Molecular Phylogeny and Evolution of the Tuerkayana (Decapoda: Brachyura: Gecarcinidae) Genus Based on Whole Mitochondrial Genome Sequences
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples and the Extraction of DNA
2.2. PCR Amplification and Sequencing
2.3. Sequence Analysis and Gene Annotation
2.4. Phylogenetic Analysis and Gene Rearrangement
2.5. Selective Pressure Detection
3. Results
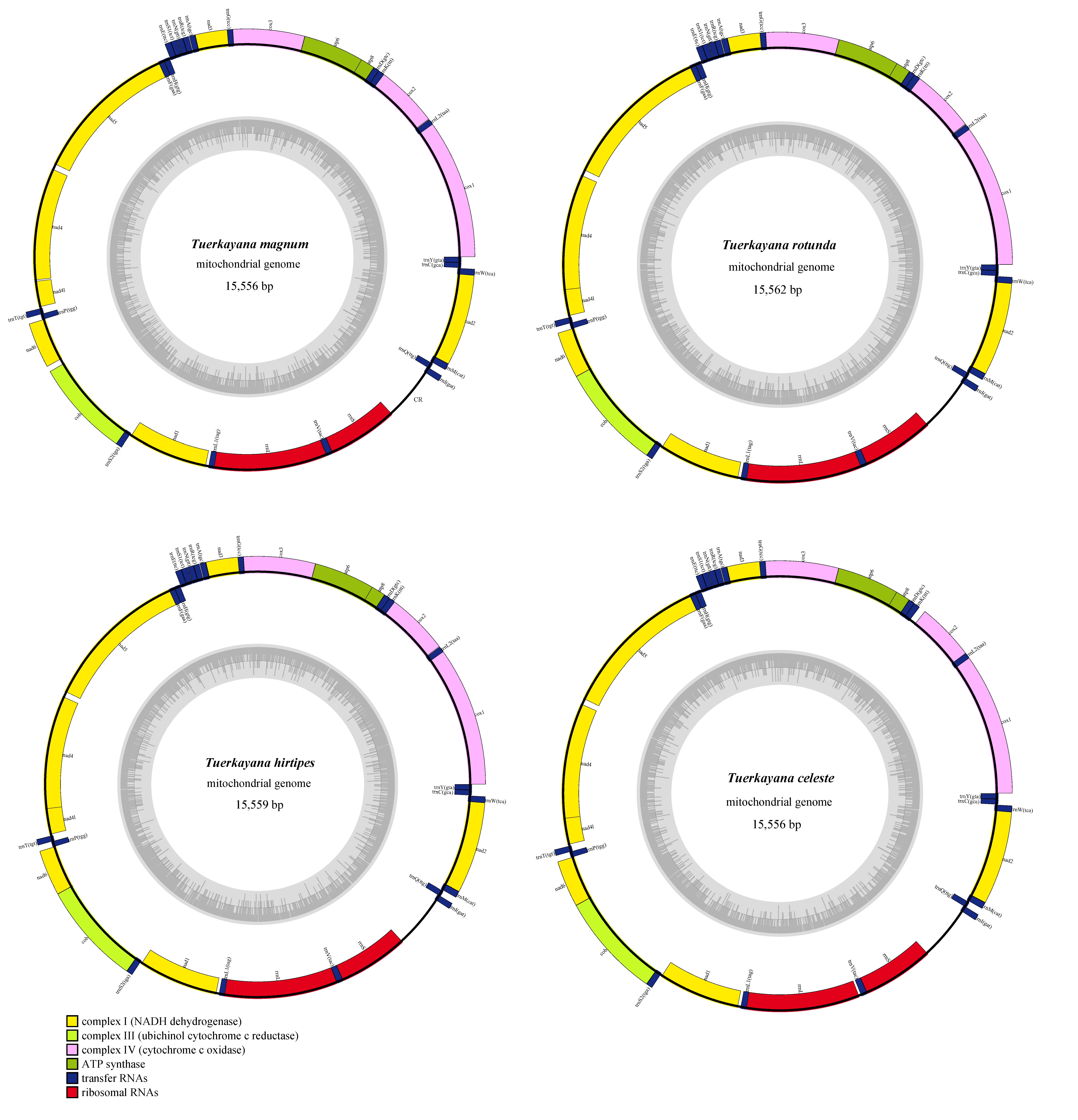
3.1. Mitogenome Organization and Base Composition
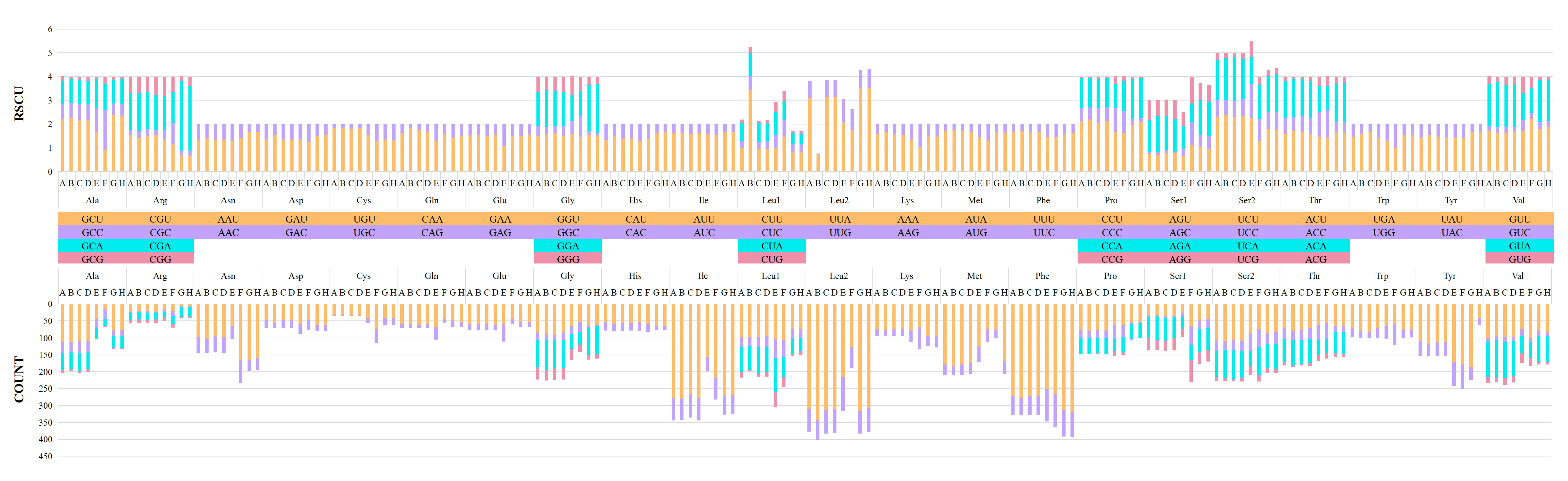
3.2. Protein-Coding Genes and Codon Usage
3.3. Transfer RNAs, Ribosomal RNAs, and CR
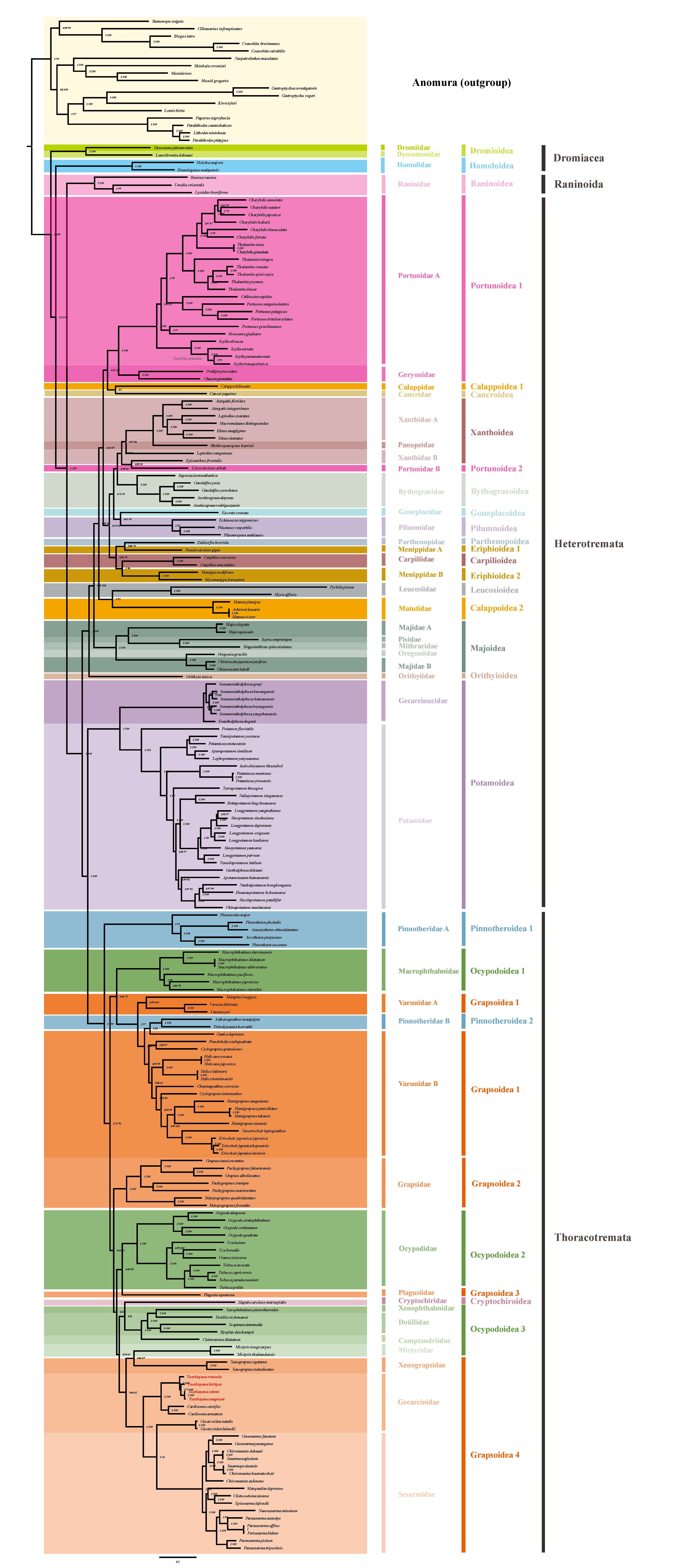
3.4. Phylogenetic Relationships
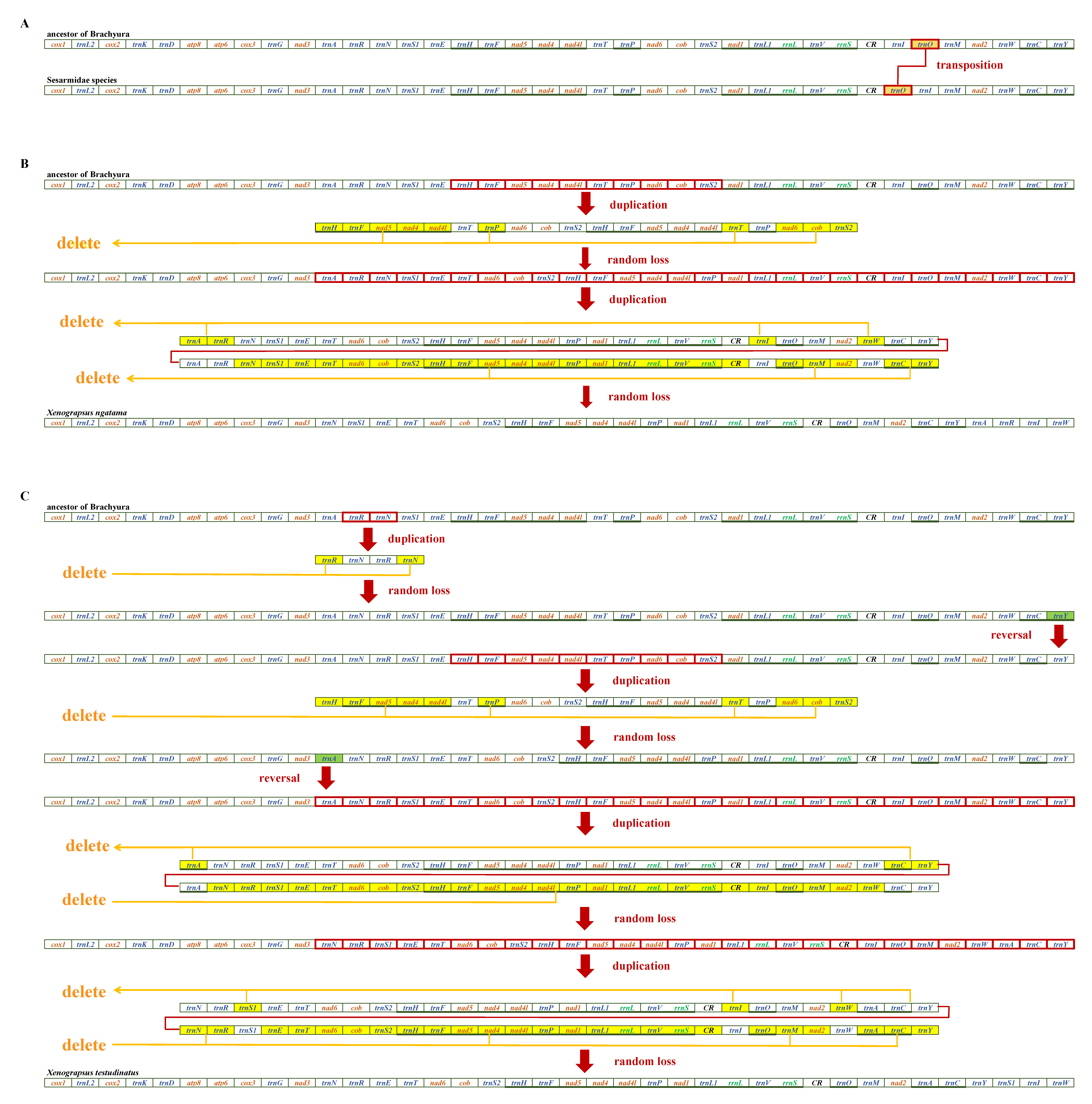
3.5. Mitogenome Gene Rearrangement
3.6. Selective Pressure in Gecarcinidae
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Broughton, R.E.; Milam, J.E.; Roe, B.A. The Complete Sequence of the Zebrafish (Danio rerio) Mitochondrial Genome and Evolutionary Patterns in Vertebrate Mitochondrial DNA. Genome Res. 2001, 11, 1958–1967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ingman, M.; Kaessmann, H.; Pääbo, S.; Gyllensten, U. Mitochondrial genome variation and the origin of modern humans. Nature 2000, 408, 708–713. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Wang, Q.; Wu, Q.; Xu, J.; Wang, J.; Wang, Z. The entire mitochondrial genome of Macrophthalmus abbreviatus reveals insights into the phylogeny and gene rearrangements of Brachyura. Biochem. Genet. 2021, 59, 617–636. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, J.; Wu, Q.; Xu, X.; Wang, P.; Wang, Z. Insights into the evolution of Brachyura (Crustacea: Decapoda) from mitochondrial sequences and gene order rearrangements. Int. J. Biol. Macromol. 2021, 170, 717–727. [Google Scholar] [CrossRef]
- Guinot, D.; Ng, N.K.; Moreno, P.A.R. Review of grapsoid families for the establishment of a new family for Leptograpsodes Montgomery, 1931, and a new genus of Gecarcinidae H. Milne Edwards, 1837 (Crustacea, Decapoda, Brachyura, Grapsoidea MacLeay, 1838). Zoosystema 2018, 40, 547–604. [Google Scholar] [CrossRef]
- Jiménez, C.; Rubio, A.O.; Cárdenas, S.A.; Arnaud, G. Ecological aspects of the land crab Gecarcinus planatus (Decapoda: Gecarcinidae) in Socorro Island, Mexico. Biol. Conserv. 1994, 69, 9–13. [Google Scholar] [CrossRef]
- Green, P.T. Burrow Dynamics of the Red Land Crab Gecarcoidea natalis (Brachyura, Gecarcinidae) in Rain Forest on Christmas Island (Indian Ocean). J. Crustac. Biol. 2004, 24, 340–349. [Google Scholar] [CrossRef] [Green Version]
- Hartnoll, R.G.; Clark, P.F. A mass recruitment event in the land crab Gecarcinus ruricola (Linnaeus, 1758) (Brachyura: Grapsoidea: Gecarcinidae), and a description of the megalop. Zool. J. Linn. Soc. 2010, 146, 149–164. [Google Scholar] [CrossRef] [Green Version]
- Adamczewska, A.M.; Morris, S. Ecology and behavior of Gecarcoidea natalis, the Christmas Island red crab, during the annual breeding migration. Biol. Bull. 2001, 200, 305–320. [Google Scholar] [CrossRef]
- Farrelly, C.A.; Greenaway, P. Land crabs with smooth lungs: Grapsidae, Gecarcinidae, and Sundathelphusidae ultrastructure and vasculature. J. Morphol. 1993, 215, 245–260. [Google Scholar] [CrossRef]
- Cuesta, J.A.; Anger, K. Larval morphology and salinity tolerance of a land crab from West Africa, Cardisoma armatum (Brachyura: Grapsoidea: Gecarcinidae). J. Crustac. Biol. 2005, 25, 640–654. [Google Scholar] [CrossRef] [Green Version]
- Allardyce, B.J.; Linton, S.M. Functional morphology of the gastric mills of carnivorous, omnivorous, and herbivorous land crabs. J. Morphol. 2010, 271, 61–72. [Google Scholar] [CrossRef]
- Mendes, R.A.S.; Silva, J.R.F.; Neto, J.S.; Hazin, F.H.V. Reproductive biology of the land crab Cardisoma guanhumi (Decapoda: Gecarcinidae) in north-eastern Brazil. J. Mar. Biol. Assoc. UK 2013, 93, 761–768. [Google Scholar] [CrossRef]
- Schubart, C.D.; Diesel, R.; Hedges, S.B. Rapid evolution to terrestrial life in Jamaican crabs. Nature 1998, 393, 363–365. [Google Scholar] [CrossRef]
- Watson-Zink, V.M. Making the grade: Physiological adaptations to terrestrial environments in decapod crabs. Arthropod Struct. Dev. 2021, 64, 101089. [Google Scholar] [CrossRef] [PubMed]
- López-Barneo, J.; Pardal, R.; Ortega-Sáenz, P. Cellular mechanism of oxygen sensing. Annu. Rev. Physiol. 2001, 63, 259–287. [Google Scholar] [CrossRef]
- Farrelly, C.A.; Greenaway, P. Morphology and ultrastructure of the gills of terrestrial crabs (Crustacea, Gecarcinidae and Grapsidae): Adaptations for air-breathing. Zoomorphology 1992, 112, 39–49. [Google Scholar] [CrossRef]
- Simon, C.; Buckley, T.R.; Frati, F.; Stewart, J.B.; Beckenbach, A.T. Incorporating Molecular Evolution into Phylogenetic Analysis, and a New Compilation of Conserved Polymerase Chain Reaction Primers for Animal Mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 2006, 37, 545–579. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Sudhir, K.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Zhou, L.; Zhou, X.; Yang, W.; Zhang, J.; Zhang, X.; Wang, Y.; Gui, J. Unusual AT-skew of Sinorhodeus microlepis mitogenome provides new insights into mitogenome features and phylogenetic implications of bitterling fishes. Int. J. Biol. Macromol. 2019, 129, 339–350. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lohse, M.; Drechsel, O.; Bock, R. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007, 52, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Sudhir, K.; Glen, S.; Koichiro, T. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Xia, X. DAMBE6: New Tools for Microbial Genomics, Phylogenetics, and Molecular Evolution. J. Hered. 2017, 108, 431–437. [Google Scholar] [CrossRef] [Green Version]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F.; Teslenko, M.; Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Trifinopoulos, J.; Nguyen, L.; Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W. The Molecular Detection of Corynespora Cassiicola on Cucumber by PCR Assay Using DNAman Software and NCBI. In Proceedings of the International Conference on Computer and Computing Technologies in Agriculture, Beijing, China, 27–30 September 2015; Springer: Cham, Switzerland, 2015; pp. 248–258. [Google Scholar]
- Bernt, M.; Merkle, D.; Ramsch, K.; Fritzsch, G.; Perseke, M.; Bernhard, D.; Schlegel, M.; Stadler, P.F.; Middendorf, M. CREx: Inferring genomic rearrangements based on common intervals. Bioinformatics 2007, 23, 2957–2958. [Google Scholar] [CrossRef] [Green Version]
- Ohta, T. The nearly neutral theory of molecular evolution. Annu. Rev. Ecol. Syst. 1992, 23, 263–286. [Google Scholar] [CrossRef]
- Yang, Z.; Nielsen, R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 2000, 17, 32–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cha, S.Y.; Yoon, H.J.; Lee, E.M.; Yoon, M.H.; Hwang, J.S.; Jin, B.R.; Han, Y.S.; Kim, I. The complete nucleotide sequence and gene organization of the mitochondrial genome of the bumblebee, Bombus ignitus (Hymenoptera: Apidae). Gene 2007, 392, 206–220. [Google Scholar] [CrossRef]
- Reyes, A.; Gissi, C.; Pesole, G.; Saccone, C. Asymmetrical directional mutation pressure in the mitochondrial genome of mammal. Mol. Biol. Evol. 2018, 15, 957–966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef]
- Lavrov, D.V.; Brown, W.M.; Boore, J.L. A novel type of RNA editing occurs in the mitochondrial tRNAs of the centipede Lithobius forficatus. Proc. Natl. Acad. Sci. USA 2000, 97, 13738–13742. [Google Scholar] [CrossRef]
- Kilpert, F.; Podsiadlowski, L. The complete mitochondrial genome of the common sea slater, Ligia oceanica (Crustacea, Isopoda) bears a novel gene order and unusual control region features. BMC Genom. 2006, 7, 241. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Wang, Z.; Tang, D.; Xu, X.; Tao, Y.; Ji, C.; Wang, Z. Characterization and comparison of the mitochondrial genomes from two Alpheidae species and insights into the phylogeny of Caridea. Genomics 2020, 112, 65–70. [Google Scholar] [CrossRef]
- Yuan, M.L.; Zhang, Q.L.; Guo, Z.L.; Wang, J.; Shen, Y.Y. Comparative mitogenomic analysis of the superfamily Pentatomoidea (Insecta: Hemiptera: Heteroptera) and phylogenetic implications. BMC Genom. 2015, 16, 460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taanman, J. The mitochondrial genome: Structure, transcription, translation and replication. Biochim. Biophys. Acta (BBA)-Bioenerg. 1999, 1410, 103–123. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.E.; Jiang, W.; Yuan, Z.; Sha, Z. Mitogenomes Provide Insights Into the Evolution of Thoracotremata (Brachyura: Eubrachyura). Front. Mar. Sci. 2022, 9, 848203. [Google Scholar] [CrossRef]
- Zhang, Y.; Gong, L.; Lu, X.; Miao, Z.; Jiang, L.; Liu, B.; Liu, L.; Li, P.; Zhang, X.; Lü, Z. Comparative mitochondrial genome analysis of Varunidae and its phylogenetic implications. Acta Oceanol. Sin. 2022, 41, 119–131. [Google Scholar] [CrossRef]
- Zhang, Y.; Gong, L.; Lu, X.; Jiang, L.; Liu, B.; Liu, L.; Lü, Z.; Li, P. Gene rearrangements in the mitochondrial genome of Chiromantes eulimene (Brachyura: Sesarmidae) and phylogenetic implications for Brachyura. Int. J. Biol. Macromol. 2020, 162, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gao, Y.; Gong, L.; Lu, X.; Jiang, L.; Liu, B.; Liu, L.; Lü, Z.; Li, P. Mitochondrial Genome of Episesarma lafondii (Brachyura: Sesarmidae) and Comparison with Other Sesarmid Crabs. J. Ocean. Univ. China 2021, 20, 1545–1556. [Google Scholar] [CrossRef]
- Basso, A.; Babbucci, M.; Pauletto, M.; Riginella, E.; Patarnello, T.; Negrisolo, E. The highly rearranged mitochondrial genomes of the crabs Maja crispata and Maja squinado (Majidae) and gene order evolution in Brachyura. Sci. Rep. 2017, 7, 4096. [Google Scholar] [CrossRef] [Green Version]
- Tsang, L.M.; Ahyong, S.T.; Shih, H.; Ng, P.K. Further polyphyly of pinnotheroid crabs: The molecular phylogenetic position of the polychaete-associated Aphanodactylidae. Invertebr. Syst. 2018, 32, 92–99. [Google Scholar] [CrossRef]
- Kobayashi, G.; Itoh, H.; Fukuda, H.; Kojima, S. The complete mitochondrial genome of the sand bubbler crab Scopimera globosa and its phylogenetic position. Genomics 2021, 113, 831–839. [Google Scholar] [CrossRef]
- Ng, P.K.; Guinot, D.; Davie, P.J. Systema Brachyurorum: Part I. An annotated checklist of extant brachyuran crabs of the world. Raffles Bull. Zool. 2008, 17, 1–286. [Google Scholar]
- Davie, P.J.; Guinot, D.; Ng, P.K. Systematics and classification of Brachyura. In Treatise on Zoology-Anatomy, Taxonomy, Biology; Brill Academic Publisher: Leiden, The Netherlands, 2015; Volume 9, pp. 1049–1130. [Google Scholar]
- Xin, Z.Z.; Liu, Y.; Zhang, D.Z.; Chai, X.Y.; Wang, Z.F.; Zhang, H.B.; Zhou, C.L.; Tang, B.P.; Liu, Q.N. Complete mitochondrial genome of Clistocoeloma sinensis (Brachyura: Grapsoidea): Gene rearrangements and higher-level phylogeny of the Brachyura. Sci. Rep. 2017, 7, 4128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Wang, Z.; Shi, X.; Wu, Q.; Tao, Y.; Guo, H.; Ji, C.; Bai, Y. Complete mitochondrial genome of Parasesarma affine (Brachyura: Sesarmidae): Gene rearrangements in Sesarmidae and phylogenetic analysis of the Brachyura. Int. J. Biol. Macromol. 2018, 118 Pt A, 31–40. [Google Scholar] [CrossRef]
- Ng, N.K.; Davie, P.J.F.; Schubart, C.D.; Ng, P.K.L. Xenograpsidae, a new family of grapsoid crabs (Crustacea: Brachyura) associated with shallow water hydrothermal vents. Raffles Bull. Zool. 2007, 16, 233–256. [Google Scholar]
- Ki, J.S.; Dahms, H.U.; Hwang, J.S.; Lee, J.S. The complete mitogenome of the hydrothermal vent crab Xenograpsus testudinatus (Decapoda, Brachyura) and comparison with brachyuran crabs. Comp. Biochem. Physiol. Part D Genom. Proteom. 2009, 4, 290–299. [Google Scholar] [CrossRef]
- Tan, M.H.; Gan, H.M.; Lee, Y.P.; Linton, S.; Grandjean, F.; Bartholomei-Santos, M.L.; Miller, A.D.; Austin, C.M. ORDER within the chaos: Insights into phylogenetic relationships within the Anomura (Crustacea: Decapoda) from mitochondrial sequences and gene order rearrangements. Mol. Phylogenet. Evol. 2018, 127, 320–331. [Google Scholar] [CrossRef]
- Gan, H.M.; Linton, S.M.; Austin, C.M. Two reads to rule them all: Nanopore long read-guided assembly of the iconic Christmas Island red crab, Gecarcoidea natalis (Pocock, 1888), mitochondrial genome and the challenges of AT-rich mitogenomes. Mar. Genom. 2019, 45, 64–71. [Google Scholar] [CrossRef]
- Lombardo, G.; Migliore, N.R.; Colombo, G.; Capodiferro, M.R.; Formenti, G.; Caprioli, M.; Moroni, E.; Caporali, L.; Lancioni, H.; Secomandi, S.; et al. The Mitogenome Relationships and Phylogeography of Barn Swallows (Hirundo rustica). Mol. Biol. Evol. 2022, 39, msac113. [Google Scholar] [CrossRef]
- Tang, P.; Zhu, J.C.; Zheng, B.Y.; Wei, S.J.; Sharkey, M.J.; Chen, X.X.; Vogler, A. Mitochondrial phylogenomics of the Hymenoptera. Mol. Phylogenet. Evol. 2019, 131, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Attardi, G. The mtDNA-encoded ND6 subunit of mitochondrial NADH dehydrogenase is essential for the assembly of the membrane arm and the respiratory function of the enzyme. EMBO J. 1998, 17, 4848–4858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kushnareva, Y.; Murphy, A.N.; Andreyev, A. Complex I-mediated reactive oxygen species generation: Modulation by cytochrome c and NAD(P)+ oxidation–reduction state. Biochem. J. 2002, 368, 545–553. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Yang, T.; Baur, J.A.; Perez, E.; Matsui, T.; Carmona, J.J.; Lamming, D.W.; Souza-Pinto, N.C.; Bohr, V.A.; Rosenzweig, A.; et al. Nutrient-Sensitive Mitochondrial NAD+ Levels Dictate Cell Survival. Cell 2007, 130, 1095–1107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakahata, Y.; Kaluzova, M.; Grimaldi, B.; Sahar, S.; Hirayama, J.; Chen, D.; Guarente, L.P.; Sassone-Corsi, P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 2008, 134, 329–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, N.; Zhang, L.; Gao, W.; Huang, C.; Huber, P.E.; Zhou, X.; Li, C.; Shen, G.; Zou, B. NAD+ metabolism: Pathophysiologic mechanisms and therapeutic potential. Signal Transduct. Target. Ther. 2020, 5, 227. [Google Scholar] [CrossRef] [PubMed]
- Ning, T.; Xiao, H.; Li, J.; Hua, S.; Zhang, Y.P. Adaptive evolution of the mitochondrial ND6 gene in the domestic horse. Genet. Mol. Res. 2010, 1, 144–150. [Google Scholar] [CrossRef]
| Gene | Model | lnL | 2lnL | p Level | Parameters | Positive Selected Sites (Posterior Probabilities) |
|---|---|---|---|---|---|---|
| atp8 | Ge | |||||
| ma | −14,486.44134 | ω0 = 0.087 ω1 = 1.0 ω2 = 1.0 | 19 I 0.852; | |||
| ma0 | −14,486.44134 | 0 | 1 | ω0 = 0.087 ω1 = 1.0 ω2 = 1.0 | ||
| cox2 | Ge | |||||
| ma | −39,595.89288 | ω0 = 0.018 ω1 = 1.0 ω2 = 1.628 | ||||
| ma0 | −39,595.89284 | −7.4 × 10−5 | 1 | ω0 = 0.018 ω1 = 1.0 ω2 = 1.0 | ||
| cox3 | Ge | |||||
| ma | −44,309.9257 | ω0 = 0.016 ω1 = 1.0 ω2 = 1.605 | ||||
| ma0 | −44,309.92564 | −0.00012 | 1 | ω0 = 0.016 ω1 = 1.0 ω2 = 1.0 | ||
| cob | Ge | |||||
| ma | −69,109.49107 | ω0 = 0.017 ω1 = 1.0 ω2 = 1.0 | ||||
| ma0 | −69,109.49113 | 0.00012 | 0.991259787 | ω0 = 0.017 ω1 = 1.0 ω2 = 1.0 | ||
| cox1 | Ge | |||||
| ma | −77,798.40391 | ω0 = 0.008 ω1 = 1.0 ω2 = 2.421 | ||||
| ma0 | −77,798.40376 | −0.000292 | 1 | ω0 = 0.008 ω1 = 1.0 ω2 = 1.0 | ||
| atp6 | Ge | |||||
| ma | −42,775.44489 | ω0 = 0.02 ω1 = 1.0 ω2 = 11.004 | ||||
| ma0 | −42,775.44974 | 0.009696 | 0.921560467 | ω0 = 0.02 ω1 = 1.0 ω2 = 1.0 | ||
| nad4l | Ge | |||||
| ma | −19,516.44828 | ω0 = 0.023 ω1 = 1.0 ω2 = 1.0 | ||||
| ma0 | −19,514.55719 | −3.782182 | 1 | ω0 = 0.023 ω1 = 1.0 ω2 = 1.0 | ||
| nad1 | Ge | |||||
| ma | −55,412.74594 | ω0 = 0.017 ω1 = 1.0 ω2 = 1.0 | ||||
| ma0 | −55,412.74594 | 0 | 1 | ω0 = 0.017 ω1 = 1.0 ω2 = 1.0 | ||
| nad3 | Ge | |||||
| ma | −24,126.5674 | ω0 = 0.026 ω1 = 1.0 ω2 = 1.0 | ||||
| ma0 | −24,126.5674 | 2 × 10−6 | 0.998871621 | ω0 = 0.026 ω1 = 1.0 ω2 = 1.0 | ||
| nad2 | Ge | |||||
| ma | −90,455.6106 | ω0 = 0.042 ω1 = 1.0 ω2 = 1.0 | ||||
| ma0 | −90,455.6106 | 0 | 1 | ω0 = 0.042 ω1 = 1.0 ω2 = 1.0 | ||
| nad5 | Ge | |||||
| ma | −123,998.0812 | ω0 = 0.034 ω1 = 1.0 ω2 = 1.0 | ||||
| ma0 | −123,998.0812 | 0 | 1 | ω0 = 0.034 ω1 = 1.0 ω2 = 1.0 | ||
| nad4 | Ge | |||||
| ma | −91,823.48875 | ω0 = 0.03 ω1 = 1.0 ω2 = 1.0 | 386 I 0.728; | |||
| ma0 | −91,823.48875 | 0 | 1 | ω0 = 0.03 ω1 = 1.0 ω2 = 1.0 | ||
| nad6 | Ge | |||||
| ma | −45,717.39912 | ω0 = 0.043 ω1 = 1.0 ω2 = 137.937 | 28 L 0.686; 36 V 0.556; 88 I 0.997; | |||
| ma0 | −45,721.31165 | 7.825062 | 0.005152668 | ω0 = 0.043 ω1 = 1.0 ω2 = 1.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Zheng, Y.; Zhao, X.; Xu, X.; Xu, Z.; Cui, C. Molecular Phylogeny and Evolution of the Tuerkayana (Decapoda: Brachyura: Gecarcinidae) Genus Based on Whole Mitochondrial Genome Sequences. Biology 2023, 12, 974. https://doi.org/10.3390/biology12070974
Wang Z, Zheng Y, Zhao X, Xu X, Xu Z, Cui C. Molecular Phylogeny and Evolution of the Tuerkayana (Decapoda: Brachyura: Gecarcinidae) Genus Based on Whole Mitochondrial Genome Sequences. Biology. 2023; 12(7):974. https://doi.org/10.3390/biology12070974
Chicago/Turabian StyleWang, Zhengfei, Yuqing Zheng, Xinyue Zhao, Xinyi Xu, Zhiwen Xu, and Chong Cui. 2023. "Molecular Phylogeny and Evolution of the Tuerkayana (Decapoda: Brachyura: Gecarcinidae) Genus Based on Whole Mitochondrial Genome Sequences" Biology 12, no. 7: 974. https://doi.org/10.3390/biology12070974
APA StyleWang, Z., Zheng, Y., Zhao, X., Xu, X., Xu, Z., & Cui, C. (2023). Molecular Phylogeny and Evolution of the Tuerkayana (Decapoda: Brachyura: Gecarcinidae) Genus Based on Whole Mitochondrial Genome Sequences. Biology, 12(7), 974. https://doi.org/10.3390/biology12070974








