Biol. 2019, 20, 9. [Google Scholar] [CrossRef]
- Fan, H.; Zhu, J.-H.; Yao, X.-Q. Knockdown of long non-coding RNA PVT1 reverses multidrug resistance in colorectal cancer cells. Mol. Med. Rep. 2018, 17, 8309–8315. [Google Scholar] [CrossRef]
- Xiao, Y.; Yurievich, U.A.; Yosypovych, S.V. Long noncoding RNA XIST is a prognostic factor in colorectal cancer and inhibits 5-fluorouracil-induced cell cytotoxicity through promoting thymidylate synthase expression. Oncotarget 2017, 8, 83171–83182. [Google Scholar] [CrossRef]
- Wang, L.; Cho, K.B.; Li, Y.; Tao, G.; Xie, Z.; Guo, B. Long Noncoding RNA (lncRNA)-Mediated Competing Endogenous RNA Networks Provide Novel Potential Biomarkers and Therapeutic Targets for Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 5758. [Google Scholar] [CrossRef]
- Wang, M.; Han, D.; Yuan, Z.; Hu, H.; Zhao, Z.; Yang, R.; Jin, Y.; Zou, C.; Chen, Y.; Wang, G.; et al. Long non-coding RNA H19 confers 5-Fu resistance in colorectal cancer by promoting SIRT1-mediated autophagy. Cell Death Dis. 2018, 9, 1149. [Google Scholar] [CrossRef]
- LI, Q.-W.; LEI, W.; CHEN, C.; GUO, W. Recent advances of long noncoding RNAs involved in the development of multiple sclerosis. Chin. J. Nat. Med. 2020, 18, 36–46. [Google Scholar] [CrossRef]
- Jin, S.-J.; Jin, M.-Z.; Xia, B.-R.; Jin, W.-L. Long Non-coding RNA DANCR as an Emerging Therapeutic Target in Human Cancers. Front. Oncol. 2019, 9, 1225. [Google Scholar] [CrossRef]
- Titze-de-Almeida, S.S.; de Paula Brandão, P.R.; Faber, I.; Titze-de-Almeida, R. Leading RNA Interference Therapeutics Part 1: Silencing Hereditary Transthyretin Amyloidosis, with a Focus on Patisiran. Mol. Diagn. Ther. 2020, 24, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.R.; Abd-Aziz, N.; Affendi, S.; Poh, C.L. Role of microRNAs in antiviral responses to dengue infection. J. Biomed. Sci. 2020, 27, 4. [Google Scholar] [CrossRef] [PubMed]
- Hanna, J.; Hossain, G.S.; Kocerha, J. The Potential for microRNA Therapeutics and Clinical Research. Front. Genet. 2019, 10, 478. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-T.; Han, C.; Sun, Y.-M.; Chen, T.-Q.; Chen, Y.-Q. Noncoding RNAs in cancer therapy resistance and targeted drug development. J. Hematol. Oncol. 2019, 12, 55. [Google Scholar] [CrossRef]
- Liang, G.; Zhu, Y.; Ali, D.J.; Tian, T.; Xu, H.; Si, K.; Sun, B.; Chen, B.; Xiao, Z. Engineered exosomes for targeted co-delivery of miR-21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer. J. Nanobiotechnol. 2020, 18, 10. [Google Scholar] [CrossRef]
- Hong, X.; Dong, T.; Yi, T.; Hu, J.; Zhang, Z.; Lin, S.; Niu, W. Impact of 5-Fu/oxaliplatin on mouse dendritic cells and synergetic effect with a colon cancer vaccine. Chin. J. Cancer Res. 2018, 30, 197–208. [Google Scholar] [CrossRef]
- Van Der Kraak, L.; Goel, G.; Ramanan, K.; Kaltenmeier, C.; Zhang, L.; Normolle, D.P.; Freeman, G.J.; Tang, D.; Nason, K.S.; Davison, J.M.; et al. 5-Fluorouracil upregulates cell surface B7-H1 (PD-L1) expression in gastrointestinal cancers. J. Immunother. Cancer 2016, 4, 65. [Google Scholar] [CrossRef]
- Goel, G.; Ramanan, K.; Kaltenmeier, C.; Zhang, L.; Freeman, G.J.; Normolle, D.P.; Tang, D.; Lotze, M.T. Effect of 5-fluorouracil on membranous PD-L1 expression in colon cancer cells. J. Clin. Oncol. 2016, 34, 592. [Google Scholar] [CrossRef]
- Ieranò, C.; D’Alterio, C.; Napolitano, M.; Portella, L.; Rea, G.; Monaco, G.; Maiolino, P.; Luciano, A.; Barbieri, A.; Palma, G.; et al. Abstract 4103: Blockade of intrinsic PD-1 promotes human colon cancer cells survival, chemo resistance and in vivo tumor growth of human colon cancer cells. Cancer Res. 2019, 79, 4103. [Google Scholar] [CrossRef]

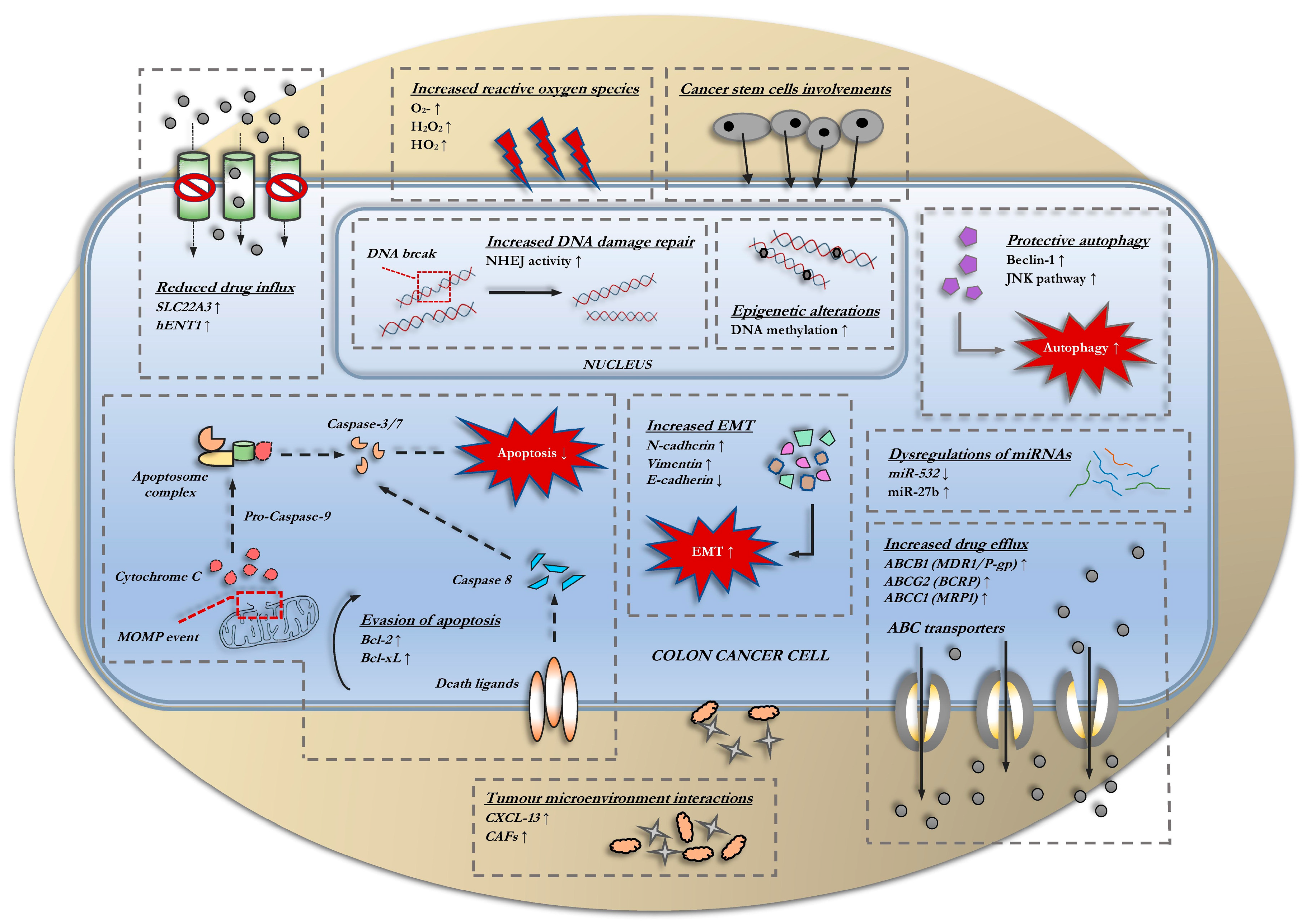
| miRNA | Resistance Expression | Mechanism | Ref |
|---|---|---|---|
| miR-361 | Underexpressed | Upregulation of ABCC10/5 expression regulated by FOXM1 | [197] |
| miR-27a | Overexpressed | Modulation of DPYD expression | [198] |
| miR-200c | Underexpressed | Enhanced Bcl-2 expression | [199] |
| miR-587 | Overexpressed | Downregulation of PPP2R1B expression that increases AKT phosphorylation and XIAP expression | [200] |
| miR-22 | Underexpressed | Promotes autophagy via BTG1 upregulation | [201] |
| miR-224 | Underexpressed | Suppressed apoptosis via repressed E2F activity | [94] |
| miR-375 | Underexpressed | Upregulate TYMS expression | [202] |
| miR-31 | Overexpressed | Downregulation of KANK1 that abrogate CXXC5-mediated apoptosis | [203] |
| miR-204 | Underexpressed | Directly targets HGMA2 that activates the PI3K/Akt signaling pathway | [204] |
| miR-214 | Underexpressed | Downregulation of HSP27 | [205] |
| miR-543 | Overexpressed | Upregulation of PTEN that activates the PI3K/AKT signaling pathway | [206] |
| miR-15b | Underexpressed | Downregulations of NF-κB1 and IKK-α targets | [207] |
| miR-106a | Overexpressed | Downregulation of TGFβR2 that promotes EMT and inhibit apoptosis | [208] |
| miR-206 | Underexpressed | Enhanced Bcl-2 expression | [209] |
| miR-532 | Underexpressed | Promotes Wnt/β-catenin signaling activation | [194] |
| miR-494 | Underexpressed | Upregulation of TOP2α that facilitates DNA-damage repair | [195] |
| miR-125b | Underexpressed | Induction of Sp1/CD248 expression | [210] |
| miR-377 | Underexpressed | Induction of BRD4 expression | [211] |
| miR-139 | Underexpressed | Modulation of NOTCH1, Bcl-2, and AMFR expression | [212] |
| miR-27b | Underexpressed | Upregulation of ATG10 that promotes autophagy | [213] |
| Enzyme | Resistance Mechanism | Recent Evidence | Outcome |
|---|---|---|---|
| Thymidylate synthase | TYMS overexpression via gene amplification through CNV and tandem repeats |
| Restored level of dTMP |
| Thymidine phosphorylase | TYMP suppression via loss-of-function |
| Reduced intra-tumoral concentration of active 5-FU metabolite |
| Dihydropyrimidine dehydrogenase | DPD upregulation via polymorphism |
| |
| Methylenetetrahydrofolate reductase | MTHFR upregulation via polymorphism |
| SNP | Gene | Genotype | Outcome | Remark | Ref |
|---|---|---|---|---|---|
| c.1905 + 1 | DYPD*2A | G > A |
|
| [15,264] |
| c.2846 | D949V | A > T |
|
| [265,266] |
| c.1679 | DYPD*13 | T > G |
|
| [267] |
| c.1129–5923 | HapB3 | C > G |
|
| [267,268] |
| c.1236 | G > A | ||||
| DPYD*5 | T85C | T > C |
|
| [226] |
| DPYD*9A | A1627G | AG + -GG |
|
| [226] |
| Small Molecule Inhibitor | Target | Mechanism | Ref |
|---|---|---|---|
| Depsipeptide | HDAC | Elevation of MHC class II expression and cellular apoptosis | [275] |
| Valproic acid (VPA) | Modulation of TYMS and TYMP expression | [276] | |
| Suberanilohydroxamic acid (SAHA) | Decreased TYMS mRNA and protein expression | [288] | |
| Cobimetinib | MEK | Decreased TYMS expression | [280] |
| Selumetinib | Abrogation of TK1 expression | [289] | |
| U0126 | Generation of more γH2AX foci, diminishing ERCC1 and TYMS expression | [282] | |
| Thiostrepton | FOXM1 | Suppression of TYMS expression and the regulation of TK-1 and TYMPS expression | [281] |
| Luminespib | HSP90 | Downregulation of TYMS | [277,278] |
| Ganetespib | Not stated | [279] | |
| Apatorsen | HSP27 | Accelerated apoptosis | [290] |
| Ibrutinib | BTK | Inhibition of TGFB1 protective response and induction of pro-apoptotic E2F expression | [291] |
| AVL-292 | |||
| Trimethylglycine | STAT6 | Increased E-cadherin marker and decreased ERCC1 expression | [292] |
| JTE-013 | S1PR2 | Downregulation of DPD expression | [236] |
| Gimeracil | DPD | Downregulation of DPD expression | [293] |
| Quinacrine | Nrf2 | Increases the susceptibility of tumor cells to 5-FU under hypoxic conditions | [285] |
| Tipiracil | TYMP | Downregulations of TYMP | [118] |
| F5446 | SUV39H1 | Increased Fas expression and FasL-induced apoptosis | [293] |
| Aminooxyacetic acid (AOAA) | Hydrogen sulfide | Downregulations of TYMS and EREG expression | [294] |
| SLC-0111 | Carbonic anhydrase IX | Not stated | [295] |
| AT7519 | CDK | Not stated | [296] |
| Ribavirin | Eif4E | Increased cell cycle arrest at G2/M phase via increased cyclin B1, p-histone (Ser10), and Mad2 expression | [297] |
| Diethylaminobenzaldehyde (DEAB) | ALDH1 | Not stated | [126] |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azwar, S.; Seow, H.F.; Abdullah, M.; Faisal Jabar, M.; Mohtarrudin, N. Recent Updates on Mechanisms of Resistance to 5-Fluorouracil and Reversal Strategies in Colon Cancer Treatment. Biology 2021, 10, 854. https://doi.org/10.3390/biology10090854
Azwar S, Seow HF, Abdullah M, Faisal Jabar M, Mohtarrudin N. Recent Updates on Mechanisms of Resistance to 5-Fluorouracil and Reversal Strategies in Colon Cancer Treatment. Biology. 2021; 10(9):854. https://doi.org/10.3390/biology10090854
Chicago/Turabian StyleAzwar, Shamin, Heng Fong Seow, Maha Abdullah, Mohd Faisal Jabar, and Norhafizah Mohtarrudin. 2021. "Recent Updates on Mechanisms of Resistance to 5-Fluorouracil and Reversal Strategies in Colon Cancer Treatment" Biology 10, no. 9: 854. https://doi.org/10.3390/biology10090854
APA StyleAzwar, S., Seow, H. F., Abdullah, M., Faisal Jabar, M., & Mohtarrudin, N. (2021). Recent Updates on Mechanisms of Resistance to 5-Fluorouracil and Reversal Strategies in Colon Cancer Treatment. Biology, 10(9), 854. https://doi.org/10.3390/biology10090854






