Simple Summary
Chalk5 is an important gene used in rice breeding to improve rice chalkiness, which greatly affects rice quality. For this study, the first exon of Chalk5 was edited, and two different knockout mutants were obtained. These two lines showed a decreased percentage of grains with chalkiness, chalkiness degree, and seed setting ratio. Moreover, their chalkiness was insensitive to temperature during the grain-filling stage, and the head milled rice rate was improved even under high-temperature conditions. The two lines showed significantly reduced PGWC and DEC without changes in other agronomic traits in the hybrid background. These results indicate that Chalk5 gene-edited lines are useful in indica hybrid rice breeding for a high yield and superior grain quality in high-temperature growing environments.
Abstract
Chalkiness is an important grain quality trait in rice. Chalk5, encoding a vacuolar H+-translocating pyrophosphatase, is a major gene affecting both the percentage of grains with chalkiness (PGWC) and chalkiness degree (DEC) in rice. Reducing its expression can decrease both PGEC and DEC. In this study, the first exon of Chalk5 was edited in the elite restorer line 9311 using the CRISPR/Cas9 system and two knockout mutants were obtained, one of which did not contain the exogenous Cas9 cassette. PGWC and DEC were both significantly reduced in both mutants, while the seed setting ratio (SSR) was also significantly decreased. Staggered sowing experiments showed that the chalkiness of the mutants was insensitive to temperature during the grain-filling stage, and the head milled rice rate (HMRR) could be improved even under high-temperature conditions. Finally, in the hybrid background, the mutants showed significantly reduced PGWC and DEC without changes in other agronomic traits. The results provide important germplasm and allele resources for breeding high-yield rice varieties with superior quality, especially for high-yield indica hybrid rice varieties with superior quality in high-temperature conditions.
1. Introduction
Rice is one of the most important staple crops and is a carbohydrate source for more than half of the world’s population [1]. In Asia, it is the staple food for about 90% of the population. With the improvement in living standards, people are paying increasing attention to high-quality rice [2]. Thus, superior quality has become a key breeding target that is equal to a high yield. Rice quality parameters include milling quality, appearance quality, cooking and eating quality, and nutritional quality. Among them, chalkiness is an important appearance quality trait that directly affects the market value of rice. Chalkiness results from incomplete filling during grain development [3], and can be classified as white belly, white core, and white back. It is usually measured using the percentage of grains with chalkiness (PGWC) and chalkiness degree (DEC). High chalkiness not only reduces the head milled rice yield, but also affects palatability. Thus, breeding rice varieties with low chalkiness is crucial to meet the demand for superior quality rice.
Chalkiness is a typical quantitative trait controlled by multiple genes [4]. In recent years, researchers have identified numerous quantitative trait loci (QTLs) affecting chalkiness and have cloned several genes using various genetic populations and different strategies [2,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18]. Among them, Chalk5 is a major gene controlling both PGWC and DEC, which has been cloned using a population derived from Zhenshan97 with high chalkiness and H94 with low chalkiness [19]. Chalk5 encodes a vacuolar H+-translocating pyrophosphatase, which affects protein body formation and vesicle-like structure accumulation through disrupting the pH homeostasis of the endomembrane system during seed development, leading to the formation of air spaces in the endosperm and ultimately causing chalkiness. The expression level of Chalk5 is significantly correlated with the degree of chalkiness, and a higher expression results in more chalkiness. A haplotype analysis showed that two SNPs (C/T and A/T) at 721 and 485 upstream of the start codon of Chalk5 were significantly associated with grain chalkiness.
Chalk5 is an important gene for improving rice appearance quality in molecular breeding. The introgression of the low-chalkiness indica allele of Chalk5 significantly reduced chalkiness and further optimized the milling quality of indica–japonica hybrid rice [20]. Moreover, different combinations of alleles of GS3, Chalk5, and Chalk7 had large effects on grain chalkiness [21]. A favorable allele combination of the three genes could greatly reduce rice chalkiness without negatively affecting the grain yield.
In addition to genetic factors, the temperature during the grain-filling stage also significantly affects chalkiness in rice [22]; in particular, the temperature during the initial 15 days of grain filling has a significant impact on chalkiness. When the average temperature exceeds 26 °C, chalkiness increases significantly. Under high-temperature conditions, the expression of Chalk5 increases, while the expression of starch synthesis-related genes decreases and the expression of sucrose and starch degradation-related genes increases, leading to reduced accumulation of sucrose and starch and ultimately resulting in chalkiness [23].
CRISPR/Cas9 is an effective gene editing technique that has emerged in recent years, which can precisely modify target genes to create new mutations [24]. Compared to marker-assisted breeding, CRISPR/Cas9 can precisely modify target genes and avoid negative effects through the introduction of other negative genes into the background; therefore, it has wide application prospects in functional studies and crop improvement. For example, Chalk5 is closely linked to the genes GS5 and qSW5 that control grain width [25,26,27]. This linkage of different genes results in a positive correlation between chalkiness and grain width. However, if we want to decrease chalkiness, the reduction in grain width will lead to a decrease in thousand grain weight, ultimately affecting the rice yield. Therefore, editing Chalk5 using the CRISPR/Cas9 system can improve quality without reducing the yield.
The functional divergency of Chalk5 in cultivars is mainly caused by the polymorphism sites in the promoter. A strong functional allele of Chalk5 is highly expressed at the mRNA level with high chalkiness. A recent study editing the promoter of Chalk5 using this CRISPR/Cas9 system also validated it [28]. The expression of Chalk5 is suppressed with considerable effect, but whether the knockout of Chalk5 would produce a more valuable effect for high-quality rice breeding is still unknown. In this study, we used the CRISPR/Cas9 system to edit the first exon of Chalk5 in the backbone restorer line 9311, and we evaluated the yield and chalkiness of the knockout lines under different filling temperatures to assess their sensitivity to the temperature. Further, yield and quality traits were evaluated in the hybrid backgrounds through crossing the gene editing lines with two-line sterile lines. The results provide important germplasm and allele resources for breeding high-yield rice varieties with superior quality, especially for high yields with superior quality indica hybrid rice in high-temperature conditions.
2. Materials and Methods
2.1. Materials
The elite two-line restorer line 9311 was used as the transgenic recipient and the control. In addition, hybrid combinations were made between two photo–thermo-sensitive genic male sterile lines, LX03S and Huhan82S, and the edited lines. LX03S was bred by our group and Huhan82S was introduced from the Shanghai Agrobiological Gene Center, China.
2.2. Gene Editing
Based on the Chalk5 sequence provided by the TIGR database (http://rice.plantbiology.msu.edu/, accessed on 10 November 2022), a 20 bp target site was designed in the first exon of Chalk5, with the sequence ATGACGGAGTACAAGTACCT and with the protospacer adjacent motif “GGG” according to a previous study [29]. The ligation reaction conditions were 37 °C for 20 min, 5 cycles; 37 °C for 10 min, 20 °C for 10 min, 50 cycles; 37 °C for 20 min; and 80 °C for 5 min. The successfully ligated products were transformed into chemically competent E. coli cells using heat shock, and the transformed E. coli cells were plated on LB solid medium containing kanamycin to screen for positive colonies. The plasmids extracted from the positive colonies were introduced into EHA105 Agrobacterium tumefaciens competent cells, and the constructed pBWA(V)H_cas9i2-Chalk5 plasmid was then transformed into the rice materials via Agrobacterium-mediated transformation, with positive mutant lines selected using hygromycin. The gene editing in this experiment was finished by Wuhan BioRun Bioscience Co., Ltd. (Wuhan, China).
After obtaining the transgenic seedlings, the mutation sites were identified using sequencing. Leaf samples were collected, and DNA was extracted using the CTAB method [4]. The PCR amplification and sequencing were performed according to the method described by Qiu et al. [30], using the primers F: 5′-CGCCGTGCTCCAGTGGTAC and R: 5′-TGAACGCCGCGTTCGCCAG. The primers F: 5′-ATCCAGCACGCCATCTCCGTTGG and R: 5′-CGTAGTAGAGCCCGAACGCGTTCA were used to detect whether the vector in homozygous mutants was present.
2.3. Field Experiment and Trait Evaluation
In summer 2020, three T0-edited seedlings were planted at the Yangtze University experimental farm in Jingzhou (30.18° E, 112.15° E), Hubei. After a sequencing analysis, seeds from two positive plants were harvested. In winter 2020, 20 T1 plants per line were planted in a phytotron at Yangtze University. After a sequencing analysis and phenotypic evaluation, two homozygous plants with agronomic traits similar to 9311 were selected, and their seeds were harvested. In summer 2021, two T2 lines (TC1 and TC5) were planted in the Yangtze University experimental field. Materials were sowed on 15 May, and each line was planted in 5 rows with 8 plants per row at a spacing of 20 cm × 20 cm. Field management followed standard practices.
After maturity, single-plant harvesting was conducted, and 10 plants with identical growth were selected to evaluate the panicle number (PN), spikelet number per panicle (SN), filled grain number per panicle (GN), seed setting ratio (SSR), thousand grain weight (KGW), and grain yield (GY). After a yield evaluation, seeds from two plants were mixed and stored at room temperature for 3 months before evaluating the milling quality and appearance quality, including brown rice rate (BRR), milled rice rate (MRR), head milled rice rate (HMRR), grain length (GL), grain width (GW), length-to-width ratio (LWR), percentage of grains with chalkiness (PGWC), and chalkiness degree (DEC). These traits were measured as described by Qiu et al. [9,31].
In 2022, the original 9311 and the two edited lines were sown in four staggered sowings, with an interval of 15 days between each sowing, starting from 27 April with two replicates for each batch of sowing. The remaining planting methods were the same as in summer 2021. The heading date of each line and the temperatures during the first 15 days of grain filling were recorded. After the seeds were fully matured, two plants from each line were randomly mix-harvested. After storing at room temperature for 3 months, the HMRR, PGWC, and DEC were measured.
Furthermore, in summer 2021, 9311 and TC1 (Cas9 backbone free) were test-crossed with LX03S and Huhan82S, respectively. In summer 2022, the generated F1 hybrids were planted at the Yangtze University experimental farm in Jingzhou, Hubei. They were seeding on 27 April, following the same methods as in summer 2021. After maturity, 10 plants with identical growth were selected from each line to evaluate the PN, SN, GN, SSR, KGW, and GY values. Seeds from two plants were mixed and stored at room temperature for 3 months and then measured for the HMRR, PGWC, and DEC values.
2.4. Data Analysis
The raw data were organized using Excel 2016, and a statistical description and t-tests were performed using the Statistica 5.5 software [4].
3. Results
3.1. Generation of Chalk5 Knockout Homozygous Mutants
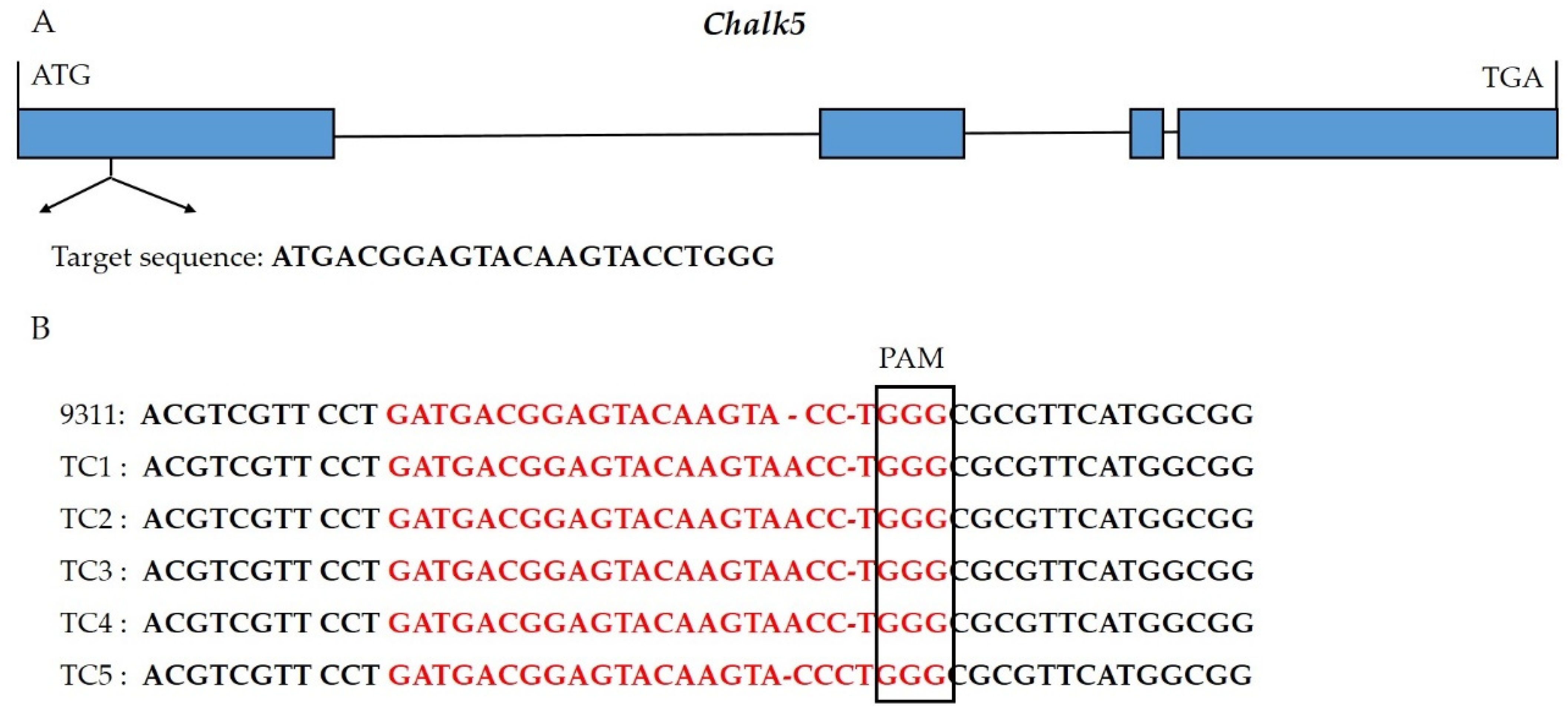
Using the CRISPR/Cas9 system, the first exon of Chalk5 was edited in 9311 and three transgenic materials were obtained. Through target site sequencing, two mutants were identified and harvested. The Chalk5 alleles in the two mutants encoded 98 and 273 amino acids (Supplementary File S1). Two homozygous mutants (TC1 and TC5) with agronomic traits similar to 9311 were selected from T1 lines (Figure 1). Among them, TC1 had an inserted A, resulting in the premature termination of amino acid coding. TC5 had an inserted C, leading to a frameshift mutation. Additionally, we performed genotyping using a Cas9-specific marker on the two positive transgenic single plants and found that TC1 did not contain the foreign Cas9 backbone.
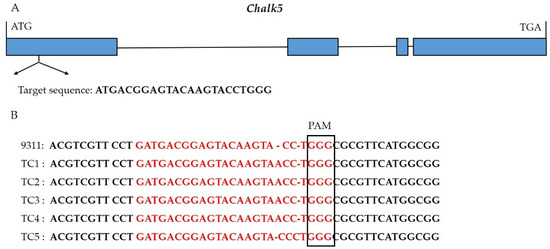
Figure 1.
Identification and generation of the knockout lines of Chalk5. (A) The gene structure of Chalk5 and position of the target sequences in the first exon. Blue boxes and black lines show the exons and introns, respectively; (B) identification of mutants in Chalk5 using sequencing of the target site in T1 transgenic individuals. PAM represents the protospacer adjacent motif. In red, base sequences were target sites.
3.2. Chalk5 Knockout Homozygous Mutants Showed Significantly Reduced Chalkiness
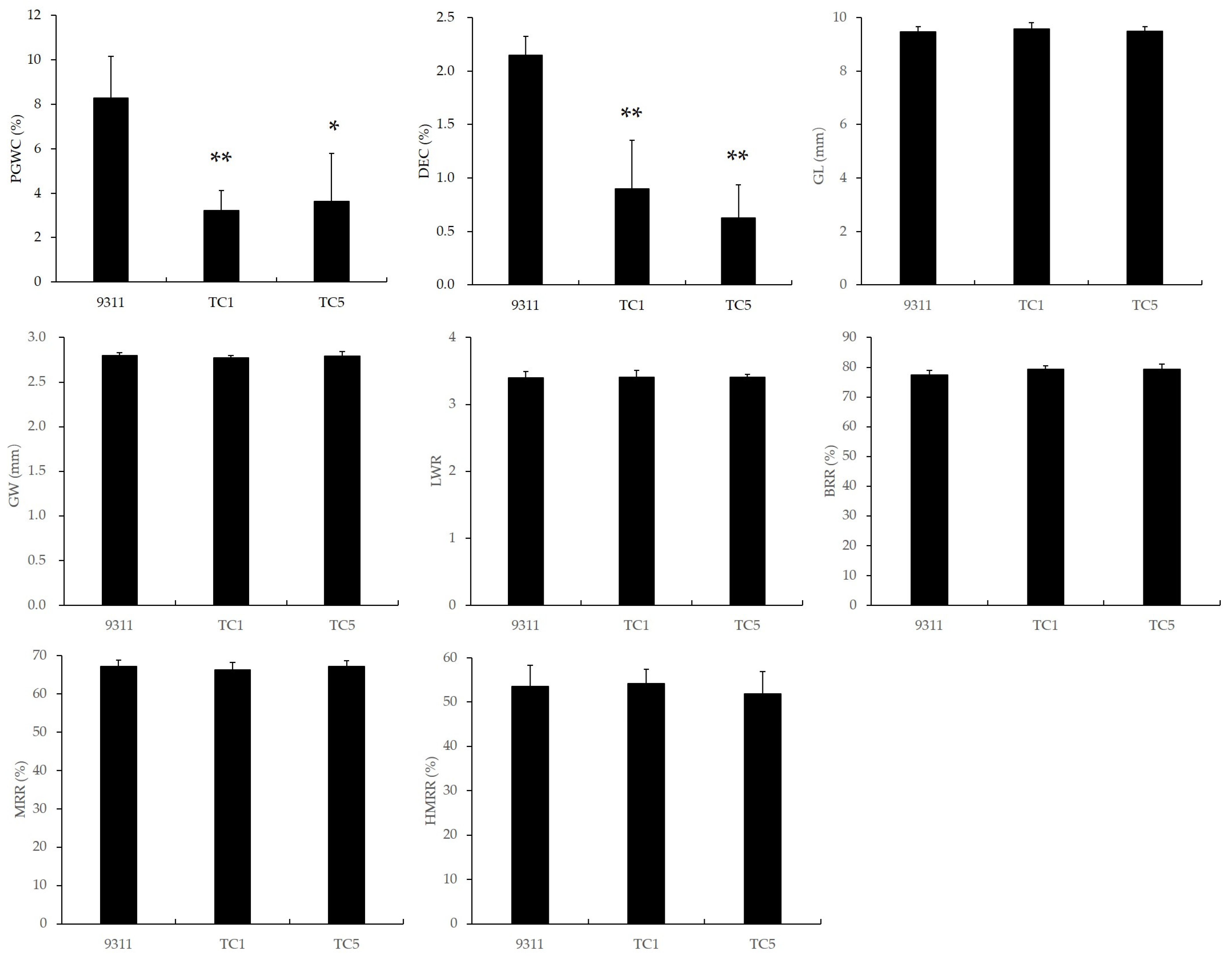
The milling and appearance quality of the two Chalk5 knockout homozygous mutants are shown in Figure 2 and Supplementary Figure S1. For chalkiness, the PGWC in the knockout lines was 3.23% and 3.63%, which were significantly reduced from the wild-type (8.28%). The DEC in TC1 and TC5 was only 0.90% and 0.63%, respectively, which was also significantly lower than the wild-type (2.15%). Thus, the knockout of Chalk5 reduced both the PGWC and DEC values efficiently. The grain shape is an important appearance quality trait and influences chalkiness. In two knockout lines, the GL, GW, and LWR values had no significant difference from the wild-type, and both are typically indica slender grain. For milling quality traits, the BRRs were 79.27% and 79.30%, the MRRs were 66.26% and 67.19%, and the HMRRs were 54.15% and 51.83%, which were similar to 9311. These results indicated that the functional loss of Chalk5 significantly reduced the PGWC and DEC values without affecting other appearance quality and milling quality traits.
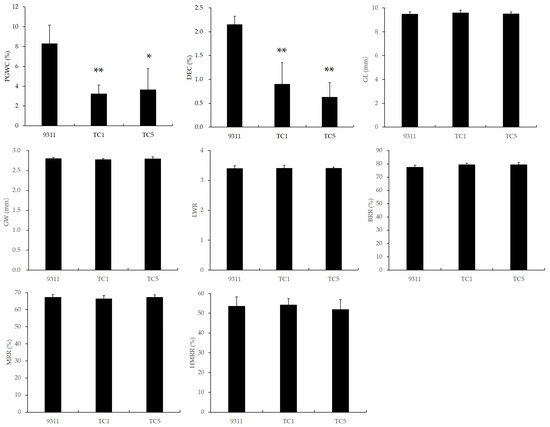
Figure 2.
The appearance and milling qualities of two knockout lines of Chalk5 and 9311. PGWC, percentage of grains with chalkiness; DEC, chalkiness degree; GL, grain length; GW, grain width; LWR, length-to-width ratio; BRR, brown rice rate; MRR, milled rice rate; and HMRR, head milled rice rate. * and ** represent significant level at p < 0.05 and 0.01, respectively, when compared to 9311 using a t-test. Error bars represent the standard deviation for each line (n = 20).
The yield-related traits of the two Chalk5 knockout homozygous mutants are shown in Supplementary Table S1. The PN and KGW values of the mutants were not significantly different from 9311, while the SSR was significantly decreased in mutants. The SN was higher in TC5, and the GN and GY values were significantly decreased only in TC1, but these trait variations were not common characteristics of knockout lines. These results suggested that the functional loss of Chalk5 led to a decrease in the SSR, but might not affect other yield-related traits.
3.3. Chalkiness of Chalk5 Knockout Homozygous Mutants Was Insensitive to Temperature
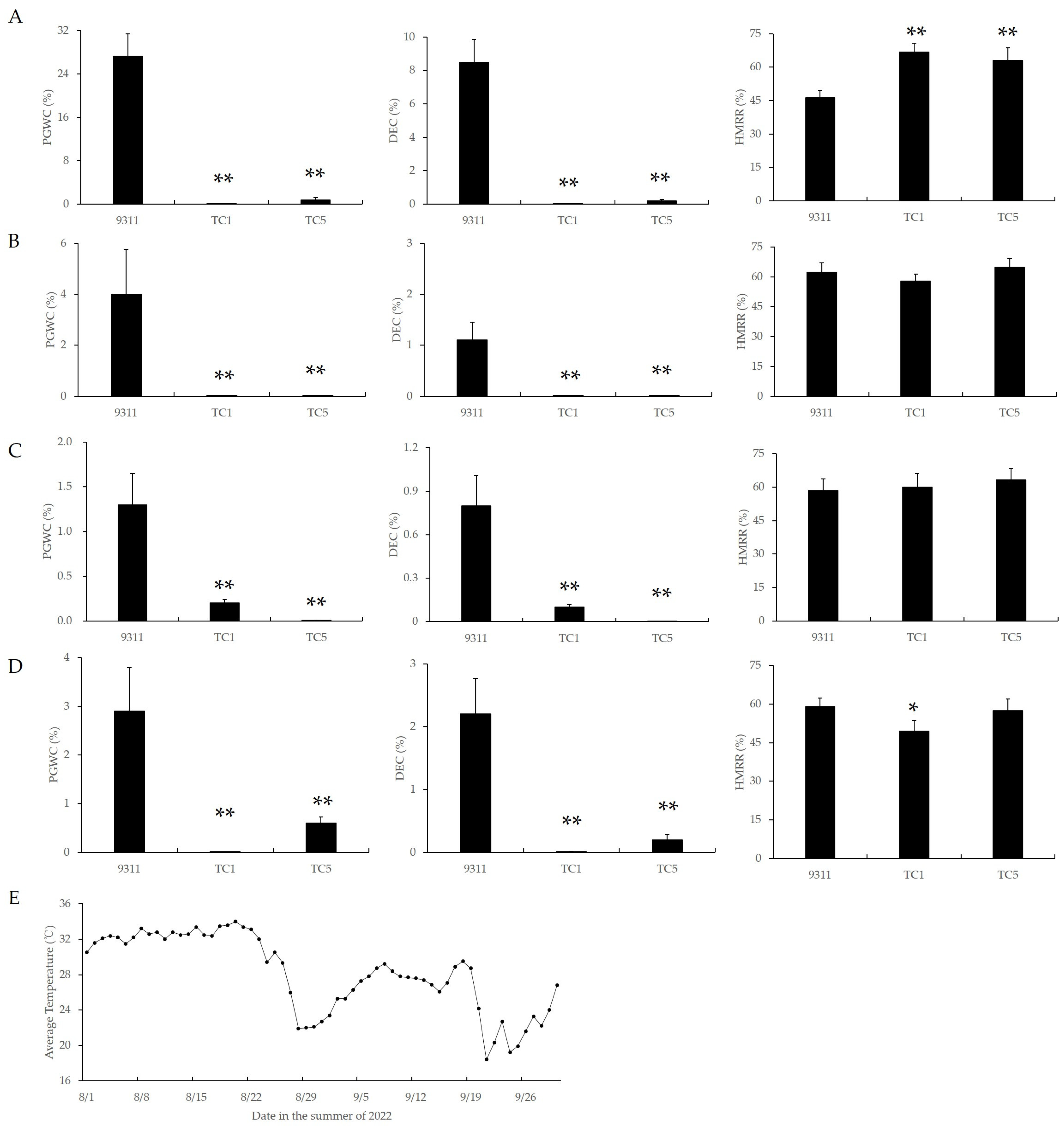
To investigate the temperature sensitivity of chalkiness in Chalk5 knockout homozygous mutants, a staggered sowing experiment was conducted. Four staggered sowings were set with an interval of 15 days between each sowing, starting from 27 April, and their heading dates were about 10 August, 20 August, 29 August, and 8 September. As the sowing date was delayed, the average temperature during the first 15 days of grain filling gradually decreased (Figure 3E). Accordingly, the PGWC and DEC values in 9311 also gradually decreased (Figure 3A–D). In all four sowing periods, the PGWC and DEC values in TC1 were significantly lower than in 9311, and there were no significant differences among different sowing periods within the same mutant. These results indicated that the chalkiness in 9311 was sensitive to temperature during the grain-filling stage, while the chalkiness of the Chalk5 knockout mutants was insensitive to temperature during this stage, suggesting that the functional loss of Chalk5 led to temperature insensitivity in chalkiness.
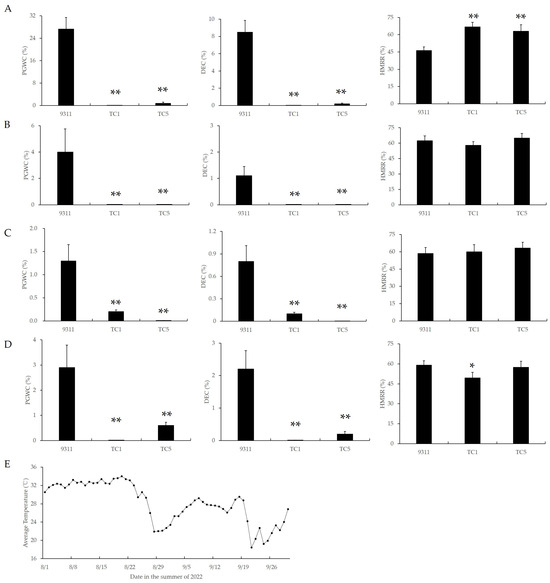
Figure 3.
The appearance and milling qualities of two knockout lines of Chalk5 and 9311 during different sowing stages. (A–D) PGWC, DEC, and HMRR values detected during the four sowing stages; (E) average temperature from 1 August to 30 September in summer 2022. The average temperatures of the first, second, third, and fourth sowing stages were 31.80 °C, 27.22 °C, 27.14 °C, and 25.49 °C during the first 15 days of the grain-filling stage. PGWC, percentage of grains with chalkiness; DEC, chalkiness degree; and HMRR, head milled rice rate. * and ** represent significance at p < 0.05 and 0.01, respectively. Error bars represent the standard deviation for each line (n = 20).
Furthermore, in the first sowing period, the HMRR value in mutants was significantly higher than that of 9311; in the second and third periods, the HMRR value in mutants was similar to 9311; and in the fourth period, the HMRR value in TC1 was slightly reduced (Figure 3; Supplementary Figure S2). These results suggested that the functional loss of Chalk5 could improve the HMRR value under high-temperature conditions.
3.4. Chalk5 Knockout Homozygous Mutants Significantly Decreased Chalkiness in Hybrid Background
To evaluate the effects of Chalk5 knockout homozygous mutants on milled rice and the appearance quality and yield traits in the hybrid background, TC1 was selected and crossed with LX03S and Huhan82S. The PGWC and DEC values of LX03S/TC1 and Huhan82S/TC1 were both significantly lower than those of LX03S/9311 and Huhan82S/9311 (Table 1), while other agronomic traits showed no significant differences. Interestingly, the HMRR value of LX03S/TC1 and Huhan82S/TC1 was similar to that of LX03S/9311 and Huhan82S/9311. These results indicated that although the functional loss of Chalk5 led to a decrease in the SSR and GY values, the hybrid combinations derived from the two-line sterile lines LX03S, Huhan82S, and TC1 showed a similar SSR, thus resulting in almost the same GY value, probably due to a complementary effect between sterile lines and the Chalk5 knockout homozygous mutants in the hybrid background.

Table 1.
The effect of Chalk5 knockout on appearance quality, milling quality, and yield-related traits in two hybrid backgrounds.
4. Discussion
In recent years, people have been paying increasing attention to rice quality. During the past two decades, the DEC value of Chinese-approved indica rice varieties has gradually decreased [32]. However, of all the quality traits of the 479 Chinese-approved indica hybrid rice varieties, the variance in DEC is the most significant, indicating that chalkiness is becoming an important trait affecting rice quality. Chalkiness is one of the most challenging qualities for hybrid rice, and very limited advances have been made in recent decades using the conventional breeding method. Chalk5 encodes a vacuolar H+-translocating pyrophosphatase that disrupts the pH homeostasis of the endomembrane system during seed development, affecting protein body formation and vesicle-like structure accumulation, leading to the formation of air spaces in the endosperm and causing chalkiness. Transgenic experiments have shown a significant positive correlation between the expression level of Chalk5 and chalkiness. Therefore, chalkiness can be reduced through decreasing the expression of Chalk5. Liu et al. [33] edited Chalk5 in the japonica rice variety Nanjing9108, and found that the mutants had significantly reduced chalkiness. Gann et al. [28] edited the promoter of Chalk5 in the japonica rice variety Nipponbare, and the expression levels of the mutants were significantly decreased, leading to significantly reduced chalkiness. In this study, we used the CRISPR/Cas9 system to edit the first exon of Chalk5 in the indica rice variety 9311 and obtained two types of Chalk5 homozygous mutants. Chalk5 encodes a peptide of 768 amino acids in length with an H+ PPase domain spanning from the 18th to the 756th amino acid. TC1 had an inserted A, resulting in a premature stop codon and producing a truncated peptide of 98 amino acids, while TC5 had an inserted C, causing a frameshift mutation and producing a truncated peptide of 273 amino acids. Both types of mutation disrupt the H+ PPase domain, leading to the functional loss of Chalk5. The PGWC value in the mutants was 3.23% and 3.63%, and the DEC value was 0.90% and 0.63%, with almost no chalkiness. These results demonstrated that we could develop a superior quality germplasm with almost no chalkiness through editing the promoter or coding region of Chalk5 to reduce its expression to extremely low levels or cause a complete loss of function.
Chalkiness is a typical quantitative trait controlled by multiple genes and is easily affected by the environment. A high temperature during grain filling has a significant impact on chalkiness. When the average temperature during the first 15–20 days of grain filling exceeds 26 °C, the PGWC and DEC values increase significantly [22]. In some varieties, the expression of Chalk5 is induced by a high temperature, leading to increased chalkiness [28]. In this study, we used the CRISPR/Cas9 system to create two types of Chalk5 loss-of-function mutants, which is equivalent to reducing the expression of Chalk5 to zero. By using staggered sowing, we allowed 9311 and the mutants to undergo grain filling under different temperature conditions. When the average temperature during the filling stage was 26–32 °C, the PGWC and DEC values in 9311 ranged from 1.30% to 27.32% and 0.83% to 8.49%, respectively, with a noticeable increase when the filling temperature was 32.9 °C. Therefore, the Chalk5 allele in 9311 is temperature sensitive. In contrast, the PGWC and DEC values in the mutants ranged from 0% to 0.80% and 0% to 0.2%, respectively (Figure 3), without significant differences at different grain-filling temperatures. Thus, the Chalk5 loss-of-function allele exhibits temperature insensitivity. Moreover, the mutants can improve the HMRR value under high-temperature conditions. The Yangtze River Valley is one of the most important rice production areas in China [34], where high temperatures in summer not only reduce the rice yield but also lead to increased chalkiness and decreased HMRR. As the chalkiness of the mutants is insensitive to temperature during the grain-filling stage and the HMRR value does not decrease at high temperatures, these mutants are important germplasm resources for improving the quality of parental lines in the Yangtze River Valley.
The edited lines showed a significant decrease in the SSR, thus resulting in a significant decrease in the GY value. Therefore, breeders should be aware of the negative impacts of quality improvement on the yield. In the hybrid background, the PGWC and DEC values were significantly reduced compared to 9311, while the SSR showed no significant difference. This means that the sterile line background can compensate for the negative effect of the decreased SSR of the edited line. Therefore, these mutants can serve as important germplasm resources for improving the appearance quality of rice, especially for hybrid varieties without a yield penalty in the Yangtze River Valley, where the temperature is normally high during the grain-filling stage. Most probably, the hybrid combination with a high yield and superior grain quality will be developed using Chalk5 gene-edited lines.
5. Conclusions
Two types of loss-of-function mutants were obtained through editing the first exon of Chalk5 using the CRISPR/Cas9 system. The PGWC and DEC values, as well as the SSR, were all significantly reduced in the mutants. The PGWC and DEC values in the mutants were insensitive to high temperatures during the grain-filling stage, and the HMRR value was significantly improved under high-temperature conditions. A hybrid combination showed a significant decrease in PGWC and DEC values, while its yield-related traits showed no significant changes. These results indicate that Chalk5 gene-edited lines are useful in indica hybrid rice breeding for high yields and superior grain quality in high-temperature growing environments.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/biology13080617/s1. Supplementary File S1: amino Acid sequences of two knockout lines of Chalk5 and 9311; Supplementary Figure S1: the appearance quality performance of two knockout lines of Chalk5 and 9311; Supplementary Figure S2: the appearance and milling quality performance of two knockout lines of Chalk5 and 9311 under high-temperature conditions; and Supplementary Table S1: the yield-related traits of two knockout lines of Chalk5 and 9311.
Author Contributions
G.F. and J.J.: Methodology, Investigation; Y.L., R.W., F.L., H.L. and J.X., Investigation; X.Q.: Writing—original draft, Writing—review and editing. Z.L.: Conceptualization, Methodology, Data curation, Writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (32171993), the Natural Science Foundation of Hubei Province (2023AFA022), The Key Project of Science and Technology Innovation 2025 in Ningbo City (2021Z001), the Key Research and Development Program of the Hubei Province (2023BBB102), the Special Project for High-quality Development of Hubei Seed Industry (HBZY2023B001-10), and open funds of the Engineering research center of ecology and agricultural use of wetland, Ministry of education (KFT202301).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The supporting data involved in this article are all original and can be provided upon request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Xing, Y.Z.; Zhang, Q.F. Genetic and molecular bases of rice yield. Ann. Rev. Plant Biol. 2010, 61, 421–442. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Yang, J.; Zhang, F.; Niu, Y.; Zhao, X.; Shen, C.; Chen, K.; Teng, S.; Xu, J. Genetic dissection of rice appearance quality and cooked rice elongation by genome-wide association study. Crop J. 2021, 9, 1470–1480. [Google Scholar] [CrossRef]
- Qiu, X.; Yuan, Z.; He, W.; Liu, H.; Xu, J.; Xing, D. Progress in genetic and breeding research on rice chalkiness. J. Plant Genet. Resour. 2014, 15, 992–998. [Google Scholar]
- Qiu, X.; Chen, K.; Lv, W.; Ou, X.; Zhu, Y.; Xing, D.; Yang, L.; Fan, F.; Yang, J.; Xu, J.; et al. Examining two sets of introgression lines reveals background-independent and stably expressed QTL that improve grain appearance quality in rice (Oryza sativa L.). Theor. Appl. Genet. 2017, 130, 951–967. [Google Scholar] [CrossRef] [PubMed]
- He, P.; Li, S.G.; Qian, Q.; Ma, Y.Q.; Li, J.Z.; Wang, W.M.; Chen, Y.; Zhu, L.H. Genetic analysis of rice grain quality. Theor. Appl. Genet. 1999, 98, 502–508. [Google Scholar] [CrossRef]
- Zhou, L.; Chen, L.; Jiang, L.; Zhang, W.; Liu, L.; Liu, X.; Zhao, Z.; Liu, S.; Zhang, L.; Wang, J.; et al. Fine mapping of the grain chalkiness QTL qPGWC-7 in rice (Oryza sativa L.). Theor. Appl. Genet. 2008, 118, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Liu, X.; Wan, X.; Weng, J.; Liu, S.; Liu, X.; Chen, M.; Li, J.; Su, N.; Wu, F.; et al. Identification of a stable quantitative trait locus for percentage grains with white chalkiness in rice (Oryza sativa). J. Integr. Plant Biol. 2011, 53, 598–607. [Google Scholar] [CrossRef]
- Wang, S.; Wu, K.; Yuan, Q.; Liu, X.; Liu, Z.; Lin, X.; Zeng, R.; Zhu, H.; Dong, G.; Qian, Q.; et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 2012, 44, 950–954. [Google Scholar] [CrossRef]
- Qiu, X.; Pang, Y.; Yuan, Z.; Xing, D.; Xu, J.; Dingkuhn, M.; Li, Z.; Ye, G. Genome-wide association study of grain appearance and milling quality in a worldwide collection of indica rice germplasm. PLoS ONE 2015, 10, e0145577. [Google Scholar] [CrossRef]
- Zhao, X.; Daygon, V.D.; McNally, K.L.; Hamilton, R.S.; Xie, F.; Reinke, R.F.; Fitzgerald, M.A. Identification of stable QTLs causing chalk in rice grains in nine environments. Theor. Appl. Genet. 2016, 129, 141–153. [Google Scholar] [CrossRef]
- Wang, X.; Pang, Y.; Wang, C.; Chen, K.; Zhu, Y.; Shen, C.; Ali, J.; Xu, J.; Li, Z. New candidate genes affecting rice grain appearance and milling quality detected by genome-wide and gene-based association analyses. Front. Plant Sci. 2017, 7, 1998. [Google Scholar] [CrossRef]
- Zhu, A.; Zhang, Y.; Zhang, Z.; Wang, B.; Xue, P.; Cao, Y.; Chen, Y.; Li, Z.; Liu, Q.; Cheng, S.; et al. Genetic dissection of qPCG1 for a quantitative trait locus for percentage of chalky grain in rice (Oryza sativa L.). Front. Plant Sci. 2018, 9, 1173. [Google Scholar] [CrossRef]
- Wu, B.; Xia, D.; Zhou, H.; Cheng, S.; Wang, Y.; Li, M.; Gao, G.; Zhang, Q.; Li, X.; He, Y. Fine mapping of qWCR7, a grain chalkiness QTL in rice. Mol. Breed. 2021, 41, 68. [Google Scholar] [CrossRef]
- Yang, W.; Liang, J.; Hao, Q.; Luan, X.; Tan, Q.; Lin, S.; Zhu, H.; Liu, G.; Liu, Z.; Bu, S.; et al. Fine mapping of two grain chalkiness QTLs sensitive to high temperature in rice. Rice 2021, 14, 33. [Google Scholar] [CrossRef]
- Wu, B.; Yun, P.; Zhou, H.; Xia, D.; Gu, Y.; Li, P.; Yao, J.; Zhou, Z.; Chen, J.; Liu, R.; et al. Natural variation in WHITE-CORE RATE 1 regulates redox homeostasis in rice endosperm to affect grain quality. Plant Cell 2022, 34, 1912–1932. [Google Scholar] [CrossRef]
- Zhao, D.; Zhang, C.; Li, Q.; Liu, Q. Genetic control of grain appearance quality in rice. Biotechnol. Adv. 2022, 60, 108014. [Google Scholar] [CrossRef]
- Yang, W.; Xiong, L.; Liang, J.; Hao, Q.; Luan, X.; Tan, Q.; Lin, S.; Zhu, H.; Liu, G.; Liu, Z.; et al. Substitution mapping of two closely linked QTLs on chromosome 8 controlling grain chalkiness in rice. Rice 2021, 14, 85. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.S.; Li, Q.F.; Zhang, C.Q.; Zhang, C.; Yang, Q.Q.; Pan, L.X.; Ren, X.Y.; Lu, J.; Gu, M.H.; Liu, Q.Q. GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat. Commun. 2018, 9, 1240. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fan, C.; Xing, Y.; Yun, P.; Luo, L.; Yan, B.; Peng, B.; Xie, W.; Wang, G.; Li, X.; et al. Chalk5 encodes a vacuolar H(+)-translocating pyrophosphatase influencing grain chalkiness in rice. Nat. Genet. 2014, 46, 398–404. [Google Scholar] [CrossRef]
- Wang, X. Analysis of the application value of Chalk5 gene in the breeding system from Liaoing Province. North Rice 2017, 47, 20–22. [Google Scholar] [CrossRef]
- Wang, D.; Wang, J.; Sun, W.; Qiu, X.; Yuan, Z.; Yu, S. Verifying the breeding value of a rare haplotype of Chalk7, GS3, and Chalk5 to improve grain appearance quality in rice. Plants 2022, 11, 1470. [Google Scholar] [CrossRef] [PubMed]
- Terao, T.; Hirose, T. Temperature-dependent QTLs in indica alleles for improving grain quality in rice: Increased prominence of QTLs responsible for reduced chalkiness under high-temperature conditions. Mol. Breed. 2018, 38, 52. [Google Scholar] [CrossRef]
- Lo, P.C.; Hu, L.; Kitano, H.; Matsuoka, M. Starch metabolism and grain chalkiness under high temperature stress. Natl. Sci. Rev. 2016, 3, 280–282. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.-L.; et al. Targeted genome modification of crop plants using a CRISPR/Cas system. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar] [CrossRef] [PubMed]
- Shomura, A.; Izawa, T.; Ebana, K.; Ebitani, T.; Kanegae, H.; Konishi, S.; Yano, M. Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 2008, 40, 1023–1028. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fan, C.; Xing, Y.; Jiang, Y.; Luo, L.; Sun, L.; Shao, D.; Xu, C.; Li, X.; Xiao, J.; et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 2011, 43, 1266–1269. [Google Scholar] [CrossRef]
- Duan, P.G.; Xu, J.S.; Zeng, D.L.; Zhang, B.L.; Geng, M.F.; Zhang, G.Z.; Huang, K.; Huang, L.J.; Xu, R.; Ge, S.; et al. Natural variation in the promoter of GSE5 contributes to grain size diversity in rice. Mol. Plant 2017, 10, 685–694. [Google Scholar] [CrossRef]
- Gann, P.J.I.; Dharwadker, D.; Cherati, S.R.; Vinzant, K.; Khodakovskaya, M.; Srivastava, V. Targeted mutagenesis of the vacuolar H+ translocating pyrophosphatase gene reduces grain chalkiness in rice. Plant J. 2023, 115, 1261–1276. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Li, M.; Wu, H.; Liao, Y.; Xin, J.; Yuan, X.; Li, Y.; Wei, A.; Zou, X.; Guo, D.; et al. Generation of aroma in three-line hybrid rice through CRISPR/Cas9 editing of BETAINE ALDEHYDE DEHYDROGENASE2 (OsBADH2). Physiol. Plant. 2024, 176. [Google Scholar] [CrossRef]
- Qiu, X.; Gong, R.; Tan, Y.; Yu, S. Mapping and characterization of the major quantitative trait locus qSS7 associated with increased length and decreased width of rice seeds. Theor. Appl. Genet. 2012, 125, 1717–1726. [Google Scholar] [CrossRef]
- Hu, H.; Lv, W.; Li, Q.; Ou, X.; Xu, J.; Li, Z.; Xing, D.; Yang, L.; Xu, J.; Qiu, X.; et al. Characterization of main effects, epistatic effects and genetic background effects on QTL for yield related traits by two sets of reciprocal introgression lines in rice (Orysa sativa). Int. J. Agric. Biol. 2018, 20, 2125–2132. [Google Scholar] [CrossRef]
- Jia, Q.; Wu, X.; Qian, K.; Peng, Z.; Cai, Y.; Ye, R. Analysis on quality characteristics of rice varieties approved in China from 2000 to 2020. Hubei Agric. Sci. 2022, 61, 11–18. [Google Scholar]
- Liu, P.; He, L.; Mei, L.; Zhai, W.; Chen, X.; Ma, B. Rapid and directional improvement of elite rice variety via combination of genomics and multiplex genome editing. J. Agric. Food Chem. 2022, 70, 6156–6167. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Xiong, W.; Ju, H.; Cao, Y.; Yang, D. Changes of high temperature events during rice growth period in MLRYR under RCP scenarios. Chin. J. Agrometeorol. 2015, 36, 383–392. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).