Ascorbate-Glutathione Cycle Genes Families in Euphorbiaceae: Characterization and Evolutionary Analysis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Retrieval of APX, MDAR, DHAR, and GR Proteins
2.2. Phylogenetic Analysis
2.3. Chromosomal Positions and Synthenic Analysis
2.4. Calculation of Ka/Ks and Divergence Time
2.5. Structural Analysis of APX, MDAR, DHAR, and GR Proteins
2.6. Prediction of Potential Cis-Regulatory Elements
2.7. RNAseq and Expression Pattern Analysis
2.8. Plants, Growth Conditions, and Drought Stress Experimental Design
2.9. Total RNA Isolation and Quantitative PCR (RT-qPCR) Analysis
2.10. RT-PCR
2.11. miRNA Target Prediction in Castor Bean APX, MDAR, DHAR, and GR Genes
3. Results and Discussion
3.1. Identification and Phylogenetic Analysis of APX, MDAR, DHAR, and GR Genes
3.2. Structural Organization of APX, MDAR, DHAR, and GR Genes
3.3. Protein Sequence Analyses of APX, MDAR, DHAR, and GR
3.4. Analysis of Alternatively Spliced mRNA Variants of Castor Bean chl/mitAPX
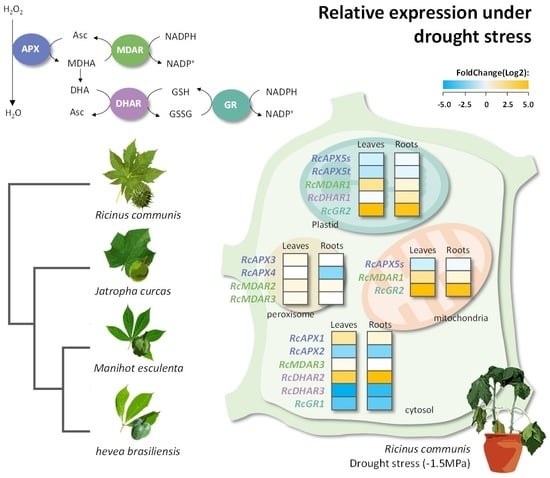
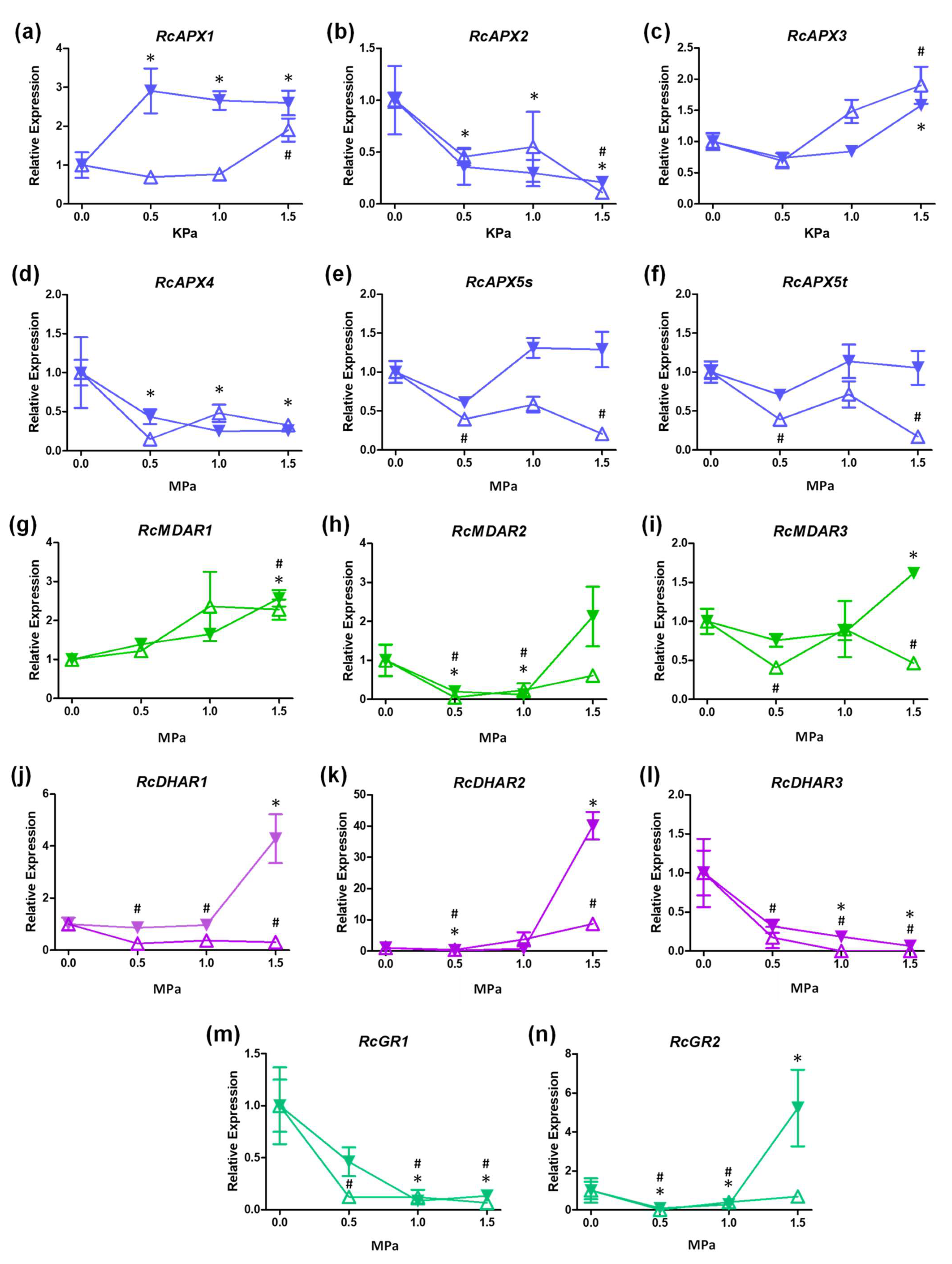
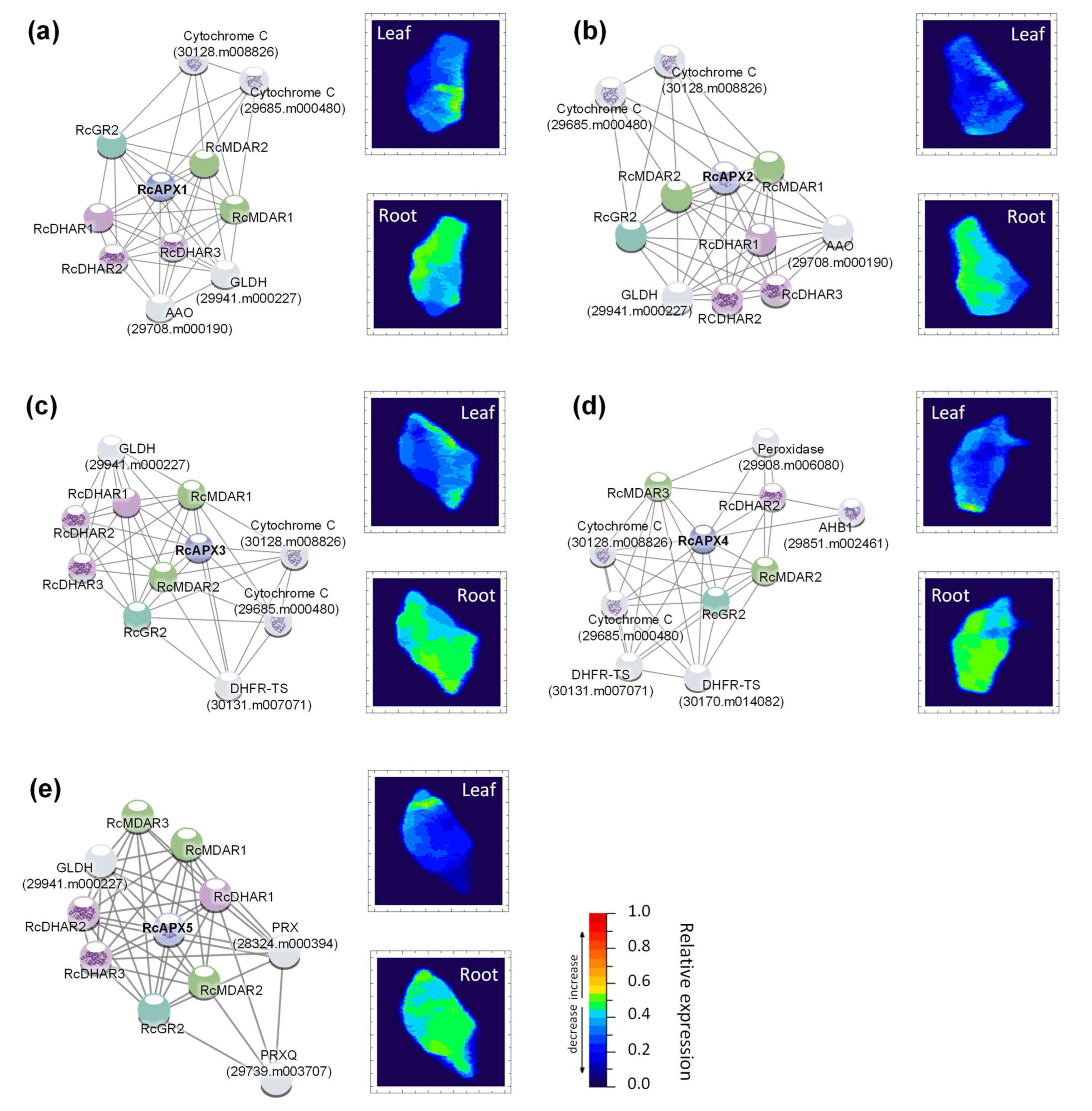
3.5. Transcriptional Profiles of Castor Bean APX, MDAR, DHAR, and GR Genes
3.6. Analysis of Putative Cis-Regulatory Elements and Scanning for miRNA Targeting in APX, MDAR, DHAR, and GR Genes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fridovich, I. Superoxide dismutase. Annu. Rev. Biochem. 1975, 44, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Scandalios, J.G. The rise of ROS. Trends Biochem. Sci. 2002, 27, 483–486. [Google Scholar] [CrossRef] [PubMed]
- Mullineaux, P.; Karpinski, S. Signal transduction in response to excess light: Getting out of the chloroplast. Curr. Opin. Plant Biol. 2002, 5, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef] [PubMed]
- Chovanová, K.; Böhmer, M.; Poljovka, A.; Budiš, J.; Harichová, J.; Szemeš, T.; Zámocký, M. Parallel Molecular Evolution of Catalases and Superoxide Dismutases—Focus on Thermophilic Fungal Genomes. Antioxidants 2020, 9, 1047. [Google Scholar] [CrossRef] [PubMed]
- Takeda, T.; Yoshimura, K.; Ishikawa, T.; Shigeoka, S. Purification and characterization of ascorbate peroxidase in Chlorella vulgaris. Biochimie 1998, 80, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Sano, S.; Ueda, M.; Kitajima, S.; Takeda, T.; Shigeoka, S.; Kurano, N.; Miyachi, S.; Miyake, C.; Yokota, A. Characterization of Ascorbate Peroxidases from Unicellular Red Alga Galdieria partita. Plant Cell Physiol. 2001, 42, 433–440. [Google Scholar] [CrossRef] [Green Version]
- Shigeoka, S.; Nakano, Y.; Kitaoka, S. Metabolism of hydrogen peroxide in Euglena gracilis Z by L-ascorbic acid peroxidase. Biochem. J. 1980, 186, 377–380. [Google Scholar] [CrossRef] [Green Version]
- Noctor, G.; Foyer, C.H. Ascorbate glutathione: Keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 249–279. [Google Scholar] [CrossRef]
- Negi, Y.K. Comparative in silico analysis of ascorbate peroxidase protein sequences from different plant species. J. Bioengg. Biomed. Sci. 2011, 1, 103. [Google Scholar]
- Miyake, C.; Asada, K. Inactivation mechanism of ascorbate peroxidase at low concentrations of ascorbate; hydrogen peroxide decomposes compound I of ascorbate peroxidase. Plant Cell Physiol. 1996, 37, 423–430. [Google Scholar] [CrossRef]
- Tanaka, M.; Takahashi, R.; Hamada, A.; Terai, Y.; Ogawa, T.; Sawa, Y.; Ishikawa, T.; Maruta, T. Distribution and functions of monodehydroascorbate reductases in plants: Comprehensive reverse genetic analysis of Arabidopsis thaliana enzymes. Antioxidants 2021, 10, 1726. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Han, X.M.; Ren, L.L.; Yang, H.L.; Zeng, Q.Y. Functional divergence of the glutathione S-transferase supergene family in Physcomitrella patens reveals complex patterns of large gene family evolution in land plants. Plant Physiol. 2012, 161, 773–786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.J.; Wang, W.; Yang, H.L.; Li, Y.; Kang, X.Y.; Wang, X.R.; Yang, Z.L. Molecular properties and functional divergence of the Dehydroascorbate reductase gene family in lower and higher plants. PLoS ONE 2015, 10, e0145038. [Google Scholar] [CrossRef] [Green Version]
- Rouhier, N.; Couturier, J.; Jacquot, J.P. Genome-wide analysis of plant glutaredoxin systems. J. Exp. Bot. 2006, 57, 1685–1696. [Google Scholar] [CrossRef] [Green Version]
- Ondarza, R.N.; Rendón, J.L.; Ondarza, M. Glutathione reductase in evolution. J. Mol. Evol. 1983, 19, 371–375. [Google Scholar] [CrossRef]
- Jardim-Messeder, D.; Zámocký, M.; Sachetto-Martins, G.; Margis-Pinheiro, M. Chloroplastic ascorbate peroxidases targeted to stroma or thylakoid membrane: The chicken or egg dilemma. FEBS Lett. 2022, 596, 2989–3004. [Google Scholar] [CrossRef]
- Lunde, C.; Baumann, U.; Shirley, N.J.; Drew, D.P.; Fincher, G.B. Gene structure and expression pattern analysis of three monodehydroascorbate reductase (Mdhar) genes in Physcomitrella patens: Implications for the evolution of the MDHAR family in plants. Plant Mol. Biol. 2006, 60, 259–275. [Google Scholar] [CrossRef] [Green Version]
- Bartoli, C.G.; Buet, A.; Grozeff, G.G.; Galatro, A.; Simontacchi, M. Ascorbate-Glutathione Cycle and Abiotic Stress Tolerance in Plants. In Ascorbic Acid in Plant Growth, Development and Stress Tolerance; Hossain, M., Munné-Bosch, S., Burritt, D., Diaz-Vivancos, P., Fujita, M., Lorence, A., Eds.; Springer: Cham, Switzerland, 2017; pp. 177–200. [Google Scholar]
- Kuźniak, E.; Kopczewski, T.; Chojak-Koźniewska, J. Ascorbate-Glutathione Cycle and Biotic Stress Tolerance in Plants. In Ascorbic Acid in Plant Growth, Development and Stress Tolerance; Hossain, M., Munné-Bosch, S., Burritt, D., Diaz-Vivancos, P., Fujita, M., Lorence, A., Eds.; Springer: Cham, Switzerland, 2017; pp. 201–231. [Google Scholar]
- Anjani, K. Castor genetic resources: A primary gene pool for exploitation. Ind. Crop. Prod. 2012, 35, 1–14. [Google Scholar] [CrossRef]
- Nikneshan, P.; Tadayyon, A.; Javanmard, M. Evaluating drought tolerance of castor ecotypes in the center of Iran. Heliyon 2019, 5, e01403. [Google Scholar] [CrossRef] [Green Version]
- Shrirame, H.Y.; Panwar, N.L.; Bamniya, B.R. Bio diesel from castor oil—A green energy option. Low Carbon Econ. 2011, 2, 1–6. [Google Scholar] [CrossRef]
- Ogunniyi, D.S. Castor oil: A vital industrial raw material. Bioresour. Technol. 2006, 97, 1086–1091. [Google Scholar] [CrossRef] [PubMed]
- Metzger, J.O.; Bornscheuer, U. Lipids as renewable resources: Current state of chemical and biotechnological conversion and diversification. Appl. Microbiol. Biotechnol. 2006, 71, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, B.S.; Cossar, R.D. Castor yield in response to planting date at four locations in the south-central United States. Ind. Crops Prod. 2009, 29, 316–319. [Google Scholar] [CrossRef]
- Karyotou, K.; Donaldson, R.P. Ascorbate peroxidase, a scavenger of hydrogen peroxide in glyoxysomal membranes. Arch. Biochem. Biophys. 2005, 434, 248–257. [Google Scholar] [CrossRef]
- Lopes, A.M.; Bobrowski, V.L.; Silva, S.D.D.A.E.; Deuner, S. Morphophysiological and biochemical alterations in Ricinus communis L. seeds submitted to cobalt 60 gamma radiation. An. Acad. Bras. Cienc. 2017, 89, 1925–1933. [Google Scholar] [CrossRef]
- Lima Neto, M.C.; Cerqueira, J.V.A.; da Cunha, J.R.; Ribeiro, R.V.; Silveira, J.A.G. Cyclic electron flow, NPQ and photorespiration are crucial for the establishment of young plants of Ricinus communis and Jatropha curcas exposed to drought. Plant Biol. 2017, 19, 650–659. [Google Scholar] [CrossRef]
- Chang, L.; Sun, H.; Yang, H.; Wang, X.; Su, Z.; Chen, F.; Wei, W. Over-expression of dehydroascorbate reductase enhances oxidative stress tolerance in tobacco. Electron. J. Biotechnol. 2017, 25, 1–8. [Google Scholar] [CrossRef]
- You, J.; Chan, Z. ROS Regulation During Abiotic Stress Responses in Crop Plants. Front. Plant Sci. 2015, 6, 1092. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Lei, L.; Lai, J.; Zhao, H.; Song, W. Effects of drought stress and water recovery on physiological responses and gene expression in maize seedlings. BMC Plant Biol. 2018, 18, 68. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q. Strategies for developing Green Super Rice. Proc. Natl. Acad. Sci. USA 2007, 104, 16402–16409. [Google Scholar] [CrossRef] [PubMed]
- Eckardt, N.A.; Cominelli, E.; Galbiati, M.; Tonelli, C. The future of science: Food and water for life. Plant Cell 2009, 21, 368–372. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2018, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0; A Toolkit Incorporating Gamma-Series Methods and Sliding Window Strategies. Genomis Proteomics Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef] [Green Version]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [Green Version]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The proteomics protocols handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA; pp. 571–607.
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–2012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chow, C.N.; Lee, T.Y.; Hung, Y.C.; Li, G.Z.; Tseng, K.C.; Liu, Y.H.; Kuo, P.L.; Zheng, H.Q.; Chang, W.C. PlantPAN3.0: A new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucleic Acids Res. 2019, 47, D1155–D1163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, A.P.; Kroon, J.T.M.; Swarbreck, D.; Febrer, M.; Larson, T.R.; Graham, I.A.; Caccamo, M.; Slabas, A.R. Tissue-Specific Whole Transcriptome Sequencing in Castor, Directed at Understanding Triacylglycerol Lipid Biosynthetic Pathways. PLoS ONE 2012, 7, e30100. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2018, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Castro, M.A.; Filho, J.L.; Dalmolin, R.J.; Sinigaglia, M.; Moreira, J.C.; Mombach, J.C.; de Almeida, R.M. ViaComplex: Software for landscape analysis of gene expression networks in genomic context. Bioinformatics 2019, 25, 1468–1469. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using realtime quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Li, J.; Feng, J.; Liu, B.; Feng, L.; Yu, X.; Li, G.; Zhai, J.; Meyers, B.C.; Xia, R. sRNAanno—A database repository of uniformly annotated small RNAs in plants. Hortic. Res. 2021, 8, 45. [Google Scholar] [CrossRef]
- Teixeira, F.K.; Menezes-Benavente, L.; Galvão, V.C.; Margis, R.; Margis-Pinheiro, M. Rice ascorbate peroxidase gene family encodes functionally diverse isoforms localized in different subcellular compartments. Planta 2006, 224, 300–314. [Google Scholar] [CrossRef] [PubMed]
- Morgante, C.V.; Rodrigues, R.A.; Marbach, P.A.; Borgonovi, C.M.; Moura, D.S.; Silva-Filho, M.C. Conservation of dual-targeted proteins in Arabidopsis and rice points to a similar pattern of gene-family evolution. Mol. Genet. Genomics. 2009, 281, 525–538. [Google Scholar] [CrossRef] [PubMed]
- Chew, O.; Whelan, J.; Millard, H. Molecular definition of the Ascorbate-Glutathione Cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J. Biol. Chem. 2003, 278, 46869–46877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, L.; Carrie, C.; Law, S.R.; Murcha, M.W.; Whelan, J. Acquisition, Conservation, and Loss of Dual-Targeted Proteins in Land Plants. Plant Physiol. 2013, 161, 644–662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leterrier, M.; Corpas, F.J.; Barroso, J.B.; Sandalio, L.M.; del Rio, L.A. Peroxisomal monodehydroascorbate reductase. Genomic clone characterization and functional analysis under environmental stress conditions. Plant Physiol. 2005, 138, 2111–2123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lisenbee, C.S.; Lingard, M.J.; Trelease, R.N. Arabidopsis peroxisomes possess functionally redundant membrane and matrix isoforms of monodehydroascorbate reductase. Plant J. 2005, 43, 900–914. [Google Scholar] [CrossRef]
- Lazzarotto, F.; Menguer, P.K.; Del-Bem, L.E.; Zámocký, M.; Margis-Pinheiro, M. Ascorbate Peroxidase Neofunctionalization at the Origin of APX-R and APX-L: Evidence from Basal Archaeplastida. Antioxidants 2021, 10, 597. [Google Scholar] [CrossRef]
- Shikanai, T.; Takeda, T.; Yamauchi, Y.; Sano, S.; Tomizawa, K.; Yokota, A.; Shigeoka, S. Inhibition of ascorbate peroxidase under oxidative stress in tobacco having bacterial catalase in chloroplasts. FEBS Lett. 1998, 428, 47–51. [Google Scholar] [CrossRef] [Green Version]
- Patterson, W.R.; Poulos, T.L. Crystal structure of recombinant pea cytosolic ascorbate peroxidase. Biochemistry 1995, 34, 4331–4341. [Google Scholar] [CrossRef]
- Sharp, K.H.; Mewies, M.; Moody, P.C.; Raven, E.L. Crystal structure of the ascorbate peroxidase-ascorbate complex. Nat. Struct. Biol. 2003, 10, 303–307. [Google Scholar] [CrossRef]
- Kim, J.J.; Kim, Y.S.; Park, S.I.; Mok, J.E.; Kim, Y.H.; Park, H.M.; Kim, I.S.; Yoon, H.S. Cytosolic monodehydroascorbate reductase gene affects stress adaptation and grain yield under paddy field conditions in Oryza sativa L. japonica. Mol. Breed. 2017, 37, 118. [Google Scholar] [CrossRef]
- Tahmasebi, A.; Aram, F.; Ebrahimi, M.; Mohammadi-Dehcheshmeh, M.; Ebrahimie, E. Genome-wide analysis of cytosolic and chloroplastic isoforms of glutathione reductase in plant cells. Plant Omics 2012, 5, 94–102. [Google Scholar]
- Qiu, Y.; Tay, Y.V.; Ruan, Y.; Adams, K.L. Divergence of duplicated genes by repeated partitioning of splice forms and subcellular localization. New Phytol. 2020, 225, 1011–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, M.; Suzuki, K.; Tanabe, N.; Suzuki, T.; Nishizawa-Yokoi, A.; Shigeoka, S.; Yoshimura, K. An oligonucleotide/oligosaccharide-binding-fold protein enhances the alternative splicing event producing thylakoid membrane-bound ascorbate peroxidase in Nicotiana tabacum. G3 2022, 25, 12. [Google Scholar] [CrossRef] [PubMed]
- Gerhardt, B.P.; Beevers, H. Developmental studies on glyoxysomes in Ricinus endosperm. J. Cell Biol. 1970, 44, 94–102. [Google Scholar] [CrossRef]
- Graham, I.A.; Eastmond, P.J. Pathways of straight and branched chain fatty acid catabolism in higher plants. Prog. Lipid Res. 2002, 41, 156–181. [Google Scholar] [CrossRef]
- Gallie, D.R. The role of L-ascorbic acid recycling in responding to environmental stress and in promoting plant growth. J. Exp. Bot. 2013, 64, 433–443. [Google Scholar] [CrossRef] [Green Version]
- Bailly, C.; Audigier, C.; Ladonne, F.; Wagner, M.H.; Coste, F.; Corbineau, F.; Côme, D. Changes in oligosaccharide content and antioxidant enzyme activities in developing bean seeds as related to acquisition of drying tolerance and seed quality. J. Exp. Bot. 2001, 52, 701–708. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Wang, J.; Tissue, D.; Holaday, A.S.; Allen, R.; Zhang, H. Photosynthesis and seed production under water-deficit conditions in transgenic tobacco plants that overexpress an Arabidopsis ascorbate peroxidase gene. Crop Sci. 2003, 43, 1477–1483. [Google Scholar] [CrossRef]
- Sun, W.H.; Duan, M.; Shu, D.F.; Yang, S.; Meng, Q.W. Over-expression of StAPX in tobacco improves seed germination and increases early seedling tolerance to salinity and osmotic stresses. Plant Cell Rep. 2010, 29, 917–926. [Google Scholar] [CrossRef]
- Singh, N.; Mishra, A.; Jha, B. Over-expression of the peroxisomal ascorbate peroxidase (SbpAPX) gene cloned from halophyte Salicornia brachiata confers salt and drought stress tolerance in transgenic tobacco. Mar. Biotechnol. 2014, 16, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Jardim-Messeder, D.; Caverzan, A.; Rauber, R.; Cunha, J.; Carvalho, F.; Letaif-Gaeta, M.; da Fonseca, G.C.; Costa, M.; Frei, M.; Silveira, J.A.G.; et al. Thylakoidal APX modulates hydrogen peroxide content and stomatal closure in rice (Oryza sativa L.). Environ. Exp. Bot. 2018, 150, 46–56. [Google Scholar] [CrossRef]
- Eastmond, P.J. MONODEHYROASCORBATE REDUCTASE4 is required for seed storage oil hydrolysis and postgerminative growth in Arabidopsis. Plant Cell 2007, 19, 1376–1387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sumugat, M.R.; Donahue, J.L.; Cortes, D.F.; Stromberg, V.K.; Grene, R.; Shulaev, V.; Welbaum, G.E. Seed Development and Germination in an Arabidopsis thaliana Line Antisense to Glutathione Reductase 2. J. New Seeds 2010, 11, 104–126. [Google Scholar] [CrossRef]
- Silva, M.; Willadino, L.; Camara, T.; Oliveira, A. Response of Ricinus communis L. to in vitro water stress induced by polyethylene glycol. Plant Growth Regulation. 2015, 78, 195–204. [Google Scholar] [CrossRef]
- Janmohammadi, M.; Abbasi, A.; Sabaghnia, N. Influence of NaCl treatments on growth and biochemical parameters of castor bean (Ricinus communis L.). Acta Agric. Slov. 2012, 99, 31–40. [Google Scholar]
- Karuppanapandian, T.; Moon, J.C.; Kim, C.; Manoharan, K.; Kim, W. Reactive oxygen species in plants: Their generation, signal transduction, and scavenging mechanisms. Aust. J. Crop Sci. 2011, 5, 709–725. [Google Scholar]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 2012, 217037. [Google Scholar] [CrossRef] [Green Version]
- Guan, L.; Zhao, J.; Scandalios, J.G. Cis-elements and transfactors that regulate expression of the maize Cat1 antioxidant gene in response to ABA and osmotic stress: H2O2 is the likely intermediary signalling molecule for the response. Plant J. 2000, 22, 87–95. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, J. Effect of abscisic acid on active oxygen species, antioxidative defense system and oxidative damage in leaves of maize seedlings. Plant Cell Physiol. 2001, 42, 1265–1273. [Google Scholar] [CrossRef] [Green Version]
- Anderson, M.D.; Prasad, T.K.; Martin, B.A.; Stewart, C.R. Differential gene expression in chilling-acclimated maize seedlings and evidence for the involvement of abscisic acid in chilling tolerance. Plant Physiol. 1994, 105, 331–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prasad, T.K.; Anderson, M.D.; Stewart, C.R. Acclimation, hydrogen peroxide, and abscisic acid protect mitochondria against irreversible chilling injury in maize seedlings. Plant Physiol. 1994, 105, 619–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bueno, P.; Piqueras, A.; Kurepa, J.; Savoure, A.; Verbruggen, N.; Van Montagu, M.; Inze, D. Expression of antioxidant enzymes in response to abscisic acid and high osmoticum in tobacco BY-2 cell cultures. Plant Sci. 1998, 138, 27–34. [Google Scholar] [CrossRef]
- Agrawal, G.K.; Jwa, N.S.; Iwahashi, H.; Rakwal, R. Importance of ascorbate peroxidases OsAPX1 and OsAPX2 in the rice pathogen response pathways and growth and reproduction revealed by their transcriptional profiling. Gene 2003, 322, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Zilinskas, B.A. Molecular cloning and characterization of gene encoding pea cytosolic ascorbate peroxidase. J. Biol. Chem. 1992, 267, 21802–21807. [Google Scholar] [CrossRef]
- Park, S.Y.; Ryu, S.H.; Jang, L.C.; Kwon, S.Y.; Kim, J.G.; Kwak, S.S. Molecular cloning of a cytosolic ascorbate peroxidase cDNA from cell cultures of sweet potato and its expression in response to stress. Mol. Genet. Genom. 2004, 271, 339–346. [Google Scholar] [CrossRef]
- Soares, A.M.D.S.; Souza, T.F.D.; Jacinto, T.; Machado, O.L.T. Effect of methyl jasmonate on antioxidative enzyme activities and on the contents of ROS and H2O2 in Ricinus communis leaves. Braz. J. Plant Physiol. 2010, 33, 151–158. [Google Scholar] [CrossRef] [Green Version]
- Jardim-Messeder, D.; Margis-Pinheiro, M.; Sachetto-Martins, G. Salicylic acid and adenine nucleotides regulate the electron transport system and ROS production in plant mitochondria. Biochim. Biophys. Acta Bioenerg. 2022, 1863, 148559. [Google Scholar] [CrossRef]
- Liu, J.; Qiu, G.; Liu, C.; Li, H.; Chen, X.; Fu, Q.; Lin, Y.; Guo, B. Salicylic Acid, a Multifaceted Hormone, Combats Abiotic Stresses in Plants. Life 2022, 12, 886. [Google Scholar] [CrossRef]
- Hossain, M.A.; Munemasa, S.; Uraji, M.; Nakamura, Y.; Mori, I.C.; Murata, Y. Involvement of Endogenous Abscisic Acid in Methyl Jasmonate-Induced Stomatal Closure in Arabidopsis. Plant Physiol. 2011, 156, 430–438. [Google Scholar] [CrossRef] [Green Version]
- Prodhan, M.Y.; Munemasa, S.; Nahar, M.N.; Nakamura, Y.; Murata, Y. Guard Cell Salicylic Acid Signaling Is Integrated into Abscisic Acid Signaling via the Ca2+/CPK-Dependent Pathway. Plant Physiol. 2018, 178, 441–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, G.; Poethig, R.S. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 2006, 133, 3539–3547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.W.; Weigel, D.; Poethig, R.S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, J.; Yang, X.; Song, Y.; Du, Q.; Li, Y.; Chen, J.; Zhang, D. Adaptive evolution and functional innovation of Populus-specific recently evolved microRNAs. New Phytol. 2017, 213, 206–219. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Chen, J.; Zhou, J.; Yu, H.; Ge, C.; Zhang, M.; Gao, X.; Dai, X.; Yang, Z.N.; Zhao, Y. An Essential Role for miRNA167 in Maternal Control of Embryonic and Seed Development. Plant Physiol. 2019, 180, 453–464. [Google Scholar] [CrossRef] [Green Version]
- Han, H.; Zhou, Y. Function and Regulation of microRNA171 in Plant Stem Cell Homeostasis and Developmental Programing. Int. J. Mol. Sci. 2022, 23, 2544. [Google Scholar] [CrossRef]
- Montgomery, T.A.; Howell, M.D.; Cuperus, J.T.; Li, D.; Hansen, J.E.; Alexander, A.L.; Chapman, E.J.; Fahlgren, N.; Allen, E.; Carrington, J.C. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 2008, 133, 128–141. [Google Scholar] [CrossRef] [Green Version]
- Wyrzykowska, A.; Bielewicz, D.; Plewka, P.; Sołtys-Kalina, D.; Wasilewicz-Flis, I.; Marczewski, W.; Jarmolowski, A.; Szweykowska-Kulinska, Z. The MYB33, MYB65, and MYB101 transcription factors affect Arabidopsis and potato responses to drought by regulating the ABA signaling pathway. Physiol. Plant 2022, 174, e13775. [Google Scholar] [CrossRef]
- Pieczynski, M.; Marczewski, W.; Hennig, J.; Dolata, J.; Bielewicz, D.; Piontek, P.; Wyrzykowska, A.; Krusiewicz, D.; Strzelczyk-Zyta, D.; Konopka-Postupolska, D.; et al. Down-regulation of CBP80 gene expression as a strategy to engineer a drought-tolerant potato. Plant Biotechnol. J. 2013, 11, 459–469. [Google Scholar] [CrossRef]

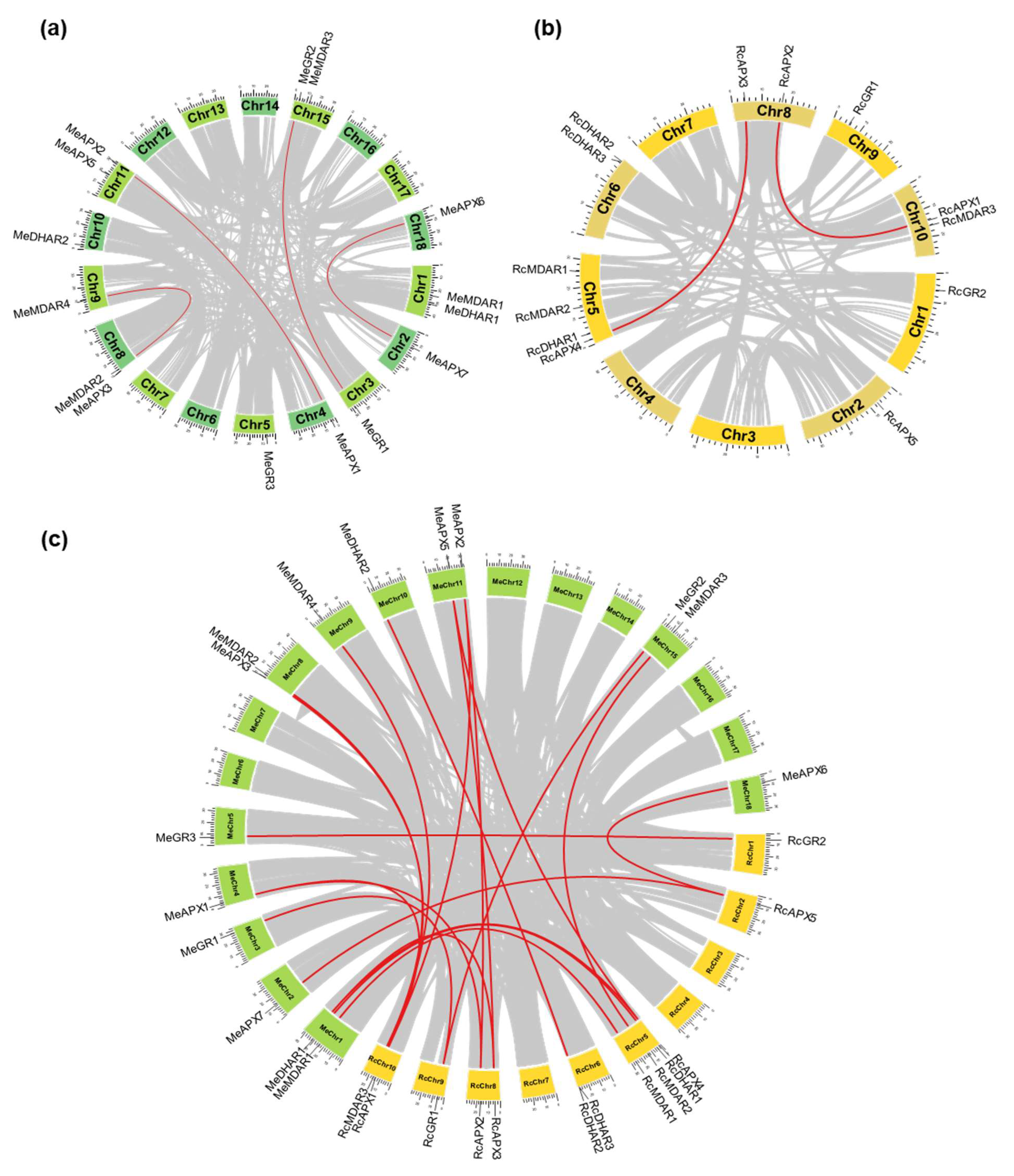

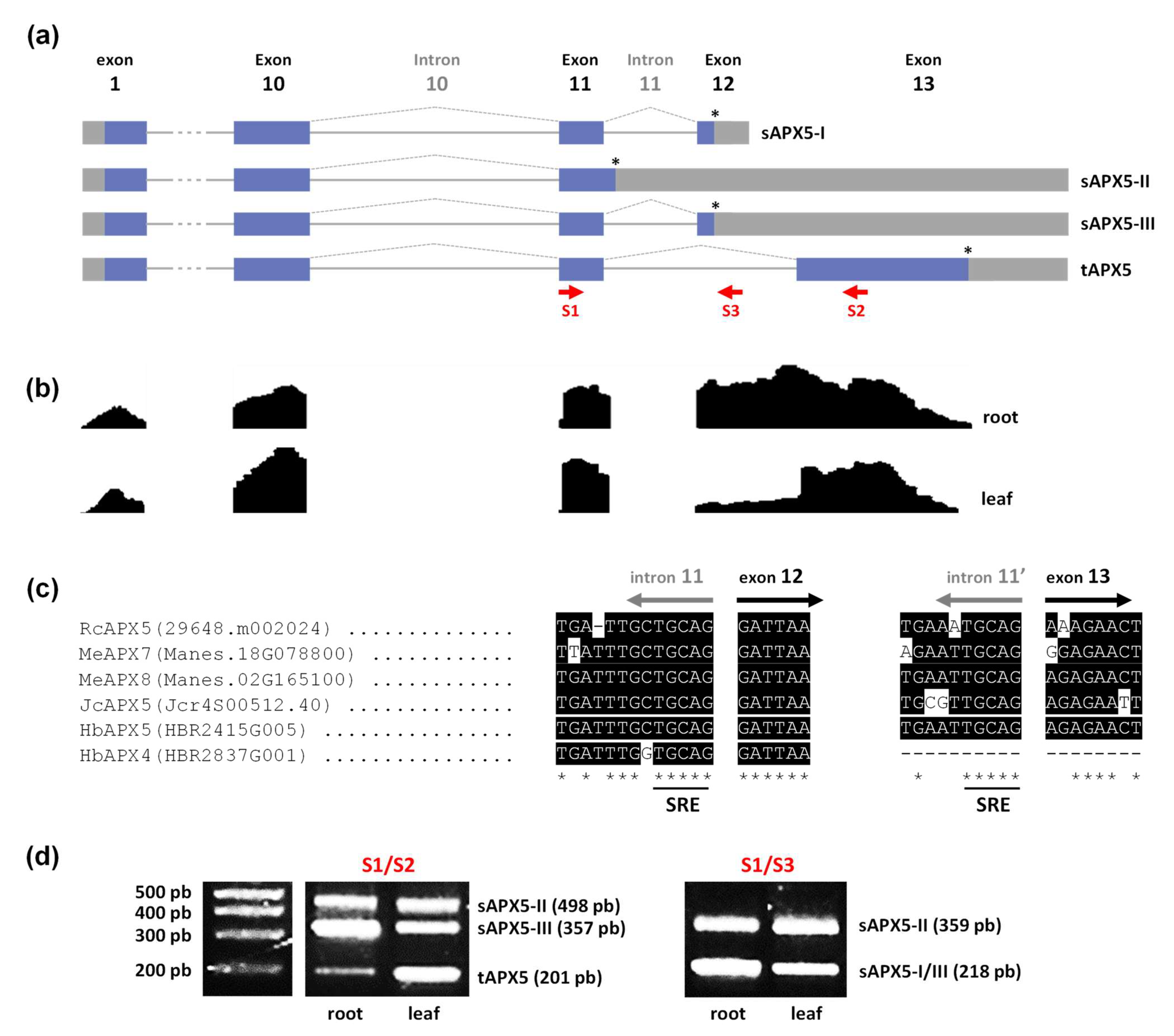
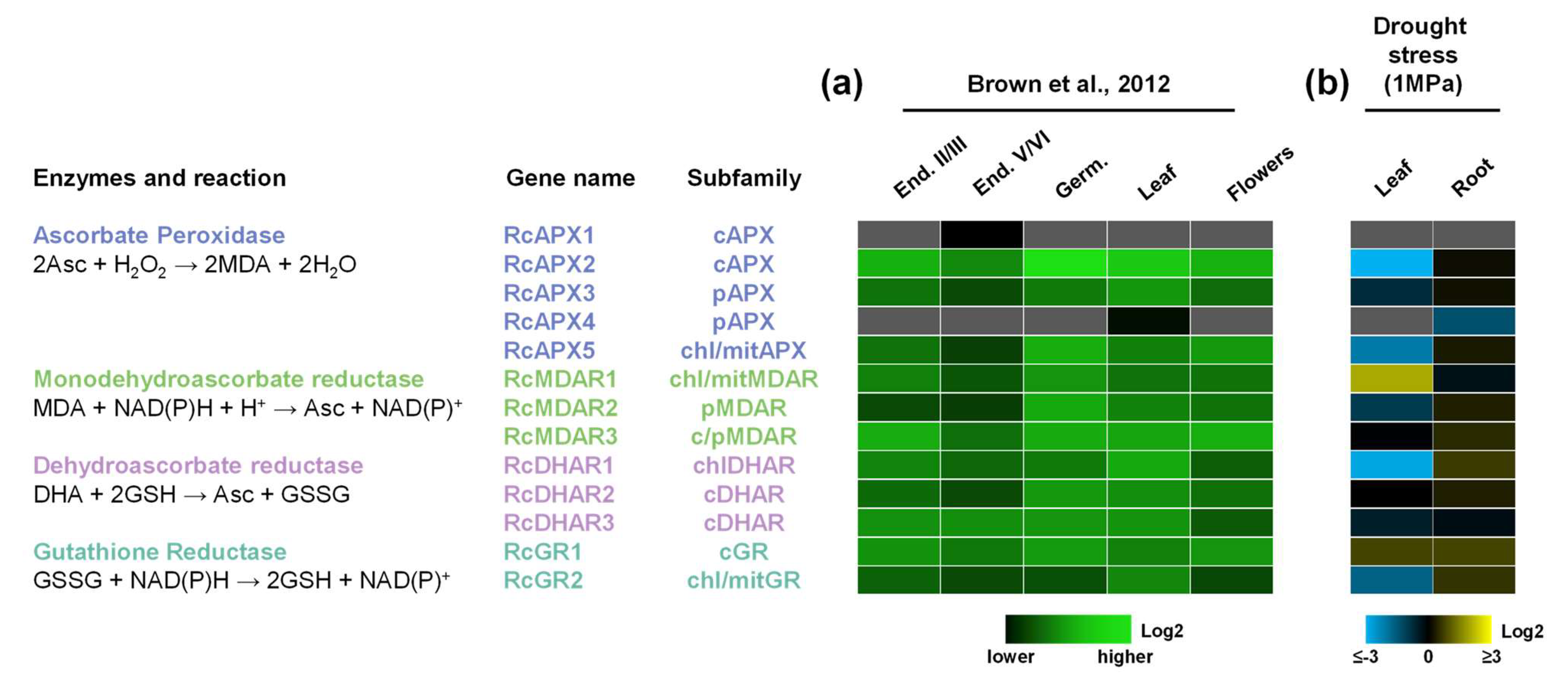
| Arabidopsis thaliana | Oryza sativa | Ricinus communis | Manihot esculenta | Jatropha curcas | Hevea brasiliensis | |
|---|---|---|---|---|---|---|
| APX | 6 | 8 | 5 | 7 | 5 | 5 |
| MDAR | 5 | 5 | 3 | 4 | 4 | 6 |
| DHAR | 4 | 2 | 3 | 2 | 2 | 3 |
| GR | 2 | 3 | 2 | 3 | 2 | 3 |
| Family | Group | Gene 1 | Gene 2 | Type | Ka | Ks | Ka/Ks | Date (MYA) |
|---|---|---|---|---|---|---|---|---|
| APX | I | RcAPX1 | RcAPX2 | Segmental | 0.3121 | 3.8168 | 0.0818 | 235.6 |
| MeAPX1 | MeAPX2 | Segmental | 0.0772 | 0.3850 | 0.2006 | 23.8 | ||
| JcAPX1 | JcAPX2 | Segmental | 0.1645 | 2.5914 | 0.0635 | 160.0 | ||
| II | RcAPX3 | RcAPX4 | Segmental | 0.2033 | 1.1992 | 0.1695 | 74.0 | |
| MeAPX4 | MeAPX5 | Segmental | 0.1685 | 1.5858 | 0.1063 | 97.9 | ||
| JcAPX3 | JcAPX4 | Segmental | 0.1436 | 1.7389 | 0.0826 | 107.3 | ||
| HbAPX2 | HbAPX3 | Segmental | 2.6165 | 1.9921 | 1.3135 | 123.0 | ||
| III | MeAPX6 | MeAPX7 | Segmental | 0.0765 | 0.3804 | 0.2011 | 23.5 | |
| HbAPX4 | HbAPX5 | Segmental | 0.0542 | 0.2556 | 0.2119 | 15.8 | ||
| MDAR | I | JcMDAR1 | JcMDAR4 | Segmental | 0.7604 | 1.3866 | 0.5484 | 85.6 |
| II | HbMDAR2 | HbMDAR3 | Segmental | 0.0388 | 0.2061 | 0.1882 | 12.7 | |
| III | HbMDAR4 | HbMDAR5 | Segmental | 0.0061 | 0.0098 | 0.6199 | 0.6 | |
| DHAR | I | RcDHAR2 | RcDHAR3 | Tandem | 0.0927 | 0.3451 | 0.2685 | 21.3 |
| GR | I | MeGR1 | MeGR2 | Segmental | 0.0934 | 0.3710 | 0.2516 | 22.9 |
| HbGR1 | HbGR2 | Segmental | 0.0330 | 0.2528 | 0.1304 | 15.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jardim-Messeder, D.; de Souza-Vieira, Y.; Lavaquial, L.C.; Cassol, D.; Galhego, V.; Bastos, G.A.; Felix-Cordeiro, T.; Corrêa, R.L.; Zámocký, M.; Margis-Pinheiro, M.; et al. Ascorbate-Glutathione Cycle Genes Families in Euphorbiaceae: Characterization and Evolutionary Analysis. Biology 2023, 12, 19. https://doi.org/10.3390/biology12010019
Jardim-Messeder D, de Souza-Vieira Y, Lavaquial LC, Cassol D, Galhego V, Bastos GA, Felix-Cordeiro T, Corrêa RL, Zámocký M, Margis-Pinheiro M, et al. Ascorbate-Glutathione Cycle Genes Families in Euphorbiaceae: Characterization and Evolutionary Analysis. Biology. 2023; 12(1):19. https://doi.org/10.3390/biology12010019
Chicago/Turabian StyleJardim-Messeder, Douglas, Ygor de Souza-Vieira, Lucas Corrêa Lavaquial, Daniela Cassol, Vanessa Galhego, Gabriel Afonso Bastos, Thais Felix-Cordeiro, Régis Lopes Corrêa, Marcel Zámocký, Márcia Margis-Pinheiro, and et al. 2023. "Ascorbate-Glutathione Cycle Genes Families in Euphorbiaceae: Characterization and Evolutionary Analysis" Biology 12, no. 1: 19. https://doi.org/10.3390/biology12010019