The Role of the MCM2-7 Helicase Subunit MCM2 in Epigenetic Inheritance
Abstract
:Simple Summary
Abstract
1. Introduction
2. The eSPAN Method to Study DNA Replication-Coupled Epigenetic Inheritance
3. The Function of Mcm2 and Its Interaction Partners in DNA Replication
4. Mcm2’s Role in Parental Histone Recycling
5. Additional Mcm2 Interaction Factors in Parental Histone Transfer
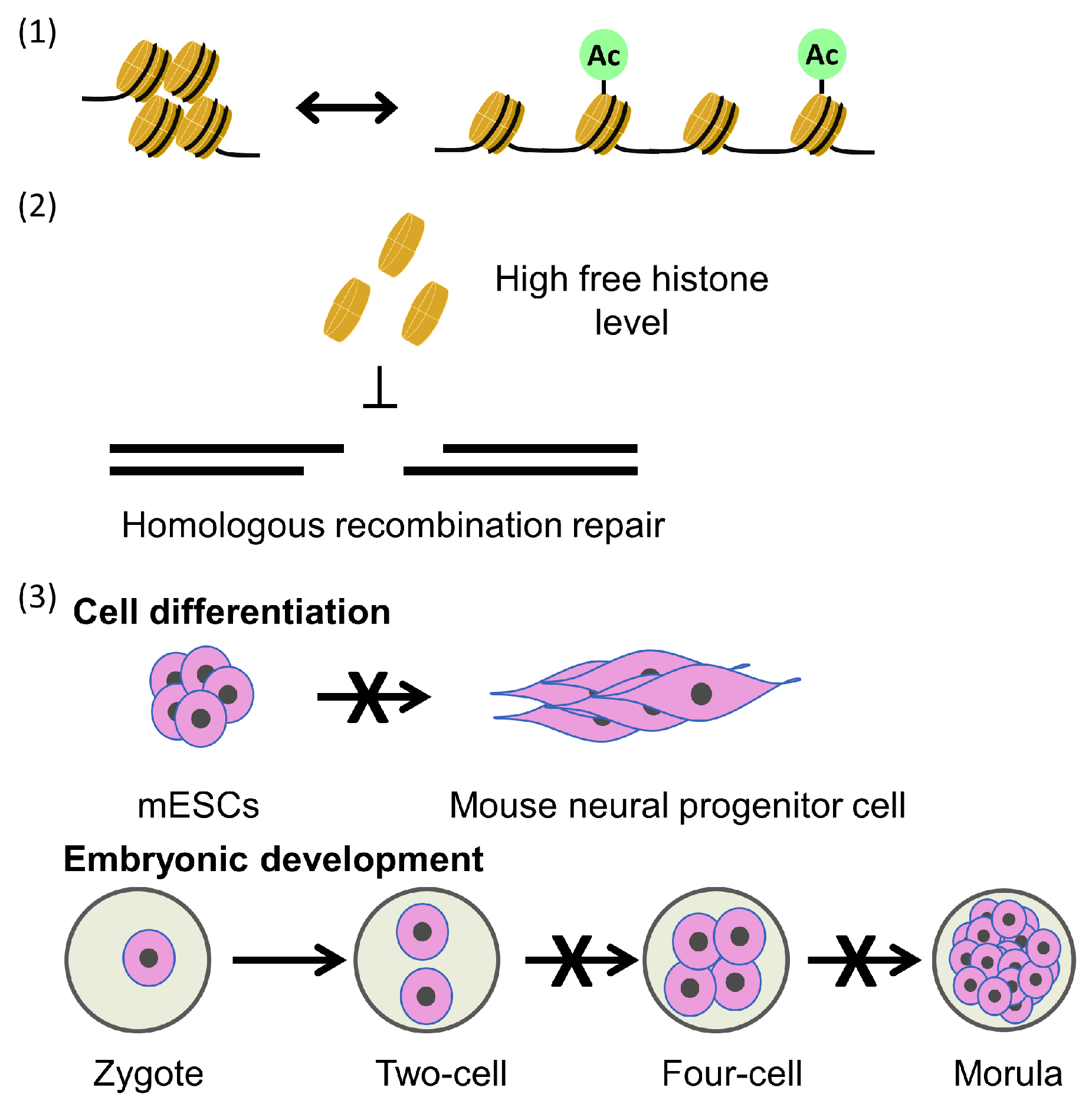
6. Consequence of Defects in Parental Histone Transfer
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bird, A. Perceptions of epigenetics. Nature 2007, 447, 396–398. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Gaullier, G.; Luger, K. Nucleosome structure and dynamics are coming of age. Nat. Struct. Mol. Biol. 2019, 26, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Serra-Cardona, A.; Zhang, Z. Replication-Coupled Nucleosome Assembly in the Passage of Epigenetic Information and Cell Identity. Trends Biochem. Sci. 2018, 43, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Feng, J.; Li, Q. The replisome guides nucleosome assembly during DNA replication. Cell Biosci. 2020, 10, 37. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Long, C.; Chen, X.; Huang, C.; Chen, S.; Zhu, B. Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science 2010, 328, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Annunziato, A.T. Split decision: What happens to nucleosomes during DNA replication? J. Biol. Chem. 2005, 280, 12065–12068. [Google Scholar] [CrossRef]
- Clement, C.; Almouzni, G. MCM2 binding to histones H3-H4 and ASF1 supports a tetramer-to-dimer model for histone inheritance at the replication fork. Nat. Struct. Mol. Biol. 2015, 22, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Flury, V.; Reveron-Gomez, N.; Alcaraz, N.; Stewart-Morgan, K.R.; Wenger, A.; Klose, R.J.; Groth, A. Recycling of modified H2A-H2B provides short-term memory of chromatin states. Cell 2023, 186, 1050–1065.e19. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Gao, Y.; Zhang, Y.; Yu, D.; Lin, J.; Feng, J.; Li, J.; Xu, Z.; Zhang, Y.; Dang, S.; et al. Parental histone transfer caught at the replication fork. Nature 2024, 627, 890–897. [Google Scholar] [CrossRef]
- Zhang, Z.; Shibahara, K.; Stillman, B. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature 2000, 408, 221–225. [Google Scholar] [CrossRef]
- Han, J.; Zhou, H.; Li, Z.; Xu, R.M.; Zhang, Z. Acetylation of lysine 56 of histone H3 catalyzed by RTT109 and regulated by ASF1 is required for replisome integrity. J. Biol. Chem. 2007, 282, 28587–28596. [Google Scholar] [CrossRef] [PubMed]
- Verreault, A.; Kaufman, P.D.; Kobayashi, R.; Stillman, B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 1996, 87, 95–104. [Google Scholar] [CrossRef]
- Quivy, J.P.; Grandi, P.; Almouzni, G. Dimerization of the largest subunit of chromatin assembly factor 1: Importance in vitro and during Xenopus early development. EMBO J. 2001, 20, 2015–2027. [Google Scholar] [CrossRef] [PubMed]
- Houlard, M.; Berlivet, S.; Probst, A.V.; Quivy, J.P.; Hery, P.; Almouzni, G.; Gerard, M. CAF-1 is essential for heterochromatin organization in pluripotent embryonic cells. PLoS Genet. 2006, 2, e181. [Google Scholar] [CrossRef] [PubMed]
- Ono, T.; Kaya, H.; Takeda, S.; Abe, M.; Ogawa, Y.; Kato, M.; Kakutani, T.; Mittelsten Scheid, O.; Araki, T.; Shibahara, K. Chromatin assembly factor 1 ensures the stable maintenance of silent chromatin states in Arabidopsis. Genes Cells 2006, 11, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Zhang, H.; Wang, Z.; Zhou, H.; Zhang, Z. A Cul4 E3 ubiquitin ligase regulates histone hand-off during nucleosome assembly. Cell 2013, 155, 817–829. [Google Scholar] [CrossRef] [PubMed]
- Tyler, J.K.; Adams, C.R.; Chen, S.R.; Kobayashi, R.; Kamakaka, R.T.; Kadonaga, J.T. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 1999, 402, 555–560. [Google Scholar] [CrossRef]
- Jackson, V.; Granner, D.K.; Chalkley, R. Deposition of histones onto replicating chromosomes. Proc. Natl. Acad. Sci. USA 1975, 72, 4440–4444. [Google Scholar] [CrossRef] [PubMed]
- Su, D.; Hu, Q.; Li, Q.; Thompson, J.R.; Cui, G.; Fazly, A.; Davies, B.A.; Botuyan, M.V.; Zhang, Z.; Mer, G. Structural basis for recognition of H3K56-acetylated histone H3-H4 by the chaperone Rtt106. Nature 2012, 483, 104–107. [Google Scholar] [CrossRef]
- Huang, S.; Zhou, H.; Katzmann, D.; Hochstrasser, M.; Atanasova, E.; Zhang, Z. Rtt106p is a histone chaperone involved in heterochromatin-mediated silencing. Proc. Natl. Acad. Sci. USA 2005, 102, 13410–13415. [Google Scholar] [CrossRef]
- Liu, S.; Xu, Z.; Leng, H.; Zheng, P.; Yang, J.; Chen, K.; Feng, J.; Li, Q. RPA binds histone H3-H4 and functions in DNA replication-coupled nucleosome assembly. Science 2017, 355, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Gan, H.; Wang, Z.; Lee, J.H.; Zhou, H.; Ordog, T.; Wold, M.S.; Ljungman, M.; Zhang, Z. RPA Interacts with HIRA and Regulates H3.3 Deposition at Gene Regulatory Elements in Mammalian Cells. Mol. Cell 2017, 65, 272–284. [Google Scholar] [CrossRef] [PubMed]
- Riley, D.; Weintraub, H. Conservative segregation of parental histones during replication in the presence of cycloheximide. Proc. Natl. Acad. Sci. USA 1979, 76, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Cusick, M.E.; DePamphilis, M.L.; Wassarman, P.M. Dispersive segregation of nucleosomes during replication of simian virus 40 chromosomes. J. Mol. Biol. 1984, 178, 249–271. [Google Scholar] [CrossRef] [PubMed]
- Annunziato, A.T. The Fork in the Road: Histone Partitioning During DNA Replication. Genes 2015, 6, 353–371. [Google Scholar] [CrossRef]
- Sirbu, B.M.; Couch, F.B.; Feigerle, J.T.; Bhaskara, S.; Hiebert, S.W.; Cortez, D. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 2011, 25, 1320–1327. [Google Scholar] [CrossRef] [PubMed]
- Alabert, C.; Bukowski-Wills, J.C.; Lee, S.B.; Kustatscher, G.; Nakamura, K.; de Lima Alves, F.; Menard, P.; Mejlvang, J.; Rappsilber, J.; Groth, A. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 2014, 16, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Zardoni, L.; Nardini, E.; Liberi, G. 2D Gel Electrophoresis to Detect DNA Replication and Recombination Intermediates in Budding Yeast. Methods Mol. Biol. 2020, 2119, 43–59. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Gan, H.; Han, J.; Zhou, Z.X.; Jia, S.; Chabes, A.; Farrugia, G.; Ordog, T.; Zhang, Z. Strand-Specific Analysis Shows Protein Binding at Replication Forks and PCNA Unloading from Lagging Strands when Forks Stall. Mol. Cell 2014, 56, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Gan, H.; Zhang, Z. Both DNA Polymerases delta and epsilon Contact Active and Stalled Replication Forks Differently. Mol. Cell Biol. 2017, 37, e00190-17. [Google Scholar] [CrossRef]
- Gan, H.; Serra-Cardona, A.; Hua, X.; Zhou, H.; Labib, K.; Yu, C.; Zhang, Z. The Mcm2-Ctf4-Polalpha Axis Facilitates Parental Histone H3-H4 Transfer to Lagging Strands. Mol. Cell 2018, 72, 140–151.e3. [Google Scholar] [CrossRef] [PubMed]
- Petryk, N.; Dalby, M.; Wenger, A.; Stromme, C.B.; Strandsby, A.; Andersson, R.; Groth, A. MCM2 promotes symmetric inheritance of modified histones during DNA replication. Science 2018, 361, 1389–1392. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Gan, H.; Serra-Cardona, A.; Zhang, L.; Gan, S.; Sharma, S.; Johansson, E.; Chabes, A.; Xu, R.M.; Zhang, Z. A mechanism for preventing asymmetric histone segregation onto replicating DNA strands. Science 2018, 361, 1386–1389. [Google Scholar] [CrossRef] [PubMed]
- Deegan, T.D.; Diffley, J.F. MCM: One ring to rule them all. Curr. Opin. Struct. Biol. 2016, 37, 145–151. [Google Scholar] [CrossRef]
- Evrin, C.; Clarke, P.; Zech, J.; Lurz, R.; Sun, J.; Uhle, S.; Li, H.; Stillman, B.; Speck, C. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc. Natl. Acad. Sci. USA 2009, 106, 20240–20245. [Google Scholar] [CrossRef] [PubMed]
- Remus, D.; Beuron, F.; Tolun, G.; Griffith, J.D.; Morris, E.P.; Diffley, J.F. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell 2009, 139, 719–730. [Google Scholar] [CrossRef]
- Speck, C.; Chen, Z.; Li, H.; Stillman, B. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat. Struct. Mol. Biol. 2005, 12, 965–971. [Google Scholar] [CrossRef]
- Chen, S.; de Vries, M.A.; Bell, S.P. Orc6 is required for dynamic recruitment of Cdt1 during repeated Mcm2-7 loading. Genes Dev. 2007, 21, 2897–2907. [Google Scholar] [CrossRef] [PubMed]
- Tye, B.K.; Sawyer, S. The hexameric eukaryotic MCM helicase: Building symmetry from nonidentical parts. J. Biol. Chem. 2000, 275, 34833–34836. [Google Scholar] [CrossRef]
- Tsuji, T.; Ficarro, S.B.; Jiang, W. Essential role of phosphorylation of MCM2 by Cdc7/Dbf4 in the initiation of DNA replication in mammalian cells. Mol. Biol. Cell 2006, 17, 4459–4472. [Google Scholar] [CrossRef]
- Tanaka, S.; Tak, Y.S.; Araki, H. The role of CDK in the initiation step of DNA replication in eukaryotes. Cell Div. 2007, 2, 16. [Google Scholar] [CrossRef] [PubMed]
- Yeeles, J.T.; Deegan, T.D.; Janska, A.; Early, A.; Diffley, J.F. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 2015, 519, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Maine, G.T.; Sinha, P.; Tye, B.K. Mutants of S. cerevisiae defective in the maintenance of minichromosomes. Genetics 1984, 106, 365–385. [Google Scholar] [CrossRef] [PubMed]
- Kahm, Y.J.; Kim, I.G.; Kim, R.K. Regulation of cancer stem cells by CXCL1, a chemokine whose secretion is controlled by MCM2. BMC Cancer 2024, 24, 319. [Google Scholar] [CrossRef] [PubMed]
- Kearsey, S.E.; Labib, K. MCM proteins: Evolution, properties, and role in DNA replication. Biochim. Biophys. Acta 1998, 1398, 113–136. [Google Scholar] [CrossRef] [PubMed]
- Mincheva, A.; Todorov, I.; Werner, D.; Fink, T.M.; Lichter, P. The human gene for nuclear protein BM28 (CDCL1), a new member of the early S-phase family of proteins, maps to chromosome band 3q21. Cytogenet. Cell Genet. 1994, 65, 276–277. [Google Scholar] [CrossRef] [PubMed]
- Foss, E.J.; Sripathy, S.; Gatbonton-Schwager, T.; Kwak, H.; Thiesen, A.H.; Lao, U.; Bedalov, A. Chromosomal Mcm2-7 distribution and the genome replication program in species from yeast to humans. PLoS Genet. 2021, 17, e1009714. [Google Scholar] [CrossRef]
- Foltman, M.; Evrin, C.; De Piccoli, G.; Jones, R.C.; Edmondson, R.D.; Katou, Y.; Nakato, R.; Shirahige, K.; Labib, K. Eukaryotic replisome components cooperate to process histones during chromosome replication. Cell Rep. 2013, 3, 892–904. [Google Scholar] [CrossRef]
- Tye, B.K. MCM proteins in DNA replication. Annu. Rev. Biochem. 1999, 68, 649–686. [Google Scholar] [CrossRef]
- Poplawski, A.; Grabowski, B.; Long, S.E.; Kelman, Z. The zinc finger domain of the archaeal minichromosome maintenance protein is required for helicase activity. J. Biol. Chem. 2001, 276, 49371–49377. [Google Scholar] [CrossRef]
- Richet, N.; Liu, D.; Legrand, P.; Velours, C.; Corpet, A.; Gaubert, A.; Bakail, M.; Moal-Raisin, G.; Guerois, R.; Compper, C.; et al. Structural insight into how the human helicase subunit MCM2 may act as a histone chaperone together with ASF1 at the replication fork. Nucleic Acids Res. 2015, 43, 1905–1917. [Google Scholar] [CrossRef] [PubMed]
- Karri, S.; Yang, Y.; Zhou, J.; Dickinson, Q.; Jia, J.; Huang, Y.; Wang, Z.; Gan, H.; Yu, C. Defective transfer of parental histone decreases frequency of homologous recombination by increasing free histone pools in budding yeast. Nucleic Acids Res. 2024, 52, 5138–5151. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hua, X.; Serra-Cardona, A.; Xu, X.; Gan, S.; Zhou, H.; Yang, W.S.; Chen, C.L.; Xu, R.M.; Zhang, Z. DNA polymerase alpha interacts with H3-H4 and facilitates the transfer of parental histones to lagging strands. Sci. Adv. 2020, 6, eabb5820. [Google Scholar] [CrossRef] [PubMed]
- Porcella, S.Y.; Koussa, N.C.; Tang, C.P.; Kramer, D.N.; Srivastava, P.; Smith, D.J. Separable, Ctf4-mediated recruitment of DNA Polymerase alpha for initiation of DNA synthesis at replication origins and lagging-strand priming during replication elongation. PLoS Genet. 2020, 16, e1008755. [Google Scholar] [CrossRef] [PubMed]
- Villa, F.; Simon, A.C.; Ortiz Bazan, M.A.; Kilkenny, M.L.; Wirthensohn, D.; Wightman, M.; Matak-Vinkovic, D.; Pellegrini, L.; Labib, K. Ctf4 Is a Hub in the Eukaryotic Replisome that Links Multiple CIP-Box Proteins to the CMG Helicase. Mol. Cell 2016, 63, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Zhang, Q.; Jia, J.; Zhou, J.; Zhang, Z.; Karri, S.; Jiang, J.; Dickinson, Q.; Yao, Y.; Tang, X.; et al. DNA polymerase delta governs parental histone transfer to DNA replication lagging strand. Proc. Natl. Acad. Sci. USA 2024, 121, e2400610121. [Google Scholar] [CrossRef] [PubMed]
- Serra-Cardona, A.; Hua, X.; McNutt, S.W.; Zhou, H.; Toda, T.; Jia, S.; Chu, F.; Zhang, Z. The PCNA-Pol delta complex couples lagging strand DNA synthesis to parental histone transfer for epigenetic inheritance. Sci. Adv. 2024, 10, eadn5175. [Google Scholar] [CrossRef] [PubMed]
- Wen, Q.; Zhou, J.; Tian, C.; Li, X.; Song, G.; Gao, Y.; Sun, Y.; Ma, C.; Yao, S.; Liang, X.; et al. Symmetric inheritance of parental histones contributes to safeguarding the fate of mouse embryonic stem cells during differentiation. Nat. Genet. 2023, 55, 1555–1566. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.J.; Whitehouse, I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature 2012, 483, 434–438. [Google Scholar] [CrossRef]
- Fu, Y.V.; Yardimci, H.; Long, D.T.; Ho, T.V.; Guainazzi, A.; Bermudez, V.P.; Hurwitz, J.; van Oijen, A.; Scharer, O.D.; Walter, J.C. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 2011, 146, 931–941. [Google Scholar] [CrossRef]
- Wang, P.; Yang, W.; Zhao, S.; Nashun, B. Regulation of chromatin structure and function: Insights into the histone chaperone FACT. Cell Cycle 2021, 20, 465–479. [Google Scholar] [CrossRef] [PubMed]
- Formosa, T.; Winston, F. The role of FACT in managing chromatin: Disruption, assembly, or repair? Nucleic Acids Res. 2020, 48, 11929–11941. [Google Scholar] [CrossRef] [PubMed]
- Sivkina, A.L.; Karlova, M.G.; Valieva, M.E.; McCullough, L.L.; Formosa, T.; Shaytan, A.K.; Feofanov, A.V.; Kirpichnikov, M.P.; Sokolova, O.S.; Studitsky, V.M. Electron microscopy analysis of ATP-independent nucleosome unfolding by FACT. Commun. Biol. 2022, 5, 2. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tang, Y.; Xu, J.; Leng, H.; Shi, G.; Hu, Z.; Wu, J.; Xiu, Y.; Feng, J.; Li, Q. The N-terminus of Spt16 anchors FACT to MCM2-7 for parental histone recycling. Nucleic Acids Res. 2023, 51, 11549–11567. [Google Scholar] [CrossRef] [PubMed]
- Foss, E.J. Tof1p regulates DNA damage responses during S phase in Saccharomyces cerevisiae. Genetics 2001, 157, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Katou, Y.; Kanoh, Y.; Bando, M.; Noguchi, H.; Tanaka, H.; Ashikari, T.; Sugimoto, K.; Shirahige, K. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 2003, 424, 1078–1083. [Google Scholar] [CrossRef]
- Xie, J.; Wooten, M.; Tran, V.; Chen, B.C.; Pozmanter, C.; Simbolon, C.; Betzig, E.; Chen, X. Histone H3 Threonine Phosphorylation Regulates Asymmetric Histone Inheritance in the Drosophila Male Germline. Cell 2015, 163, 920–933. [Google Scholar] [CrossRef] [PubMed]
- Alabert, C.; Groth, A. Chromatin replication and epigenome maintenance. Nat. Rev. Mol. Cell Biol. 2012, 13, 153–167. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, K.; Ohsumi, T.; Tada, S.; Natsume, R.; Kundu, L.R.; Nozaki, N.; Senda, T.; Enomoto, T.; Horikoshi, M.; Seki, M. Roles of histone chaperone CIA/Asf1 in nascent DNA elongation during nucleosome replication. Genes Cells 2011, 16, 1050–1062. [Google Scholar] [CrossRef]
- Groth, A.; Corpet, A.; Cook, A.J.; Roche, D.; Bartek, J.; Lukas, J.; Almouzni, G. Regulation of replication fork progression through histone supply and demand. Science 2007, 318, 1928–1931. [Google Scholar] [CrossRef]
- Huang, H.; Stromme, C.B.; Saredi, G.; Hodl, M.; Strandsby, A.; Gonzalez-Aguilera, C.; Chen, S.; Groth, A.; Patel, D.J. A unique binding mode enables MCM2 to chaperone histones H3-H4 at replication forks. Nat. Struct. Mol. Biol. 2015, 22, 618–626. [Google Scholar] [CrossRef] [PubMed]
- Ricketts, M.D.; Dasgupta, N.; Fan, J.; Han, J.; Gerace, M.; Tang, Y.; Black, B.E.; Adams, P.D.; Marmorstein, R. The HIRA histone chaperone complex subunit UBN1 harbors H3/H4- and DNA-binding activity. J. Biol. Chem. 2019, 294, 9239–9259. [Google Scholar] [CrossRef] [PubMed]
- Saxton, D.S.; Rine, J. Epigenetic memory independent of symmetric histone inheritance. eLife 2019, 8, 51421. [Google Scholar] [CrossRef]
- Ragunathan, K.; Jih, G.; Moazed, D. Epigenetics. Epigenetic inheritance uncoupled from sequence-specific recruitment. Science 2015, 348, 1258699. [Google Scholar] [CrossRef] [PubMed]
- Hansen, K.H.; Bracken, A.P.; Pasini, D.; Dietrich, N.; Gehani, S.S.; Monrad, A.; Rappsilber, J.; Lerdrup, M.; Helin, K. A model for transmission of the H3K27me3 epigenetic mark. Nat. Cell Biol. 2008, 10, 1291–1300. [Google Scholar] [CrossRef]
- Fang, Y.; Hua, X.; Shan, C.M.; Toda, T.; Qiao, F.; Zhang, Z.; Jia, S. Coordination of histone chaperones for parental histone segregation and epigenetic inheritance. Genes Dev. 2024, 38, 189–204. [Google Scholar] [CrossRef]
- Xu, X.; Hua, X.; Brown, K.; Ren, X.; Zhang, Z. Mcm2 promotes stem cell differentiation via its ability to bind H3-H4. eLife 2022, 11, 80917. [Google Scholar] [CrossRef] [PubMed]
- Wenger, A.; Biran, A.; Alcaraz, N.; Redo-Riveiro, A.; Sell, A.C.; Krautz, R.; Flury, V.; Reveron-Gomez, N.; Solis-Mezarino, V.; Volker-Albert, M.; et al. Symmetric inheritance of parental histones governs epigenome maintenance and embryonic stem cell identity. Nat. Genet. 2023, 55, 1567–1578. [Google Scholar] [CrossRef]
- Goehring, L.; Huang, T.T.; Smith, D.J. Transcription-Replication Conflicts as a Source of Genome Instability. Annu. Rev. Genet. 2023, 57, 157–179. [Google Scholar] [CrossRef]


| Gene | Deletion/Mutation | Mutation | Phenotype | Other Organisms |
|---|---|---|---|---|
| CTF4 | ctf4-4E | Point mutations: L867E, A871E, A897E and I901E | CTF4-CMG helicase and CTF4-Pol1p interactions are disrupted. parH3:H4tet show a leading strand bias | |
| Dpb3 | dpb3∆ | parH3:H4tet show a lagging strand bias | mouse | |
| Dpb4 | dpb4∆ | parH3:H4tet show a lagging strand bias | mouse | |
| Spt16 | Spt16 N-terminal deletion | Deletion of Spt16 N-terminal region | a slight a leading strand bias | |
| Mcm2 | mcm2-3A | Point mutations Y79A, Y82A, and Y91A | parH3:H4tet show a leading strand bias | Y81A, Y89A, and Y137A in humans, mouse |
| mcm2-2A | Point mutations Y79A, Y82A | parH3:H4tet show a leading strand bias | Y81A, Y89A in humans, mouse | |
| Pol1 | pol1-4A | Point mutations D141A, D142A, L144A and F147A | CTF4-Pol α interaction is disrupted. parH3:H4tet show a leading strand bias |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, J.; Yu, C. The Role of the MCM2-7 Helicase Subunit MCM2 in Epigenetic Inheritance. Biology 2024, 13, 572. https://doi.org/10.3390/biology13080572
Jia J, Yu C. The Role of the MCM2-7 Helicase Subunit MCM2 in Epigenetic Inheritance. Biology. 2024; 13(8):572. https://doi.org/10.3390/biology13080572
Chicago/Turabian StyleJia, Jing, and Chuanhe Yu. 2024. "The Role of the MCM2-7 Helicase Subunit MCM2 in Epigenetic Inheritance" Biology 13, no. 8: 572. https://doi.org/10.3390/biology13080572





