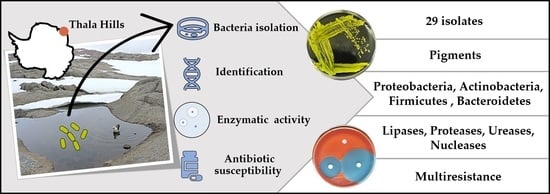
Isolation, Physiological Characterization, and Antibiotic Susceptibility Testing of Fast-Growing Bacteria from the Sea-Affected Temporary Meltwater Ponds in the Thala Hills Oasis (Enderby Land, East Antarctica)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Sites and Sampling Procedure
2.2. Bacterioplankton and Physicochemical Analysis of Water Samples
2.3. Isolation of the Fast-Growing Bacteria
2.4. Total DNA Extraction, Amplification, and Sequencing
2.5. Analysis of 16S rRNA Sequencing Data
2.6. Thermotolerance and Enzymatic Activity
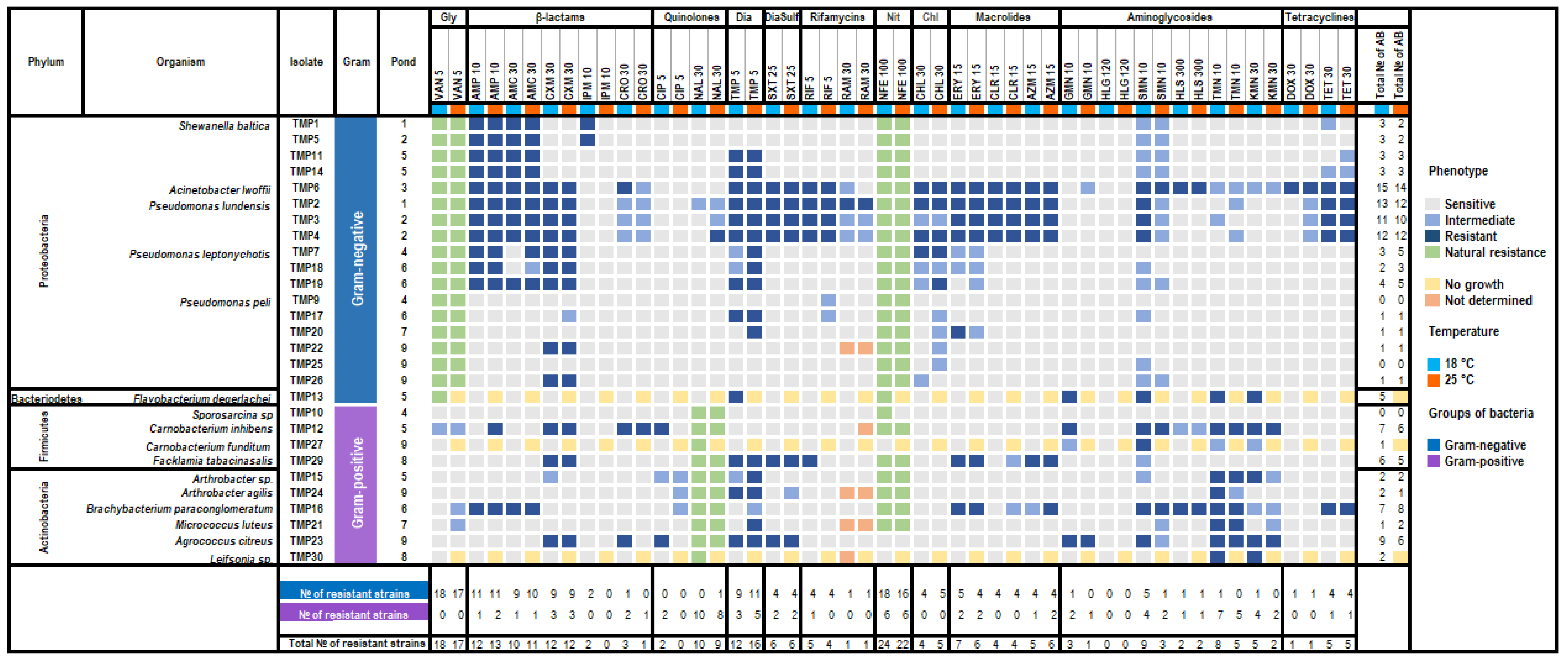
2.7. Antibiotic Susceptibility Testing
3. Results
3.1. Physicochemical Characterization of Water Samples from Meltwater Ponds
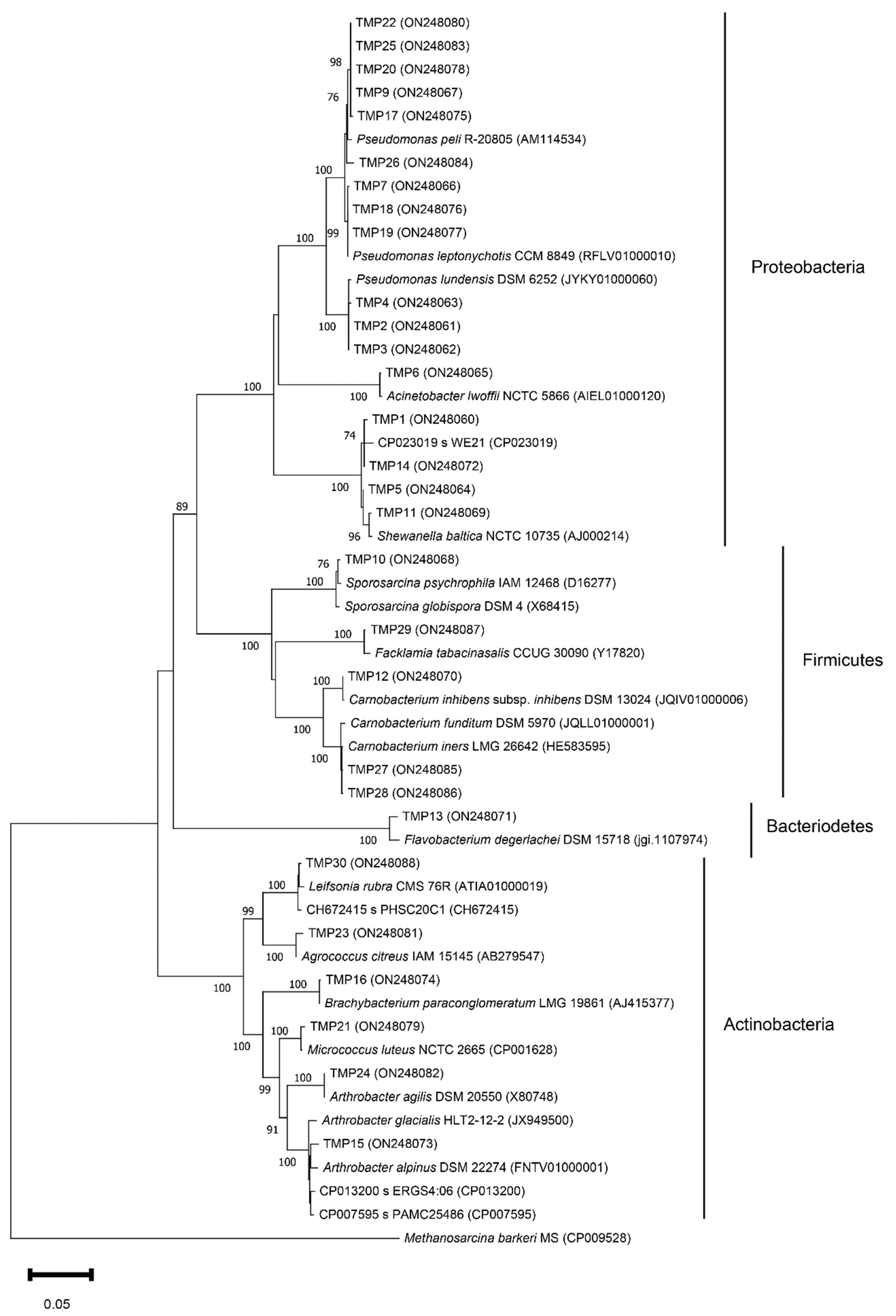
3.2. Isolation and Phylogenetic Characterization of the Meltwater Fast-Growing Bacteria
3.3. Thermotolerance and Enzymatic Activity
3.4. Antibiotic Susceptibility
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wilkins, D.; Yau, S.; Williams, T.J.; Allen, M.A.; Brown, M.V.; Demaere, M.Z.; Lauro, F.M.; Cavicchioli, R. Key microbial drivers in Antarctic aquatic environments. FEMS Microbiol. Rev. 2013, 37, 303–335. [Google Scholar] [CrossRef]
- Kakareka, S.V.; Kukharchyk, T.I.; Kokosh, Y.G.; Kudrevich, M.A.; Giginyak, Y.G.; Myamin, V.E. Chemical characteristics of antarctic lakes of the Thala Hills. Arct. Antarct. Res. 2019, 65, 422–437. [Google Scholar] [CrossRef]
- Miamin, V.E.; Nikitina, L.V.; Charniauskaya, M.I.; Zaniuk, A.A.; Titok, M.A.; Laziuk, S.K.; Sidarenka, A.V.; Valentovich, L.N.; Dolgikh, A.V. Microbiology investigation in the Vechernyy region, Tala Hills (East Antarctica). Belarus State Univ. Annu. Physiol. Biochem. Mol. Base Biosyst. Funct. 2014, 9, 58–67. [Google Scholar]
- Dieser, M.; Foreman, C.M.; Jaros, C.; Lisle, J.T.; Greenwood, M.; Laybourn-Parry, J.; Miller, P.L.; Chin, Y.-P.; McKnight, D.M. Physicochemical and biological dynamics in a coastal Antarctic lake as it transitions from frozen to open water. Antarct. Sci. 2013, 25, 663–675. [Google Scholar] [CrossRef]
- Hogg, I.D.; Cary, S.C.; Convey, P.; Newsham, K.K.; O’Donnell, A.G.; Adams, B.J.; Aislabie, J.; Frati, F.; Stevens, M.I.; Wall, D.H. Biotic interactions in Antarctic terrestrial ecosystems: Are they a factor? Soil Biol. Biochem. 2006, 38, 3035–3040. [Google Scholar] [CrossRef]
- Wait, B.; Nokes, R.; Webster-Brown, J. Freeze-thaw dynamics and the implications for stratification and brine geochemistry in meltwater ponds on the McMurdo Ice Shelf, Antarctica. Antarct. Sci. 2009, 21, 243–254. [Google Scholar] [CrossRef]
- De Mora, S.; Whitehead, R.F.; Gregory, M. The chemical composition of glacial melt water ponds and streams on the McMurdo Ice Shelf, Antarctica. Antarct. Sci. 1994, 6, 17–27. [Google Scholar] [CrossRef]
- Matsumoto, G.I.; Nakaya, S.; Murayama, H.; Masuda, N.; Kawano, T.; Watanuki, K.; Torii, T. Geochemical characteristics of Antarctic lakes and ponds. Proc. NIPR Symp. Polar Biol. 1992, 5, 125–145. [Google Scholar]
- Wait, B.; Webster-Brown, J.; Brown, K.; Healy, M.; Hawes, I. PChemistry and stratification of Antarctic meltwater ponds I: Coastal ponds near Bratina Island, McMurdo Ice Shelf. Antarct. Sci. 2006, 18, 515–524. [Google Scholar] [CrossRef]
- Jackson, E.E.; Hawes, I.; Jungblut, A.D. 16S rRNA gene and 18S rRNA gene diversity in microbial mat communities in meltwater ponds on the McMurdo Ice Shelf, Antarctica. Polar Biol. 2021, 44, 823–836. [Google Scholar] [CrossRef]
- Archer, S.D.; McDonald, I.R.; Herbold, C.W.; Lee, C.K.; Cary, C.S. Benthic microbial communities of coastal terrestrial and ice shelf Antarctic meltwater ponds. Front. Microbiol. 2015, 6, 485. [Google Scholar] [CrossRef]
- Archer, S.D.J.; McDonald, I.R.; Herbold, C.W.; Lee, C.K.; Niederberger, T.S.; Cary, C. Temporal, regional and geochemical drivers of microbial community variation in the melt ponds of the Ross Sea region, Antarctica. Polar Biol. 2016, 39, 267–282. [Google Scholar] [CrossRef]
- Archer, S.D.J.; McDonald, I.R.; Herbold, C.W.; Cary, S.C. Characterisation of bacterioplankton communities in the meltwater ponds of Bratina Island, Victoria Land, Antarctica. FEMS Microbiol. Ecol. 2014, 89, 451–464. [Google Scholar] [CrossRef]
- Smirnova, M.; Miamin, U.; Kohler, A.; Valentovich, L.; Akhremchuk, A.; Sidarenka, A.; Dolgikh, A.; Shapaval, V. Isolation and characterization of fast growing green snow bacteria from coastal East Antarctica. MicrobiologyOpen 2021, 10, e1152. [Google Scholar] [CrossRef] [PubMed]
- Kudinova, A.; Lysak, L.; Soina, V.; Mergelov, N.; Dolgikh, A.; Shorkunov, I. Bacterial communities in the soils of cryptogamic barrens of East Antarctica (the Larsemann Hills and Thala Hills oases). Eurasian Soil Sci. 2015, 48, 276–287. [Google Scholar] [CrossRef]
- Lukashanets, D.A.; Convey, P.; Borodin, O.I.; Miamin, V.Y.; Hihiniak, Y.H.; Gaydashov, A.A.; Yatsyna, A.P.; Vezhnavets, V.V.; Maysak, N.N.; Shendrik, T.V. Eukarya biodiversity in the Thala Hills, East Antarctica. Antarct. Sci. 2021, 33, 605–623. [Google Scholar] [CrossRef]
- Dolgikh, A.V.; Mergelov, N.S.; Abramov, A.A.; Lupachev, A.V.; Goryachkin, S.V. Soils of Enderby Land. In The Soils of Antarctica; Springer: Berlin/Heidelberg, Germany, 2015; pp. 45–63. [Google Scholar]
- Gribanova, E.; Miamin, V. Physiological and biochemical traits of yeasts from soils of various ecosystems of East Antarctica. Ukr. Antarct. J. 2021, 2, 106–116. [Google Scholar] [CrossRef]
- Ferrés, I.; Amarelle, V.; Noya, F.; Fabiano, E. Identification of Antarctic culturable bacteria able to produce diverse enzymes of potential biotechnological interest. Adv. Polar Sci. 2015, 26, 71–79. [Google Scholar] [CrossRef]
- Morozova, O.V.; Andreeva, I.S.; Zhirakovskiy, V.Y.; Pechurkina, N.I.; Puchkova, L.I.; Saranina, I.V.; Emelyanova, E.K.; Kamynina, T.P. Antibiotic resistance and cold-adaptive enzymes of antarctic culturable bacteria from King George Island. Polar Sci. 2022, 31, 100756. [Google Scholar] [CrossRef]
- Piegza, M.; Łaba, W.; Kačániová, M. New Arctic Bacterial Isolates with Relevant Enzymatic Potential. Molecules 2020, 25, 3930. [Google Scholar] [CrossRef]
- Ray, M.K.; Kumar, G.S.; Janiyani, K.; Kannan, K.; Jagtap, P.; Basu, M.K.; Shivaji, S. Adaptation to low temperature and regulation of gene expression in antarctic psychrotrophic bacteria. J. Biosci. 1998, 23, 423–435. [Google Scholar] [CrossRef]
- Rizzo, C.; Conte, A.; Azzaro, M.; Papale, M.; Rappazzo, A.C.; Battistel, D.; Roman, M.; Lo Giudice, A.; Guglielmin, M. Cultivable Bacterial Communities in Brines from Perennially Ice-Covered and Pristine Antarctic Lakes: Ecological and Biotechnological Implications. Microorganisms 2020, 8, 819. [Google Scholar] [CrossRef] [PubMed]
- Shawkey, M.D.; Mills, K.L.; Dale, C.; Hill, G.E. Microbial Diversity of Wild Bird Feathers Revealed throughCulture-Based and Culture-Independent Techniques. Microb. Ecol. 2005, 50, 40–47. [Google Scholar] [CrossRef]
- Yadav, A.N.; Sachan, S.G.; Verma, P.; Tyagi, S.P.; Kaushik, R.; Saxena, A.K. Culturable diversity and functional annotation of psychrotrophic bacteria from cold desert of Leh Ladakh (India). World J. Microbiol. Biotechnol. 2015, 31, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Dieser, M.; Greenwood, M.; Foreman, C.M. Carotenoid Pigmentation in Antarctic Heterotrophic Bacteria as a Strategy to Withstand Environmental Stresses. Arct. Antarct. Alp. Res. 2010, 42, 396–405. [Google Scholar] [CrossRef]
- Fong, N.J.C.; Burgess, M.L.; Barrow, K.D.; Glenn, D.R. Carotenoid accumulation in the psychrotrophic bacterium Arthrobacter agilis in response to thermal and salt stress. Appl. Microbiol. Biotechnol. 2001, 56, 750–756. [Google Scholar] [CrossRef]
- Jagannadham, M.V.; Chattopadhyay, M.K.; Subbalakshmi, C.; Vairamani, M.; Narayanan, K.; Mohan Rao, C.; Shivaji, S. Carotenoids of an Antarctic psychrotolerant bacterium, Sphingobacterium antarcticus, and a mesophilic bacterium, Sphingobacterium multivorum. Arch. Microbiol. 2000, 173, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Moyer, C.L.; Morita, R.Y. Psychrophiles and psychrotrophs. Encycl. Life Sci. 2007, 1. [Google Scholar]
- Sajjad, W.; Din, G.; Rafiq, M.; Iqbal, A.; Khan, S.; Zada, S.; Ali, B.; Kang, S. Pigment production by cold-adapted bacteria and fungi: Colorful tale of cryosphere with wide range applications. Extremophiles 2020, 24, 447–473. [Google Scholar] [CrossRef] [PubMed]
- Silva, T.R.E.; Silva, L.C.F.J.; de Queiroz, A.C.; Alexandre Moreira, M.S.; de Carvalho Fraga, C.A.; de Menezes, G.C.A.; Rosa, L.H.; Bicas, J.; de Oliveira, V.M.; Duarte, A.W.F. Pigments from Antarctic bacteria and their biotechnological applications. Crit. Rev. Biotechnol. 2021, 41, 809–826. [Google Scholar] [CrossRef]
- Gupta, V.K.; Treichel, H.; Shapaval, V.O.; de Oliveira, L.A.; Tuohy, M.G. Microbial Functional Foods and Nutraceuticals; John Wiley & Sons: Hoboken, NJ, USA, 2017. [Google Scholar]
- Martínez, J.L. Natural antibiotic resistance and contamination by antibiotic resistance determinants: The two ages in the evolution of resistance to antimicrobials. Front. Microbiol. 2012, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- Van Goethem, M.W.; Pierneef, R.; Bezuidt, O.K.I.; Van De Peer, Y.; Cowan, D.A.; Makhalanyane, T.P. A reservoir of ‘historical’ antibiotic resistance genes in remote pristine Antarctic soils. Microbiome 2018, 6, 40. [Google Scholar] [CrossRef] [PubMed]
- Jara, D.; Bello-Toledo, H.; Domínguez, M.; Cigarroa, C.; Fernández, P.; Vergara, L.; Quezada-Aguiluz, M.; Opazo-Capurro, A.; Lima, C.A.; González-Rocha, G. Antibiotic resistance in bacterial isolates from freshwater samples in Fildes Peninsula, King George Island, Antarctica. Sci. Rep. 2020, 10, 3145. [Google Scholar] [CrossRef] [PubMed]
- Hobbie, J.E.; Daley, R.J.; Jasper, S. Use of nuclepore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 1977, 33, 1225–1228. [Google Scholar] [CrossRef] [PubMed]
- Lane, D. 16S/23S rRNA sequencing. Nucleic Acid Tech. Bact. Syst. 1991, 115–175. [Google Scholar]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Hillis, D.M.; Bull, J.J. An Empirical Test of Bootstrapping as a Method for Assessing Confidence in Phylogenetic Analysis. Syst. Biol. 1993, 42, 182–192. [Google Scholar] [CrossRef]
- Bauer, A.W.; Kirby, W.M.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 12.0. 2022. Available online: http://www.eucast.org (accessed on 1 April 2022).
- CLSI: Clinical and Laboratory Standards Institute. 2020 CLSI document M100. In Performance Standards for Antimicrobial. Susceptibility Tests, 30th ed.; Pa, W., Ed.; CLSI: Wayne, PA, USA, 2020. [Google Scholar]
- Kato, S.; Yamagishi, A.; Daimon, S.; Kawasaki, K.; Tamaki, H.; Kitagawa, W.; Abe, A.; Tanaka, M.; Sone, T.; Asano, K. Isolation of previously uncultured slow-growing bacteria by using a simple modification in the preparation of agar media. Appl. Environ. Microbiol. 2018, 84, e00807–e00818. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, M.; Tafintseva, V.; Kohler, A.; Miamin, U.; Shapaval, V. Temperature- and Nutrients-Induced Phenotypic Changes of Antarctic Green Snow Bacteria Probed by High-Throughput FTIR Spectroscopy. Biology 2022, 11, 890. [Google Scholar] [CrossRef] [PubMed]
- López-Archilla, A.I.; Moreira, D.; López-García, P.; Guerrero, C. Phytoplankton diversity and cyanobacterial dominance in a hypereutrophic shallow lake with biologically produced alkaline pH. Extremophiles 2004, 8, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Lecomte, K.L.; Vignoni, P.A.; Córdoba, F.E.; Chaparro, M.A.E.; Kopalová, K.; Gargiulo, J.D.; Lirio, J.M.; Irurzun, M.A.; Böhnel, H.N. Hydrological systems from the Antarctic Peninsula under climate change: James Ross archipelago as study case. Environ. Earth Sci. 2016, 75, 623. [Google Scholar] [CrossRef]
- George, S.F.; Fierer, N.; Levy, J.S.; Adams, B. Antarctic Water Tracks: Microbial Community Responses to Variation in Soil Moisture, pH, and Salinity. Front. Microbiol. 2021, 12, 616730. [Google Scholar] [CrossRef] [PubMed]
- Antony, R.; Sanyal, A.; Kapse, N.; Dhakephalkar, P.K.; Thamban, M.; Nair, S. Microbial communities associated with Antarctic snow pack and their biogeochemical implications. Microbiol. Res. 2016, 192, 192–202. [Google Scholar] [CrossRef]
- Núñez-Montero, K.; Barrientos, L. Advances in Antarctic Research for Antimicrobial Discovery: A Comprehensive Narrative Review of Bacteria from Antarctic Environments as Potential Sources of Novel Antibiotic Compounds Against Human Pathogens and Microorganisms of Industrial Importance. Antibiotics 2018, 7, 90. [Google Scholar] [CrossRef]
- Joint, I.; Mühling, M.; Querellou, J. Culturing marine bacteria—An essential prerequisite for biodiscovery. Microb. Biotechnol. 2010, 3, 564–575. [Google Scholar] [CrossRef] [PubMed]
- Leiva, S.; Alvarado, P.; Huang, Y.; Wang, J.; Garrido, I. Diversity of pigmented Gram-positive bacteria associated with marine macroalgae from Antarctica. FEMS Microbiol. Lett. 2015, 362, fnv206. [Google Scholar] [CrossRef]
- Rego, A.; Raio, F.; Martins, T.P.; Ribeiro, H.; Sousa, A.G.G.; Séneca, J.; Baptista, M.S.; Lee, C.K.; Cary, S.C.; Ramos, V.; et al. Actinobacteria and Cyanobacteria Diversity in Terrestrial Antarctic Microenvironments Evaluated by Culture-Dependent and Independent Methods. Front. Microbiol. 2019, 10, 1018. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Huang, Q.; Deng, S.; Dong, H.; Yu, B. Planktonic actinobacterial diversity along a salinity gradient of a river and five lakes on the Tibetan Plateau. Extremophiles 2010, 14, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Antibus, D.E.; Leff, L.G.; Hall, B.L.; Baeseman, J.L.; Blackwood, C.B. Cultivable bacteria from ancient algal mats from the McMurdo Dry Valleys, Antarctica. Extremophiles 2012, 16, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Zheng, T.; Yu, Y.; Chen, B.; He, J. Relationships between Arctic and Antarctic Shewanella strains evaluated by a polyphasic taxonomic approach. Polar Biol. 2010, 33, 531–541. [Google Scholar] [CrossRef]
- Ravi, K.; Falkowski, N.R.; Scales, B.S.; Akulava, V.D.; Valentovich, L.N.; Huffnagle, G.B. The Psychrotrophic Pseudomonas lundensis, a Non-aeruginosa Pseudomonad, Has a Type III Secretion System of the Ysc Family, Which Is Transcriptionally Active at 37 °C. Mbio 2022, 13, e03869-21. [Google Scholar] [CrossRef]
- Molin, G.; Ternstrom, A.; Ursing, J. Notes: Pseudomonas lundensis, a New Bacterial Species Isolated from Meat. Int. J. Syst. Bacteriol. 1986, 36, 339–342. [Google Scholar] [CrossRef]
- Nováková, D.; Švec, P.; Zeman, M.; Busse, H.-J.; Mašlaňová, I.; Pantůček, R.; Králová, S.; Krištofová, L.; Sedláček, I. Pseudomonas leptonychotis sp. nov., isolated from Weddell seals in Antarctica. Int. J. Syst. Evol. Microbiol. 2020, 70, 302–308. [Google Scholar] [CrossRef]
- Franzmann, P.D.; Höpfl, P.; Weiss, N.; Tindall, B.J. Psychrotrophic, lactic acid-producing bacteria from anoxic waters in Ace Lake, Antarctica; Carnobacterium funditum sp. nov. and Carnobacterium alterfunditum sp. nov. Arch. Microbiol. 1991, 156, 255–262. [Google Scholar] [CrossRef]
- Van Trappen, S.; Vandecandelaere, I.; Mergaert, J.; Swings, J. Flavobacterium degerlachei sp. nov., Flavobacterium frigoris sp. nov. and Flavobacterium micromati sp. nov., novel psychrophilic bacteria isolated from microbial mats in Antarctic lakes. Int. J. Syst. Evol. Microbiol. 2004, 54, 85–92. [Google Scholar] [CrossRef]
- Bej, A.K.; Aislabie, J.; Atlas, R.M. Polar Microbiology: The Ecology, Biodiversity and Bioremediation Potential of Microorganisms in Extremely Cold Environments; CRC Press: Boca Raton, FL, USA, 2009. [Google Scholar]
- Reddy, G.S.N.; Prakash, J.S.S.; Srinivas, R.; Matsumoto, G.I.; Shivaji, S. Leifsonia rubra sp. nov. and Leifsonia aurea sp. nov., psychrophiles from a pond in Antarctica. Int. J. Syst. Evol. Microbiol. 2003, 53, 977–984. [Google Scholar] [CrossRef]
- Mindlin, S.; Petrenko, A.; Kurakov, A.; Beletsky, A.; Mardanov, A.; Petrova, M. Resistance of Permafrost and Modern Acinetobacter lwoffii Strains to Heavy Metals and Arsenic Revealed by Genome Analysis. BioMed Res. Int. 2016, 2016, 1–9. [Google Scholar] [CrossRef]
- Wang, J.-P.; Liu, B.; Liu, G.-H.; Xiao, R.-F.; Zheng, X.-F.; Shi, H.; Ge, C.-B. Draft genome sequence of Sporosarcina globispora W 25T (DSM 4), a psychrophilic bacterium isolated from soil and river water. Genome Announc. 2015, 3, e01230-15. [Google Scholar] [CrossRef]
- Nicholson, W.L.; Zhalnina, K.; De Oliveira, R.R.; Triplett, E.W. Proposal to rename Carnobacterium inhibens as Carnobacterium inhibens subsp. inhibens subsp. nov. and description of Carnobacterium inhibens subsp. gilichinskyi subsp. nov., a psychrotolerant bacterium isolated from Siberian permafrost. Int. J. Syst. Evol. Microbiol. 2015, 65, 556–561. [Google Scholar] [CrossRef]
- Kumar, R.; Singh, D.; Swarnkar, M.K.; Singh, A.K.; Kumar, S. Complete genome sequence of Arthrobacter alpinus ERGS4: 06, a yellow pigmented bacterium tolerant to cold and radiations isolated from Sikkim Himalaya. J. Biotechnol. 2016, 220, 86–87. [Google Scholar] [CrossRef]
- Zhang, D.-C.; Schumann, P.; Liu, H.-C.; Xin, Y.-H.; Zhou, Y.-G.; Schinner, F.; Margesin, R. Arthrobacter alpinus sp. nov., a psychrophilic bacterium isolated from alpine soil. Int. J. Syst. Evol. Microbiol. 2010, 60, 2149–2153. [Google Scholar] [CrossRef]
- Morita, R.Y. Psychrophilic bacteria. Bacteriol. Rev. 1975, 39, 144–167. [Google Scholar] [CrossRef]
- De Souza, M.-J.; Nair, S.; Loka Bharathi, P.A.; Chandramohan, D. Metal and antibiotic-resistance in psychrotrophic bacteria from Antarctic Marine waters. Ecotoxicology 2006, 15, 379–384. [Google Scholar] [CrossRef]
- Helmke, E.; Weyland, H. Psychrophilic versus psychrotolerant bacteria--occurrence and significance in polar and temperate marine habitats. Cell. Mol. Biol. 2004, 50, 553–561. [Google Scholar]
- Bowman, J.P.; McCammon, S.A.; Brown, M.V.; Nichols, D.S.; McMeekin, T.A. Diversity and association of psychrophilic bacteria in Antarctic sea ice. Appl. Environ. Microbiol. 1997, 63, 3068–3078. [Google Scholar] [CrossRef]
- Lo Giudice, A.; Michaud, L.; De Pascale, D.; De Domenico, M.; Di Prisco, G.; Fani, R.; Bruni, V. Lipolytic activity of Antarctic cold-adapted marine bacteria (Terra Nova Bay, Ross Sea). J. Appl. Microbiol. 2006, 101, 1039–1048. [Google Scholar] [CrossRef]
- Salwoom, L.; Raja Abd Rahman, R.; Salleh, A.; Mohd. Shariff, F.; Convey, P.; Pearce, D.; Mohamad Ali, M. Isolation, Characterisation, and Lipase Production of a Cold-Adapted Bacterial Strain Pseudomonas sp. LSK25 Isolated from Signy Island, Antarctica. Molecules 2019, 24, 715. [Google Scholar] [CrossRef] [PubMed]
- Henderson, R.J.; Olsen, R.E.; Eilertsen, H.C. Lipid composition of phytoplankton from the Barents Sea and environmental influences on the distribution pattern of carbon among photosynthetic end products. Polar Res. 1991, 10, 229–238. [Google Scholar] [CrossRef]
- Connelly, T.L.; Baer, S.E.; Cooper, J.T.; Bronk, D.A.; Wawrik, B. Urea Uptake and Carbon Fixation by Marine Pelagic Bacteria and Archaea during the Arctic Summer and Winter Seasons. Appl. Environ. Microbiol. 2014, 80, 6013–6022. [Google Scholar] [CrossRef]
- Williams, T.J.; Allen, M.A.; Demaere, M.Z.; Kyrpides, N.C.; Tringe, S.G.; Woyke, T.; Cavicchioli, R. Microbial ecology of an Antarctic hypersaline lake: Genomic assessment of ecophysiology among dominant haloarchaea. ISME J. 2014, 8, 1645–1658. [Google Scholar] [CrossRef]
- Pinchuk, G.E.; Ammons, C.; Culley, D.E.; Li, S.-M.W.; McLean, J.S.; Romine, M.F.; Nealson, K.H.; Fredrickson, J.K.; Beliaev, A.S. Utilization of DNA as a Sole Source of Phosphorus, Carbon, and Energy by Shewanella spp.: Ecological and Physiological Implications for Dissimilatory Metal Reduction. Appl. Environ. Microbiol. 2008, 74, 1198–1208. [Google Scholar] [CrossRef]
- Gushterova, A.; Vasileva-Tonkova, E.; Dimova, E.; Nedkov, P.; Haertlé, T. Keratinase Production by Newly Isolated Antarctic Actinomycete Strains. World J. Microbiol. Biotechnol. 2005, 21, 831–834. [Google Scholar] [CrossRef]
- Papale, M.; Lo Giudice, A.; Conte, A.; Rizzo, C.; Rappazzo, A.C.; Maimone, G.; Caruso, G.; La Ferla, R.; Azzaro, M.; Gugliandolo, C.; et al. Microbial Assemblages in Pressurized Antarctic Brine Pockets (Tarn Flat, Northern Victoria Land): A Hotspot of Biodiversity and Activity. Microorganisms 2019, 7, 333. [Google Scholar] [CrossRef]
- Zhang, X.-Y.; Han, X.-X.; Chen, X.-L.; Dang, H.-Y.; Xie, B.-B.; Qin, Q.-L.; Shi, M.; Zhou, B.-C.; Zhang, Y.-Z. Diversity of cultivable protease-producing bacteria in sediments of Jiaozhou Bay, China. Front. Microbiol. 2015, 6, 1021. [Google Scholar] [CrossRef]
- Lo Giudice, A.; Casella, P.; Bruni, V.; Michaud, L. Response of bacterial isolates from Antarctic shallow sediments towards heavy metals, antibiotics and polychlorinated biphenyls. Ecotoxicology 2013, 22, 240–250. [Google Scholar] [CrossRef]
- Tam, H.K.; Wong, C.M.V.L.; Yong, S.T.; Blamey, J.; González, M. Multiple-antibiotic-resistant bacteria from the maritime Antarctic. Polar Biol. 2015, 38, 1129–1141. [Google Scholar] [CrossRef]
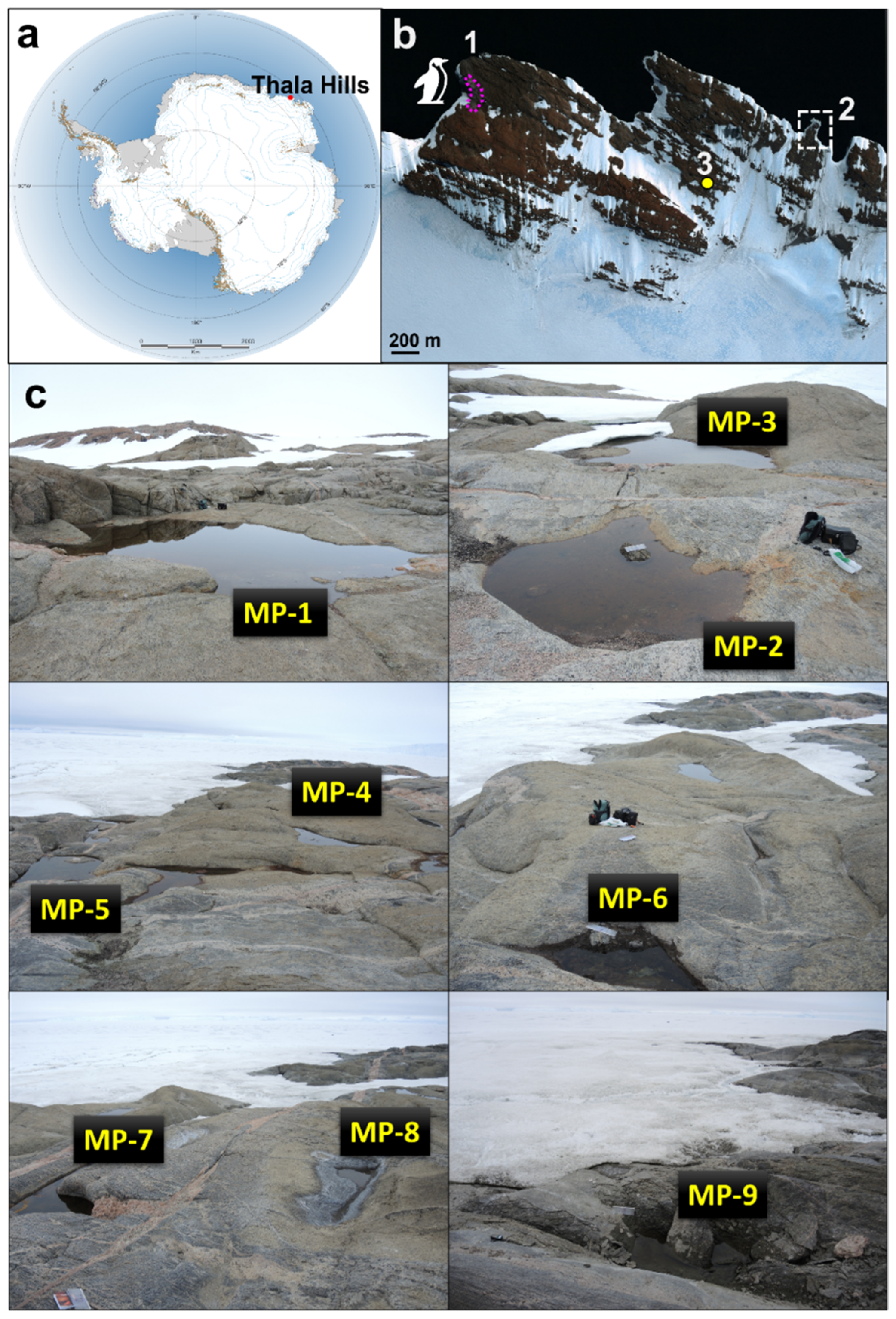

| Day | Temporary Meltwater Pond Number | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MP-1 | MP-2 | MP-3 | MP-4 | MP-5 | MP-6 | MP-7 | MP-8 | MP-9 | |||||||||||
| Physicochemical parameters | |||||||||||||||||||
| Size, m | Day 1 * | 5 × 5 | 4 × 5 | 1.5 × 2 | 2 × 2 | 3 × 2 | 1 × 0.5 | 1.5 × 1 | 0.5 × 0.5 | 1 × 1 | |||||||||
| Depth, m | 0.3 | 0.3 | 0.2 | 0.25 | 0.25 | 0.3 | 0.5 | 0.1 | 0.2 | ||||||||||
| pH | 10.1 | 9.8 | 8.0 | 10.0 | 9.6 | 9.9 | 9.5 | 8.3 | 10.0 | ||||||||||
| TDS *, mg/LT, °C | TDS | T | TDS | T | TDS | T | TDS | T | TDS | T | TDS | T | TDS | T | TDS | T | TDS | T | |
| Day 1 | 271 | 13 | 631 | 14 | 86 | 16 | 458 | 16 | 580 | 13 | 1902 | 13 | 4470 | 12 | 94,000 | 12 | 1640 | 13 | |
| Day 2 | ND | ND | 859 | 18 | 122 | 17.5 | 585 | 17.3 | 730 | 14.4 | PD * | PD | ND | ND | PD | PD | PD | PD | |
| Day 3 | 364 | 9.3 | 940 | 11.4 | 163 | 12.3 | 740 | 10.5 | 1006 | 8.9 | 5684 | 10.2 | |||||||
| Day 4 | 401 | ND | 1209 | ND | 355 | ND | 1126 | ND | 790 | ND | 6385 | ND | |||||||
| Visual observation | |||||||||||||||||||
| WB | Day 1 | + | − | − | − | − | − | + | − | − | |||||||||
| BC | B | O, B | O | O, G, B | O, G | B | O | Gr, B | − | ||||||||||
| OC | + | − | − | − | − | + | − | − | − | ||||||||||
| Skuas | + | + | − | − | − | − | − | − | − | ||||||||||
| Salt | − | − | − | − | − | − | − | + | − | ||||||||||
| Bacterioplankton parameters | |||||||||||||||||||
| BN | Day 1 | 4.52 ± 0.67 | 1.32 ± 0.17 | 1.52 ± 0.26 | 3.01 ± 0.36 | 0.95 ± 0.12 | 2.82 ± 0.41 | 2.22 ± 0.28 | 4.46 ± 0.40 | 2.02 ± 0.32 | |||||||||
| BB | 0.47 ± 0.17 | 0.16 ± 0.06 | 0.32 ± 0.12 | 0.27 ± 0.12 | 0.15 ± 0.03 | 0.50 ± 0.14 | 0.31 ± 0.11 | 0.94 ± 0.28 | 0.77 ± 0.20 | ||||||||||
| NBI | 2 | 3 | 1 | 3 | 5 | 4 | 2 | 2 | 7 | ||||||||||
| Isolate Code MP * | Collection Number | Gen Bank Accession Number | Nearest Taxonomic Neighbor by EzBioCloud Alignment | Identity (%) | Isolation Temperature (°C) | Thermotolerance (°C) | Enzymatic Activity at 18 °C |
|---|---|---|---|---|---|---|---|
| Proteobacteria | |||||||
| TMP1 1 | BIM B-1565 | ON248060 | Shewanella baltica NCTC 10735 | 100 | 5 | 4–30 | L *; Gel; D; C |
| TMP5 2 | BIM B-1557 | ON248064 | Shewanella baltica NCTC 10735 | 99.72 | 18 | 4–30 | L; Gel; D; C |
| TMP11 5 | BIM B-1561 | ON248069 | Shewanella baltica NCTC 10735 | 99.66 | 5 | 4–30 | L; Gel; D |
| TMP14 5 | BIM B-1563 | ON248072 | Shewanella WE21 | 99.38 | 5 | 4–30 | L; Gel; D |
| Shewanella baltica NCTC 10735 | 99.04 | ||||||
| TMP6 3 | BIM B-1558 | ON248065 | Acinetobacter lwoffii NCTC 5866 | 99.79 | 18 | 4–37 | L; C |
| TMP2 1 | BIM B-1554 | ON248061 | Pseudomonas lundensis DSM 6252 | 99.86 | 18 | 4–37 | Cas; Gel; U; C |
| TMP3 2 | BIM B-1555 | ON248062 | Pseudomonas lundensis DSM 6252 | 99.86 | 5 | 4–37 | Cas; Gel; U; C |
| TMP4 2 | BIM B-1556 | ON248063 | Pseudomonas lundensis DSM 6252 | 99.79 | 18 | 4–37 | Cas; Gel; U; C |
| TMP7 4 | BIM B-1559 | ON248066 | Pseudomonas leptonychotis CCM 8849 | 99.93 | 5 | 4–37 | L; Cas |
| TMP18 6 | BIM B-1568 | ON248076 | Pseudomonas leptonychotis CCM 8849 | 100 | 18 | 4–30 | L; Cas |
| TMP19 6 | BIM B-1566 | ON248077 | Pseudomonas leptonychotis CCM 8849 | 100 | 18 | 4–30 | L; Cas |
| TMP9 4 | BIM B-1560 | ON248067 | Pseudomonas peli R-20805 | 99.52 | 5 | 4–25 | L |
| TMP17 6 | BIM B-1569 | ON248075 | Pseudomonas peli R-20805 | 99.38 | 5 | 4–25 | L |
| TMP20 7 | BIM B-1546 | ON248078 | Pseudomonas peli R-20805 | 99.52 | 5 | 4–25 | L |
| TMP22 9 | BIM B-1552 | ON248080 | Pseudomonas peli R-20805 | 99.52 | 5 | 4–25 | L |
| TMP25 9 | BIM B-1542 | ON248083 | Pseudomonas peli R-20805 | 99.52 | 18 | 4–25 | L |
| TMP26 9 | BIM B-1548 | ON248084 | Pseudomonas peli R-20805 | 99.11 | 5 | 4–25 | L |
| Bacteroidetes | |||||||
| TMP13 5 | BIM B-1562 | ON248071 | Flavobacterium degerlachei DSM 15718 | 98.47 | 18 | 4–25 | C |
| Firmicutes | |||||||
| TMP10 4 | BIM B-1539 | ON248068 | Sporosarcina globispora DSM 4 | 99.59 | 18 | 4–30 | U; C |
| Sporosarcina psychrophila IAM 12468 | 99.59 | ||||||
| TMP12 5 | BIM B-1540 | ON248070 | Carnobacterium inhibens subsp. inhibens DSM 13024 | 100 | 5 | 4–30 | L |
| TMP27 9 | BIM B-1541 | ON248085 | Carnobacterium funditum DSM 5970 | 100 | 18 | 4–18 | ND |
| TMP28 9 | BIM B-1544 | ON248086 | Carnobacterium iners LMG 26642 | 99.86 | 18 | 4–18 | ND |
| TMP29 8 | BIM B-1577 | ON248087 | Facklamia tabacinasalis CCUG 30090 | 99.46 | 18 | 10–30 | ND |
| Actinobacteria | |||||||
| TMP15 5 | BIM B-1549 | ON248073 | Arthrobacter ERGS4:06 | 98.97 | 5 | 4–25 | C |
| Arthrobacter PAMC25486 | 98.90 | ||||||
| Arthrobacter alpinus DSM 22274 | 98.70 | ||||||
| Arthrobacter glacialis HLT2-12-2 | 98.70 | ||||||
| TMP24 9 | BIM B-1543 | ON248082 | Arthrobacter agilis DSM 20550 | 100 | 18 | 4–25 | L; C |
| TMP16 6 | BIM B-1571 | ON248074 | Brachybacterium paraconglomeratum LMG19861 | 99.93 | 5 | 10–37 | Cas; A; U; BG; C |
| TMP21 7 | BIM B-1545 | ON248079 | Micrococcus luteus NCTC 2665 | 99.58 | 5 | 10–37 | L; Cas; Gel; A; C |
| TMP23 9 | BIM B-1547 | ON248081 | Agrococcus citreus IAM 15145 | 99.50 | 18 | 18–25 | C |
| TMP30 8 | BIM B-1567 | ON248088 | Leifsonia PHSC20C1 | 99.59 | 18 | 4–25 | C |
| Leifsonia rubra CMS 76R | 99.45 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akulava, V.; Miamin, U.; Akhremchuk, K.; Valentovich, L.; Dolgikh, A.; Shapaval, V. Isolation, Physiological Characterization, and Antibiotic Susceptibility Testing of Fast-Growing Bacteria from the Sea-Affected Temporary Meltwater Ponds in the Thala Hills Oasis (Enderby Land, East Antarctica). Biology 2022, 11, 1143. https://doi.org/10.3390/biology11081143
Akulava V, Miamin U, Akhremchuk K, Valentovich L, Dolgikh A, Shapaval V. Isolation, Physiological Characterization, and Antibiotic Susceptibility Testing of Fast-Growing Bacteria from the Sea-Affected Temporary Meltwater Ponds in the Thala Hills Oasis (Enderby Land, East Antarctica). Biology. 2022; 11(8):1143. https://doi.org/10.3390/biology11081143
Chicago/Turabian StyleAkulava, Volha, Uladzislau Miamin, Katsiaryna Akhremchuk, Leonid Valentovich, Andrey Dolgikh, and Volha Shapaval. 2022. "Isolation, Physiological Characterization, and Antibiotic Susceptibility Testing of Fast-Growing Bacteria from the Sea-Affected Temporary Meltwater Ponds in the Thala Hills Oasis (Enderby Land, East Antarctica)" Biology 11, no. 8: 1143. https://doi.org/10.3390/biology11081143
APA StyleAkulava, V., Miamin, U., Akhremchuk, K., Valentovich, L., Dolgikh, A., & Shapaval, V. (2022). Isolation, Physiological Characterization, and Antibiotic Susceptibility Testing of Fast-Growing Bacteria from the Sea-Affected Temporary Meltwater Ponds in the Thala Hills Oasis (Enderby Land, East Antarctica). Biology, 11(8), 1143. https://doi.org/10.3390/biology11081143