The Molecular Mechanism of Cold-Stress Tolerance: Cold Responsive Genes and Their Mechanisms in Rice (Oryza sativa L.)
Abstract
Simple Summary
Abstract
1. Introduction
2. Physiological Responses to Cold Stress
3. Activation and Mechanism of Cold-Responsive TFs and QTLs
4. Signal Transduction and Membrane Stability
5. Genes Identified in Rice during Cold Stress at Various Developmental Stages
6. Application of Omics Technologies in the Identification of Cold-Stress Response Genes or Pathways
7. Rice Breeding for Cold Tolerance
8. Novel Rice Management Strategies to Induce Cold Tolerance
9. Conclusions and Outlooks
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sohag, A.A.M.; Tahjib-Ul-Arif, M.; Afrin, S.; Khan, M.K.; Hannan, M.A.; Skalicky, M.; Mortuza, M.G.; Brestic, M.; Hossain, M.A.; Murata, Y. Insights into nitric oxide-mediated water balance, antioxidant defence and mineral homeostasis in rice (Oryza sativa L.) under chilling stress. Nitric Oxide 2020, 100, 7–16. [Google Scholar] [CrossRef]
- Unan, R.; Genctan, T.; Pedroso, R.M. Cold stress reduces rice grain yield in temperate conditions. Rev. Bras. Eng. Agric. Ambient. 2022, 26, 947–952. [Google Scholar] [CrossRef]
- Rativa, A.G.; Navarro, B.B.; Gastmann, R.; Lamb, T.I.; Silva, A.S.; Dias, P.V.; Lemainski, L.E.; Mario, R.B.; Ponte, L.R.; Gindri, R.G. Decreased night temperature affects development and grain yield only in cold-susceptible rice (Oryza sativa) plants. Crop Pasture Sci. 2021, 72, 782–788. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.-K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zheng, C.; Chen, J.; Qiu, J.; Huang, Z.; Wang, Q.; Ye, Y. Cold acclimation improves photosynthesis by regulating the ascorbate–glutathione cycle in chloroplasts of Kandelia obovata. J. For. Res. 2019, 30, 755–765. [Google Scholar] [CrossRef]
- Zheng, Y.; Rimmington, G.M.; Xie, Z.; Zhang, L.; An, P.; Zhou, G.; Li, X.; Yu, Y.; Chen, L.; Shimizu, H. Responses to air temperature and soil moisture of growth of four dominant species on sand dunes of central Inner Mongolia. J. Plant Res. 2008, 121, 473–482. [Google Scholar] [CrossRef]
- Guo, J.; Liu, S.; Li, X.; Liu, F. Crop exposure to cold stress: Responses in physiological, biochemical and molecular levels. In Sustainable Crop Productivity and Quality Under Climate Change; Elsevier: Amsterdam, The Netherlands, 2022; pp. 1–19. [Google Scholar]
- Yan, T.; Sun, M.; Su, R.; Wang, X.; Lu, X.; Xiao, Y.; Deng, H.; Liu, X.; Tang, W.; Zhang, G. Transcriptomic Profiling of Cold Stress-Induced Differentially Expressed Genes in Seedling Stage of Indica Rice. Plants 2023, 12, 2675. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Zhu, J.-K.; Sunkar, R. Gene regulation during cold stress acclimation in plants. In Plant Stress Tolerance: Methods and Protocols; Springer: Berlin, Germany, 2010; pp. 39–55. [Google Scholar]
- Mittler, R.; Zandalinas, S.I.; Fichman, Y.; Van Breusegem, F. Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 663–679. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Zhou, Y.; Fan, F.; Peng, J.; Zhang, J. Coordination of light, circadian clock with temperature: The potential mechanisms regulating chilling tolerance in rice. J. Integr. Plant Biol. 2020, 62, 737–760. [Google Scholar] [CrossRef]
- Zhang, J.; Luo, W.; Zhao, Y.; Xu, Y.; Song, S.; Chong, K. Comparative metabolomic analysis reveals a reactive oxygen species-dominated dynamic model underlying chilling environment adaptation and tolerance in rice. New Phytol. 2016, 211, 1295–1310. [Google Scholar] [CrossRef]
- Kan, C.-C.; Chung, T.-Y.; Wu, H.-Y.; Juo, Y.-A.; Hsieh, M.-H. Exogenous glutamate rapidly induces the expression of genes involved in metabolism and defense responses in rice roots. BMC Genom. 2017, 18, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Dametto, A.; Sperotto, R.A.; Adamski, J.M.; Blasi, É.A.; Cargnelutti, D.; de Oliveira, L.F.; Ricachenevsky, F.K.; Fregonezi, J.N.; Mariath, J.E.; da Cruz, R.P. Cold tolerance in rice germinating seeds revealed by deep RNAseq analysis of contrasting indica genotypes. Plant Sci. 2015, 238, 1–12. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, Q.; Wang, S.; Hong, Y.; Wang, Z. Rice and cold stress: Methods for its evaluation and summary of cold tolerance-related quantitative trait loci. Rice 2014, 7, 1–12. [Google Scholar] [CrossRef]
- Zhang, Y.; Fan, L.; Zhao, M.; Chen, Q.; Qin, Z.; Feng, Z.; Fujiwara, T.; Zhao, L. Chitooligosaccharide plays essential roles in regulating proline metabolism and cold stress tolerance in rice seedlings. Acta Physiol. Plant. 2019, 41, 1–11. [Google Scholar] [CrossRef]
- Zhu, Y.; Chen, K.; Mi, X.; Chen, T.; Ali, J.; Ye, G.; Xu, J.; Li, Z. Identification and fine mapping of a stably expressed QTL for cold tolerance at the booting stage using an interconnected breeding population in rice. PLoS ONE 2015, 10, e0145704. [Google Scholar] [CrossRef]
- Vilas, J.M.; Corigliano, M.G.; Clemente, M.; Maiale, S.J.; Rodríguez, A.A. Close relationship between the state of the oxygen evolving complex and rice cold stress tolerance. Plant Sci. 2020, 296, 110488. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef]
- Wang, B.; Wang, Y.H.; Xie, L.K.; Yu, W.C.; Lan, Q.K.; Wang, Y.; Chen, C.B.; Zhang, Y. Knocking out OsNAC050 Expression Causes Low-Temperature Tolerance in Rice by Regulating Photosynthesis and the Sucrose Metabolic Pathway. Agriculture 2023, 13, 1378. [Google Scholar] [CrossRef]
- Thapa, R.; Tabien, R.E.; Johnson, C.D.; Septiningsih, E.M. Comparative transcriptomic analysis of germinating rice seedlings to individual and combined anaerobic and cold stress. BMC Genom. 2023, 24, 185. [Google Scholar] [CrossRef]
- Wu, B.; Chen, S.Y.; Cheng, S.Y.; Li, C.Y.; Li, S.H.; Chen, J.X.; Zha, W.J.; Liu, K.; Xu, H.S.; Li, P.D.; et al. Transcriptome Analysis Revealed the Dynamic and Rapid Transcriptional Reprogramming Involved in Cold Stress and Related Core Genes in the Rice Seedling Stage. Int. J. Mol. Sci. 2023, 24, 1914. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Zhang, J.X.; Wang, X.; Essemine, J.; Jin, J.; Qu, M.N.; Xiang, Y.; Chen, W.X. Cold-induced inhibition of photosynthesis-related genes integrated by a TOP6 complex in rice mesophyll cells. Nucleic Acids Res. 2023, 51, 1823–1842. [Google Scholar] [CrossRef]
- Guo, Z.H.; Ma, W.D.; Cai, L.J.; Guo, T.; Liu, H.; Wang, L.N.; Liu, J.L.; Ma, B.; Feng, Y.J.; Liu, C.X.; et al. Comparison of anther transcriptomes in response to cold stress at the reproductive stage between susceptible and resistant Japonica rice varieties. BMC Plant Biol. 2022, 22, 500. [Google Scholar] [CrossRef] [PubMed]
- He, J.P.; Yao, L.; Pecoraro, L.; Liu, C.X.; Wang, J.; Huang, L.Q.; Gao, W.Y. Cold stress regulates accumulation of flavonoids and terpenoids in plants by phytohormone, transcription process, functional enzyme, and epigenetics. Crit. Rev. Biotechnol. 2023, 43, 680–697. [Google Scholar] [CrossRef]
- Li, Z.; Chen, H.; Guan, Q.J.; Li, L.X.; Xuan, Y.H. Gibberellic acid signaling promotes resistance to saline-alkaline stress by increasing the uptake of ammonium in rice. Plant Physiol. Biochem. 2024, 207, 108424. [Google Scholar] [CrossRef]
- Tang, J.Q.; Tian, X.J.; Mei, E.Y.; He, M.L.; Gao, J.W.; Yu, J.; Xu, M.; Liu, J.L.; Song, L.; Li, X.F.; et al. WRKY53 negatively regulates rice cold tolerance at the booting stage by fine-tuning anther gibberellin levels. Plant Cell 2022, 34, 4495–4515. [Google Scholar] [CrossRef]
- Macioszek, V.K.; Jecz, T.; Ciereszko, I.; Kononowicz, A.K. Jasmonic Acid as a Mediator in Plant Response to Necrotrophic Fungi. Cells-Basel 2023, 12, 1027. [Google Scholar] [CrossRef]
- Ma, J.N.; Morel, J.B.; Riemann, M.; Nick, P. Jasmonic acid contributes to rice resistance against Magnaporthe oryzae. BMC Plant Biol. 2022, 22, 601. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.I.R.; Kumari, S.; Nazir, F.; Khanna, R.R.; Gupta, R.; Chhillar, H. Defensive role of plant hormones in advancing abiotic stress-resistant rice plants. Rice Sci. 2023, 30, 15–35. [Google Scholar] [CrossRef]
- Gopinath, I. QTL mapping for cold tolerance with specific reference to rice. J. Pharmacogn. Phytochem. 2020, 9, 146–153. [Google Scholar]
- Yan, W.; Yuan, S.; Zu, Y.; Chang, Z.; Li, Y.; Chen, Z.; Xie, G.; Chen, L.; Lu, C.; Deng, X.W. Ornithine δ-aminotransferase OsOAT is critical for male fertility and cold tolerance during rice plant development. Plant J. 2023, 114, 1301–1318. [Google Scholar] [CrossRef]
- Ma, Y.; Dai, X.; Xu, Y.; Luo, W.; Zheng, X.; Zeng, D.; Pan, Y.; Lin, X.; Liu, H.; Zhang, D. COLD1 confers chilling tolerance in rice. Cell 2015, 160, 1209–1221. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Katsura, K.; Maruyama, K.; Taji, T.; Kobayashi, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 2006, 47, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, J.; Wang, X.; Chen, J. OVP1, a vacuolar H+-translocating inorganic pyrophosphatase (V-PPase), overexpression improved rice cold tolerance. Plant Physiol. Biochem. 2011, 49, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, Z.; Chong, K.; Xu, Y. Chilling tolerance in rice: Past and present. J. Plant Physiol. 2022, 268, 153576. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.P.; Zhao, Y.; Xu, S.J.; Zhang, Z.Y.; Xu, Y.Y.; Zhang, J.Y.; Chong, K. OsMADS57 together with OsTB1 coordinates transcription of its target OsWRKY94 and D14 to switch its organogenesis to defense for cold adaptation in rice. New Phytol. 2018, 218, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.T.; Schläppi, M.R.; Mao, B.G.; Wang, W.; Wang, A.J.; Chu, C.C. The bZIP73 transcription factor controls rice cold tolerance at the reproductive stage. Plant Biotechnol. J. 2019, 17, 1834–1849. [Google Scholar] [CrossRef]
- Liu, C.; Ou, S.; Mao, B.; Tang, J.; Wang, W.; Wang, H.; Cao, S.; Schläppi, M.R.; Zhao, B.; Xiao, G. Early selection of bZIP73 facilitated adaptation of japonica rice to cold climates. Nat. Commun. 2018, 9, 3302. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.T.; Wu, Y.B.; Wang, X.P. bZIP transcription factor OsbZIP52/RISBZ5: A potential negative regulator of cold and drought stress response in rice. Planta 2012, 235, 1157–1169. [Google Scholar] [CrossRef]
- Sun, Y.; Xie, Z.Z.; Jin, L.; Qin, T.; Zhan, C.H.; Huang, J.L. Histone deacetylase OsHDA716 represses rice chilling tolerance by deacetylating OsbZIP46 to reduce its transactivation function and protein stability. Plant Cell 2024, 36, 1913–1936. [Google Scholar] [CrossRef]
- Yu, S.W.; Huang, A.N.; Li, J.; Gao, L.; Feng, Y.N.; Pemberton, E.; Chen, C.L. OsNAC45 plays complex roles by mediating POD activity and the expression of development-related genes under various abiotic stresses in rice root. Plant Growth Regul. 2018, 84, 519–531. [Google Scholar] [CrossRef]
- Huang, L.; Hong, Y.B.; Zhang, H.J.; Li, D.Y.; Song, F.M. Rice NAC transcription factor ONAC095 plays opposite roles in drought and cold stress tolerance. BMC Plant Biol. 2016, 16, 203. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xiang, Z.; Li, J.; Wang, S.; Chen, Y.; Liu, Y.; Mao, D.; Luan, S.; Chen, L. bHLH57 confers chilling tolerance and grain yield improvement in rice. Plant Cell Environ. 2023, 46, 1402–1418. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Zhang, H.; Pan, X.; Chen, X.; Zhang, Z.; Lu, X.; Huang, R. Overexpression of ethylene response factor TERF2 confers cold tolerance in rice seedlings. Transgenic Res. 2011, 20, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Chen, Y.; Wang, Y.; Shen, Y.; Yang, J.; Jia, B.; Sun, X.; Sun, M. A comprehensive investigation of the regulatory roles of OsERF096, an AP2/ERF transcription factor, in rice cold stress response. Plant Cell Rep. 2023, 42, 2011–2022. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.T.; Yang, S.H. COLD1: A cold sensor in rice. Sci. China Life Sci. 2015, 58, 409–410. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Gong, Z. One SNP in COLD1 determines cold tolerance during rice domestication. J. Genet. Genom. 2015, 4, 133–134. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Huan, Q.; Xu, Y.; Qian, W.; Chong, K.; Zhang, J. Integrated global analysis reveals a vitamin E-vitamin K1 sub-network, downstream of COLD1, underlying rice chilling tolerance divergence. Cell Rep. 2021, 36, 109397. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Xu, W.; Song, Q.; Tan, L.; Liu, J.; Zhu, Z.; Fu, Y.; Su, Z.; Sun, C. Microarray-assisted fine-mapping of quantitative trait loci for cold tolerance in rice. Mol. Plant 2013, 6, 757–767. [Google Scholar] [CrossRef]
- Saito, K.; Hayano-Saito, Y.; Maruyama-Funatsuki, W.; Sato, Y.; Kato, A. Physical mapping and putative candidate gene identification of a quantitative trait locus Ctb1 for cold tolerance at the booting stage of rice. Theor. Appl. Genet. 2004, 109, 515–522. [Google Scholar] [CrossRef]
- Saito, K.; Hayano-Saito, Y.; Kuroki, M.; Sato, Y. Map-based cloning of the rice cold tolerance gene Ctb1. Plant Sci. 2010, 179, 97–102. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Pan, Y.; Li, J.; Zhou, L.; Shi, H.; Zeng, Y.; Guo, H.; Yang, S.; Zheng, W. Natural variation in CTB4a enhances rice adaptation to cold habitats. Nat. Commun. 2017, 8, 14788. [Google Scholar] [CrossRef] [PubMed]
- Knight, H.; Trewavas, A.J.; Knight, M.R. Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell 1996, 8, 489–503. [Google Scholar] [PubMed]
- Huda, K.M.K.; Banu, M.S.A.; Yadav, S.; Sahoo, R.K.; Tuteja, R.; Tuteja, N. Salinity and drought tolerant OsACA6 enhances cold tolerance in transgenic tobacco by interacting with stress-inducible proteins. Plant Physiol. Biochem. 2014, 82, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.J.; Guo, X.Y.; Xu, Y.Y.; Li, H.; Ma, L.; Yao, X.F.; Weng, Y.X.; Guo, Y.; Liu, C.M.; Chong, K. OsCIPK7 point-mutation leads to conformation and kinase-activity change for sensing cold response. J. Integr. Plant Biol. 2019, 61, 1194–1200. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.Y.; Zhang, D.J.; Wang, Z.L.; Xu, S.J.; Batistic, O.; Steinhorst, L.; Li, H.; Weng, Y.X.; Ren, D.T.; Kudla, J.; et al. Cold-induced calreticulin OsCRT3 conformational changes promote OsCIPK7 binding and temperature sensing in rice. EMBO J. 2023, 42, e110518. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ren, Y.; Liu, X.; Luo, S.; Zhang, X.; Liu, X.; Lin, Q.; Zhu, S.; Wan, H.; Yang, Y. Transcriptional activation and phosphorylation of OsCNGC9 confer enhanced chilling tolerance in rice. Mol. Plant 2021, 14, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Lu, S.; Li, Z.; Cheng, J.; Hu, P.; Zhu, T.; Wang, X.; Jin, M.; Wang, X.; Li, L. CYCLIC NUCLEOTIDE-GATED ION CHANNELs 14 and 16 promote tolerance to heat and chilling in rice. Plant Physiol. 2020, 183, 1794–1808. [Google Scholar] [CrossRef] [PubMed]
- Sangwan, V.; Örvar, B.L.; Beyerly, J.; Hirt, H.; Dhindsa, R.S. Opposite changes in membrane fluidity mimic cold and heat stress activation of distinct plant MAP kinase pathways. Plant J. 2002, 31, 629–638. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Jiang, N.; Wang, G.-L.; Hong, Y.; Wang, Z. Advances in understanding cold sensing and the cold-responsive network in rice. Adv. Crop. Sci. Tech. 2013, 1, 104. [Google Scholar]
- Kerbler, S.M.; Wigge, P.A. Temperature Sensing in Plants. Annu. Rev. Plant Biol. 2023, 74, 341–366. [Google Scholar] [CrossRef]
- Nozawa, Y. Adaptive regulation of membrane lipids and fluidity during thermal acclimation in Tetrahymena. Proc. Jpn. Acad. Ser. 2011, 87, 450–462. [Google Scholar] [CrossRef] [PubMed]
- Tovuu, A.; Zulfugarov, I.S.; Wu, G.; Kang, I.S.; Kim, C.; Moon, B.Y.; An, G.; Lee, C.-H. Rice mutants deficient in ω-3 fatty acid desaturase (FAD8) fail to acclimate to cold temperatures. Plant Physiol. Biochem. 2016, 109, 525–535. [Google Scholar] [CrossRef] [PubMed]
- Los, D.A.; Murata, N. Membrane fluidity and its roles in the perception of environmental signals. Biochim. Biophys. Acta Biomembr. 2004, 1666, 142–157. [Google Scholar] [CrossRef]
- Song, S.-Y.; Chen, Y.; Chen, J.; Dai, X.-Y.; Zhang, W.-H. Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 2011, 234, 331–345. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Gene regulation during cold acclimation in plants. Physiol. Plant. 2006, 126, 52–61. [Google Scholar] [CrossRef]
- Xu, H.; Yang, X.; Zhang, Y.; Wang, H.; Wu, S.; Zhang, Z.; Ahammed, G.J.; Zhao, C.; Liu, H. CRISPR/Cas9-mediated mutation in auxin efflux carrier OsPIN9 confers chilling tolerance by modulating reactive oxygen species homeostasis in rice. Front. Plant Sci. 2022, 13, 967031. [Google Scholar] [CrossRef]
- Ouyang, Q.; Zhang, Y.; Yang, X.; Yang, C.; Hou, D.; Liu, H.; Xu, H. Overexpression of OsPIN9 impairs chilling tolerance via disturbing ROS homeostasis in rice. Plants 2023, 12, 2809. [Google Scholar] [CrossRef]
- Huo, X.; Xiao, J.; Peng, X.; Lin, Y.; Liu, D.; Liu, W.; Liao, Y.; Li, J.; Zhu, M.; Fu, C. The grain yield regulator NOG1 plays a dual role in latitudinal adaptation and cold tolerance during rice domestication. Front. Genet. 2022, 13, 1039677. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, R.; Huang, K.; Huang, S.; Wang, H.; Wei, Z.; Li, Z.; Bian, M.; Jiang, W.; Wu, T. The OsWRKY63–OsWRKY76–OsDREB1B module regulates chilling tolerance in rice. Plant J. 2022, 112, 383–398. [Google Scholar] [CrossRef]
- Liu, S.; Zheng, Y.; Zhao, L.; Gulam, M.; Ullah, A.; Xie, G. CALMODULIN-LIKE16 and PIN-LIKES7a cooperatively regulate rice seedling primary root elongation under chilling. Plant Physiol. 2024, 195, 1660–1680. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.-L.; Han, X.; Meng, Y.; Wang, L.; Zhang, W.; Yang, C.; Li, H.; Tang, S.; Guo, Z.; Liu, C. Acyl carrier protein OsMTACP2 confers rice cold tolerance at the booting stage. Plant Physiol. 2024, 195, 1277–1292. [Google Scholar] [CrossRef] [PubMed]
- Hoang, T.V.; Vo, K.T.X.; Rahman, M.M.; Zhong, R.; Lee, C.; Ketudat Cairns, J.R.; Ye, Z.-H.; Jeon, J.-S. SPOTTED-LEAF7 targets the gene encoding β-Galactosidase9, which functions in rice growth and stress responses. Plant Physiol. 2023, 193, 1109–1125. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Liu, H.; Peng, K.; Huang, X. Cold-upregulated glycosyltransferase gene 1 (OsCUGT1) plays important roles in rice height and spikelet fertility. J. Plant Res. 2023, 136, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, B.; Luo, W.; Xu, Y.; Wang, J.; Xue, Z.; Niu, Y.; Cheng, Z.; Ge, S.; Zhang, W. Natural variation of codon repeats in COLD11 endows rice with chilling resilience. Sci. Adv. 2023, 9, eabq5506. [Google Scholar] [CrossRef]
- Islam, F.; Khan, M.S.S.; Ahmed, S.; Abdullah, M.; Hannan, F.; Chen, J. OsLPXC negatively regulates tolerance to cold stress via modulating oxidative stress, antioxidant defense and JA accumulation in rice. Free Radic. Biol. Med. 2023, 199, 2–16. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Ding, Z.; Duan, M.; Xiong, Y.; Li, X.; Yuan, X.; Huang, J. OsLUX confers rice cold tolerance as a positive regulatory factor. Int. J. Mol. Sci. 2023, 24, 6727. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, J.; Wang, D.; Chang, J.; Chen, H.; Chen, D.; Deng, W.; Tian, C. Genome-Wide Identification and Analysis of OsSPXs Revealed Its Genetic Influence on Cold Tolerance of Dongxiang Wild Rice (DXWR). Int. J. Mol. Sci. 2023, 24, 8755. [Google Scholar] [CrossRef]
- Li, R.Q.; Song, Y.; Wang, X.Q.; Zheng, C.F.; Liu, B.; Zhang, H.L.; Ke, J.; Wu, X.J.; Wu, L.Q.; Yang, R.F.; et al. OsNAC5 orchestrates OsABI5 to fine-tune cold tolerance in rice. J. Integr. Plant Biol. 2024, 66, 660–682. [Google Scholar] [CrossRef]
- Yang, L.; Lei, L.; Wang, J.; Zheng, H.; Xin, W.; Liu, H.; Zou, D. qCTB7 positively regulates cold tolerance at booting stage in rice. Theor. Appl. Genet. 2023, 136, 135. [Google Scholar] [CrossRef]
- Xia, C.; Liang, G.; Chong, K.; Xu, Y. The COG1-OsSERL2 complex senses cold to trigger signaling network for chilling tolerance in japonica rice. Nat. Commun. 2023, 14, 3104. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Guo, Z.; Wang, K.; Wang, R.; Fang, C. Comparative Metabolomic Analysis Reveals the Role of OsHPL1 in the Cold-Induced Metabolic Changes in Rice. Plants 2023, 12, 2032. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Zhuang, J.; Zhang, Z.; Chen, W.; Xu, H.; Zhao, M.; Ma, D. Multi-omics approach reveals the contribution of OsSEH1 to rice cold tolerance. Front. Plant Sci. 2023, 13, 1110724. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Zhang, J.; Zan, X.; Zhu, J.; Chen, H.; Li, X.; Zhou, Z.; Gao, X.; Chen, R.; Huang, Z. Overexpression of rice histone H1 gene reduces tolerance to cold and heat stress. Plants 2023, 12, 2408. [Google Scholar] [CrossRef] [PubMed]
- Lou, Q.J.; Guo, H.F.; Li, J.; Han, S.C.; Khan, N.U.; Gu, Y.S.; Zhao, W.T.; Zhang, Z.Y.; Zhang, H.L.; Li, Z.C.; et al. Cold-adaptive evolution at the reproductive stage in Geng/japonica subspecies reveals the role of OsMAPK3 and OsLEA9. Plant J. 2022, 111, 1032–1051. [Google Scholar] [CrossRef]
- Jia, M.; Meng, X.; Song, X.; Zhang, D.; Kou, L.; Zhang, J.; Jing, Y.; Liu, G.; Liu, H.; Huang, X. Chilling-induced phosphorylation of IPA1 by OsSAPK6 activates chilling tolerance responses in rice. Cell Discov. 2022, 8, 71. [Google Scholar] [CrossRef]
- Xie, Y.; Waqas, M.; Khan, M.U.; Lan, C.; Weng, P.; Zou, J.; Wu, X.; Lin, W.; Li, Z. Overexpression of the rice gene Lsi1 (low silicon gene 1) enhances plant-microbe interactions that result in improved chilling tolerance. Plant Growth Regul. 2022, 98, 525–538. [Google Scholar] [CrossRef]
- Liu, H.; Yang, L.; Xu, S.; Lyu, M.-J.; Wang, J.; Wang, H.; Zheng, H.; Xin, W.; Liu, J.; Zou, D. OsWRKY115 on qCT7 links to cold tolerance in rice. Theor. Appl. Genet. 2022, 135, 2353–2367. [Google Scholar] [CrossRef]
- Zhang, H.; Wu, T.; Li, Z.; Huang, K.; Kim, N.-E.; Ma, Z.; Kwon, S.-W.; Jiang, W.; Du, X. OsGATA16, a GATA transcription factor, confers cold tolerance by repressing OsWRKY45–1 at the seedling stage in rice. Rice 2021, 14, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zeng, Y.; Pan, Y.; Zhou, L.; Zhang, Z.; Guo, H.; Lou, Q.; Shui, G.; Huang, H.; Tian, H. Stepwise selection of natural variations at CTB2 and CTB4a improves cold adaptation during domestication of japonica rice. New Phytol. 2021, 231, 1056–1072. [Google Scholar] [CrossRef]
- Zeng, Y.; Wen, J.; Zhao, W.; Wang, Q.; Huang, W. Rational improvement of rice yield and cold tolerance by editing the three genes OsPIN5b, GS3, and OsMYB30 with the CRISPR–Cas9 system. Front. Plant Sci. 2020, 10, 1663. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Li, Z.; Wan, M.; Wu, T.; Bian, M.; Huang, K.; Zhang, X. Overexpression of OsETR4, aputative ethylene receptor increases cold tolerance in rice. Int. J. Agric. Biol 2020, 24, 969–978. [Google Scholar]
- Li, Z.; Feng, S.; Zhan, W.; Xu, L.; Fang, C.; Zhang, Z.; Lin, W. Lsi1 plays an active role in enhancing the chilling tolerance of rice roots. Plant Growth Regul. 2020, 90, 529–543. [Google Scholar] [CrossRef]
- Xu, Y.; Hu, D.; Hou, X.; Shen, J.; Liu, J.; Cen, X.; Fu, J.; Li, X.; Hu, H.; Xiong, L. OsTMF attenuates cold tolerance by affecting cell wall properties in rice. New Phytol. 2020, 227, 498–512. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, R.; Wang, Y.; Zhang, L.; Yao, S. A point mutation in LTT1 enhances cold tolerance at the booting stage in rice. Plant Cell Environ. 2020, 43, 992–1007. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Liang, H.; Gao, L.; Dai, G.; Chen, W.; Yang, X.; Qing, D.; Gao, J.; Wu, H.; Huang, J. Transcriptomic profiling of germinating seeds under cold stress and characterization of the cold-tolerant gene LTG5 in rice. BMC Plant Biol. 2020, 20, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Phan, H.; Liu, Y.; Cao, S.; Zhang, Z.; Chu, C.; Schläppi, M.R. Glycosyltransferase OsUGT90A1 helps protect the plasma membrane during chilling stress in rice. J. Exp. Bot. 2020, 71, 2723–2739. [Google Scholar] [CrossRef]
- Mao, D.; Xin, Y.; Tan, Y.; Hu, X.; Bai, J.; Liu, Z.-y.; Yu, Y.; Li, L.; Peng, C.; Fan, T. Natural variation in the HAN1 gene confers chilling tolerance in rice and allowed adaptation to a temperate climate. Proc. Natl. Acad. Sci. USA 2019, 116, 3494–3501. [Google Scholar] [CrossRef]
- Wang, P.; Xiong, Y.; Gong, R.; Yang, Y.; Fan, K.; Yu, S. A key variant in the cis-regulatory element of flowering gene Ghd8 associated with cold tolerance in rice. Sci. Rep. 2019, 9, 9603. [Google Scholar] [CrossRef]
- Moon, S.-J.; Min, M.K.; Kim, J.-A.; Kim, D.Y.; Yoon, I.S.; Kwon, T.R.; Byun, M.O.; Kim, B.-G. Ectopic expression of OsDREB1G, a member of the OsDREB1 subfamily, confers cold stress tolerance in rice. Front. Plant Sci. 2019, 10, 297. [Google Scholar] [CrossRef]
- Chen, N.; Xu, Y.; Wang, X.; Du, C.; Du, J.; Yuan, M.; Xu, Z.; Chong, K. OsRAN2, essential for mitosis, enhances cold tolerance in rice by promoting export of intranuclear tubulin and maintaining cell division under cold stress. Plant Cell Environ. 2011, 34, 52–64. [Google Scholar] [CrossRef]
- Kim, C.-Y.; Vo, K.T.X.; Nguyen, C.D.; Jeong, D.-H.; Lee, S.-K.; Kumar, M.; Kim, S.-R.; Park, S.-H.; Kim, J.-K.; Jeon, J.-S. Functional analysis of a cold-responsive rice WRKY gene, OsWRKY71. Plant Biotechnol. Rep. 2016, 10, 13–23. [Google Scholar] [CrossRef]
- Wang, S.-T.; Sun, X.-L.; Hoshino, Y.; Yu, Y.; Jia, B.; Sun, Z.-W.; Sun, M.-Z.; Duan, X.-B.; Zhu, Y.-M. MicroRNA319 positively regulates cold tolerance by targeting OsPCF6 and OsTCP21 in rice (Oryza sativa L.). PLoS ONE 2014, 9, e91357. [Google Scholar] [CrossRef]
- Guan, S.; Xu, Q.; Ma, D.; Zhang, W.; Xu, Z.; Zhao, M.; Guo, Z. Transcriptomics profiling in response to cold stress in cultivated rice and weedy rice. Gene 2019, 685, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Zhou, P.; Zhu, Y.; Liu, F.; Li, R.; Qiu, Y. Proteomic analysis of rice seedlings under cold stress. Protein J. 2017, 36, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, J.; Wang, X.; Li, R.; Chen, B. Proteomic response of hybrid wild rice to cold stress at the seedling stage. PLoS ONE 2018, 13, e0198675. [Google Scholar] [CrossRef] [PubMed]
- Bolaños, J.; Edmeades, G. Eight cycles of selection for drought tolerance in lowland tropical maize. II. Responses in reproductive behavior. Field Crops Res. 1993, 31, 253–268. [Google Scholar] [CrossRef]
- Feng, X.; Wang, C.; Nan, J.; Zhang, X.; Wang, R.; Jiang, G.; Yuan, Q.; Lin, S. Updating the elite rice variety Kongyu 131 by improving the Gn1a locus. Rice 2017, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Jiang, G.; Feng, X.; Nan, J.; Zhang, X.; Yuan, Q.; Lin, S. Updating the genome of the elite rice variety Kongyu131 to expand its ecological adaptation region. Front. Plant Sci. 2019, 10, 288. [Google Scholar] [CrossRef]
- Qi, Y.B.; Summat, P.; Panyawut, N.; Sikaewtung, K.; Ditthab, K.; Tongmark, K.; Chakhonkaen, S.; Sangarwut, N.; Wasinanon, T.; Kaewmungkun, K.; et al. Identification of Rice Accessions Having Cold Tolerance at the Seedling Stage and Development of Novel Genotypic Assays for Predicting Cold Tolerance. Plants 2023, 12, 215. [Google Scholar] [CrossRef]
- Shin, N.-H.; Han, J.-H.; Vo, K.T.X.; Seo, J.; Navea, I.P.; Yoo, S.-C.; Jeon, J.-S.; Chin, J.H. Development of a temperate climate-adapted indica multi-stress tolerant rice variety by pyramiding quantitative trait loci. Rice 2022, 15, 22. [Google Scholar] [CrossRef] [PubMed]
- Chukwu, S.C.; Rafii, M.Y.; Ramlee, S.I.; Ismail, S.I.; Oladosu, Y.; Okporie, E.; Onyishi, G.; Utobo, E.; Ekwu, L.; Swaray, S. Marker-assisted selection and gene pyramiding for resistance to bacterial leaf blight disease of rice (Oryza sativa L.). Biotechnol. Biotechnol. Equip. 2019, 33, 440–455. [Google Scholar] [CrossRef]
- Usman, M.G.; Rafii, M.Y.; Martini, M.Y.; Yusuff, O.A.; Ismail, M.R.; Miah, G. Introgression of heat shock protein (Hsp70 and sHsp) genes into the Malaysian elite chilli variety Kulai (Capsicum annuum L.) through the application of marker-assisted backcrossing (MAB). Cell Stress Chaperones 2018, 23, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Miah, G.; Rafii, M.; Ismail, M.; Sahebi, M.; Hashemi, F.; Yusuff, O.; Usman, M. Blast disease intimidation towards rice cultivation: A review of pathogen and strategies to control. J. Anim. Plant Sci. 2017, 27, 1058–1066. [Google Scholar]
- Yang, L.; Wang, J.; Han, Z.; Lei, L.; Liu, H.L.; Zheng, H.; Xin, W.; Zou, D. Combining QTL-seq and linkage mapping to fine map a candidate gene in qCTS6 for cold tolerance at the seedling stage in rice. BMC Plant Biol. 2021, 21, 278. [Google Scholar] [CrossRef]
- de Souza, E.M.; Lamb, T.I.; Lamb, T.A.; dos Santos Silva, A.; da Fré de Carvalho, S.; Nyland, V.; Lopes, M.C.B.; Grohs, M.; Marconatto, L.; dos Anjos Borges, L.G.; et al. Rhizospheric soil from rice paddy presents isolable bacteria able to induce cold tolerance in rice plants. J. Plant Nutr. 2021, 21, 1993–2006. [Google Scholar] [CrossRef]
- Tahjib-ul-Arif, M. Improving Cold Stress Tolerance in Rice Seedlings. J. Environ. Sci. Sustain. Soc. 2023, 12, MR02_p05–MR02_p08. [Google Scholar] [CrossRef]
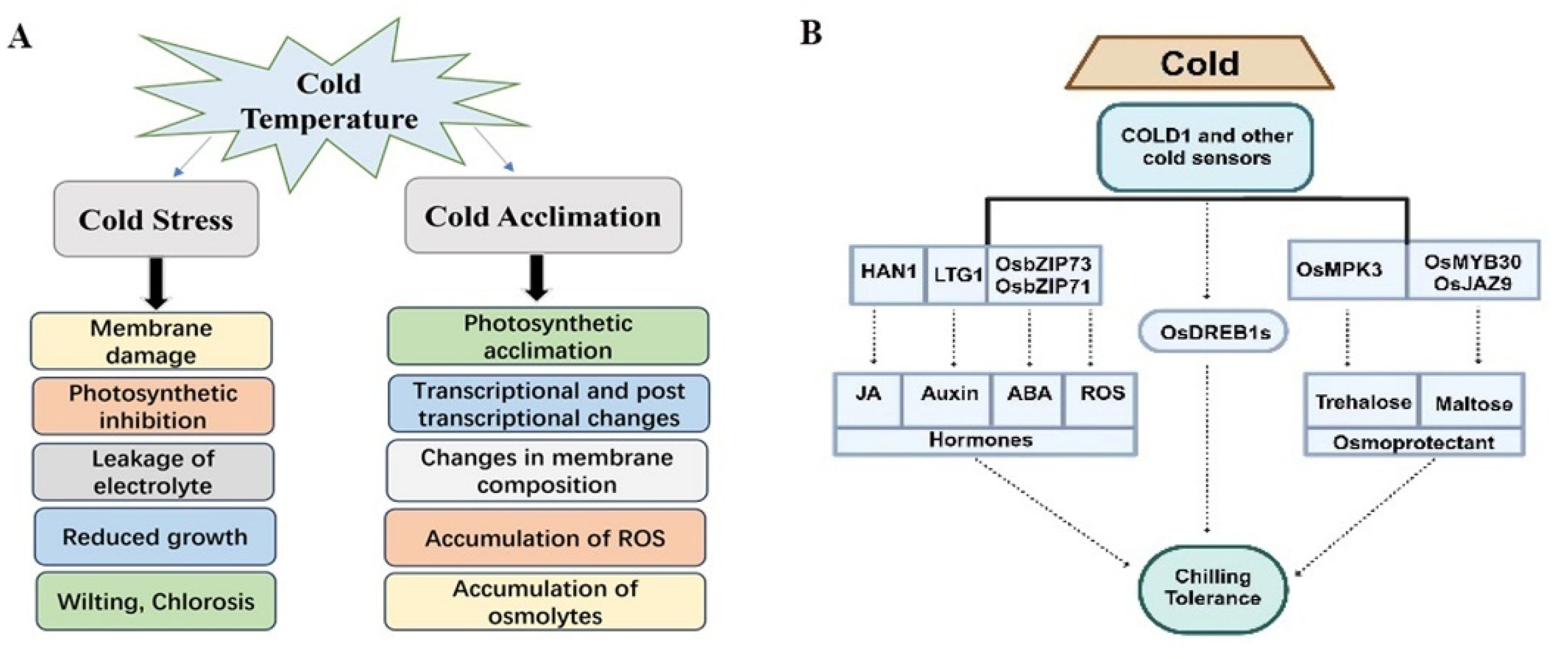
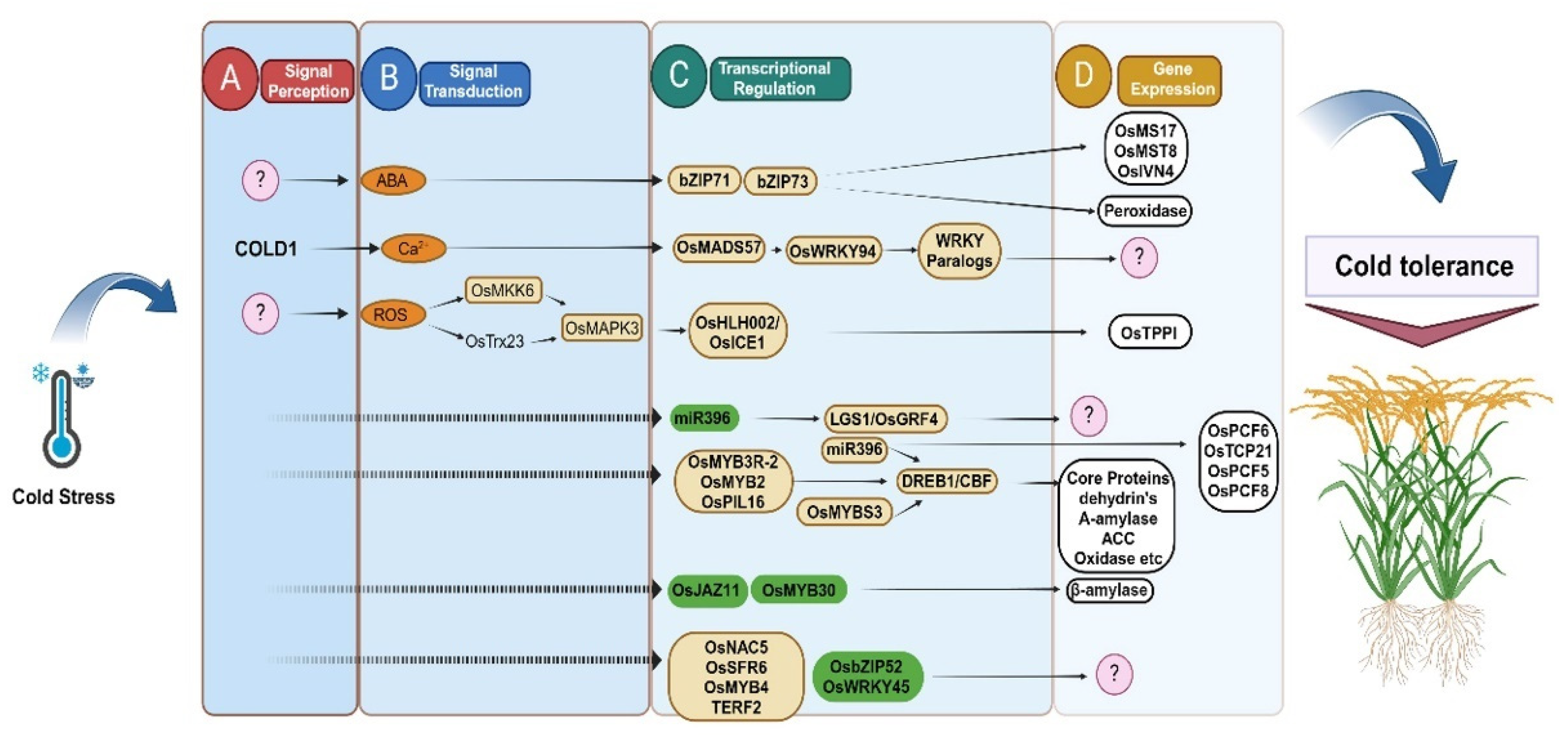
| Gene | Developmental Stages | Functions of Genes | Reference |
|---|---|---|---|
| bHLH57 | Flowering, Booting, Germination | Cold tolerance and grain yield improvement ↑ | [44] |
| OsPIN9 | Seedling stage | Regulation of auxin, ROS homeostasis and cold tolerance ↑ | [68,69] |
| NOG1 | Seedling stage | Grain number and yield during cold ↑ | [70] |
| OsDREB1B | Seedling stage | Regulate chilling tolerance ↑ | [71] |
| OsCML16, OsPILS7a | Seedling Stage | Regulate primary root elongation and cold tolerance ↑ | [72] |
| OsMTACP2 | Seedling stage | Mediated wax ester biosynthesis and cold tolerance ↑ | [73] |
| OsERF096 | Different stages | Regulation of cold stress ↑ | [46] |
| OsSPL7 | Maturity stage | Rice growth and stress responses ↑ | [74] |
| OsCUGT1 | Germination, reproductive | Rice height and spikelet fertility ↑ | [75] |
| OsCRT3 | Seedling stage | Regulator of chilling tolerance ↑ | [57] |
| COLD11 | Germination stage | Chilling tolerance ↑ | [76] |
| OsLPXC | Reproductive stage | Regulate cold tolerance ↑ | [77] |
| OsLUX | Seedling stage | Cold stress and circadian rhythm ↑ | [78] |
| OsSPXs | Different stages | Rice adaptation to cold stress ↑ | [79] |
| OsNAC5 | Germination and seedling | Cold tolerance ↑ | [80] |
| qCTB7 | Booting stage | Regulates the appearance and morphology of the anthers and pollen ↑ | [81] |
| COG1 | Germination stage | Cold tolerance ↑ | [82] |
| OsOAT | Germination stages | Male fertility, cold tolerance ↑ | [32] |
| OsHPL1 | Seedling, germination | Modulates rice metabolism ↑ | [83] |
| OsSEH1 | Seedling stage | Cold tolerance ↑ | [84] |
| OsHis1 | Germination and seedling | Tolerance to temperature stress ↑ | [85] |
| OsMAPK3 OsLEA9 | Reproductive stage | Cold tolerance ↑ | [86] |
| OsSAPK6 | Seedling stage | Activate cold resistance | [87] |
| OsLsi1 | Seedling stage | Enhances microbe-plant interactions and cold tolerance ↑ | [88] |
| OsWRKY115 | Seedling stage | Cold tolerance ↑ | [89] |
| OsGATA16 | Seedling stage | Cold tolerance ↑ | [90] |
| OsCTB2 | Booting stage | Cold adaptation ↑ | [91] |
| OsCNGC9 | Seedling stage | Enhanced cold tolerance ↑ | [58] |
| OsPIN5b, GS3, and OsMYB30 | Reproductive stage | Increased panicle length, enlarged grain size, enhanced cold tolerance ↑ | [92] |
| OsETR4 | Seedling stage | Seedling survival rate ↑ | [93] |
| OsLsi1 | Germination stages | Enhanced the antioxidant system and non-structural carbohydrates ↑ | [94] |
| OsTMF | Different stages | Regulate chilling tolerance by affecting cell wall properties | [95] |
| OsLTT1 | Booting stage | Cold tolerance Maintaining tapetum degradation and pollen development ↑ | [96] |
| OsLTG5 | Seedling stage | Cold tolerance ↑ | [97] |
| OsUGT90A1 | Seedling stage | Protect the plasma membrane and promote leaf growth ↑ | [98] |
| OsHAN1 | Different stages | Chilling tolerance ↑ | [99] |
| Ghd8 | Different stages | Flowering time, heading date ↑ | [100] |
| OsDREB1G | Seedling and germination | Cold stress response ↑ | [101] |
| OsRAN2 | Different stages | Regulate export of intranuclear tubulin and cell division ↑ | [102] |
| OsWRKY71 | Seedling stage | Functions as a transcriptional repressor ↑ | [103] |
| COLD1 | Seedling stage | Chilling tolerance ↑ | [33] |
| Osa-miR319b | Different stages | wider leaf blades and delayed development ↑ | [104] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shahzad, N.; Nabi, H.G.; Qiao, L.; Li, W. The Molecular Mechanism of Cold-Stress Tolerance: Cold Responsive Genes and Their Mechanisms in Rice (Oryza sativa L.). Biology 2024, 13, 442. https://doi.org/10.3390/biology13060442
Shahzad N, Nabi HG, Qiao L, Li W. The Molecular Mechanism of Cold-Stress Tolerance: Cold Responsive Genes and Their Mechanisms in Rice (Oryza sativa L.). Biology. 2024; 13(6):442. https://doi.org/10.3390/biology13060442
Chicago/Turabian StyleShahzad, Nida, Hafiz Ghulam Nabi, Lei Qiao, and Wenqiang Li. 2024. "The Molecular Mechanism of Cold-Stress Tolerance: Cold Responsive Genes and Their Mechanisms in Rice (Oryza sativa L.)" Biology 13, no. 6: 442. https://doi.org/10.3390/biology13060442
APA StyleShahzad, N., Nabi, H. G., Qiao, L., & Li, W. (2024). The Molecular Mechanism of Cold-Stress Tolerance: Cold Responsive Genes and Their Mechanisms in Rice (Oryza sativa L.). Biology, 13(6), 442. https://doi.org/10.3390/biology13060442





