Differential Cellular Response to Mercury in Non-Farmed Fish Species Based on Mitochondrial DNA Copy Number Variation Analysis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Area of Study
2.2. Sampling
2.3. Chemical Analysis and Mercury Bioaccumulation
2.4. DNA Extraction
2.5. mtDNAcn Evaluation
2.6. Statistical Analysis
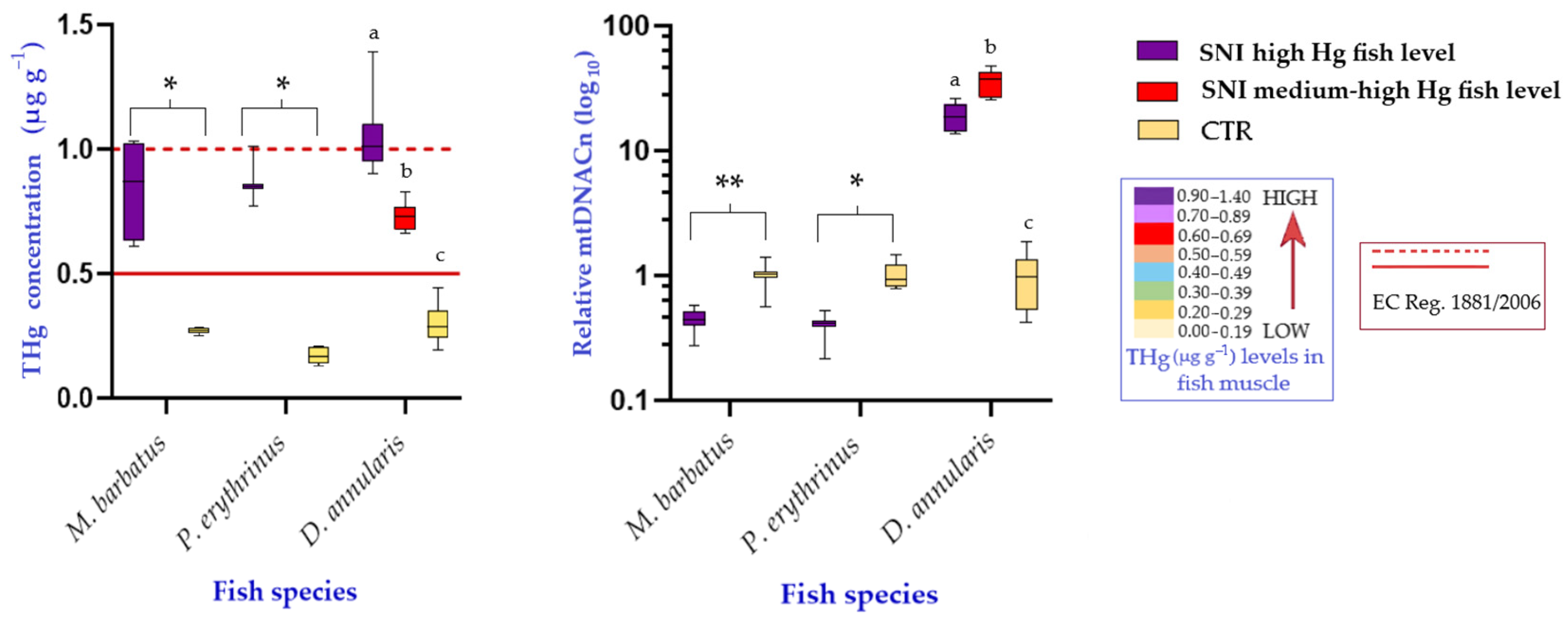
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- van der Oost, R.; Beyer, J.; Vermeulen, N.P.E. Fish bioaccumulation and biomarkers in environmental risk assessment: A review. Environ. Toxicol. Pharmacol. 2003, 57, 57–149. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, C.E.; Manuguerra, S.; Cuesta, A.; Esteban, M.A.; Santulli, A.; Messina, C.M. Sub-lethal doses of polybrominated diphenyl ethers affect some biomarkers involved in energy balance and cell cycle, via oxidative stress in the marine fish cell line SAF-1. Aquat. Toxicol. 2019, 210, 1–10. [Google Scholar] [CrossRef]
- Messina, C.M.; Espinosa Ruiz, C.; Regoli, F.; Manuguerra, S.; D’Agostino, F.; Avellone, G.; Sprovieri, M.; Santulli, A. BDE-47 exposure modulates cellular responses, oxidative stress and biotransformation related-genes in Mytilus galloprovincialis. Fish Shellfish Immunol. 2020, 107, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Authman, M.M.N.; Zaki, M.S.; Khallaf, E.A.; Abbas, H.H. Use of Fish as Bio-indicator of the Effects of Heavy Metals Pollution. Aquac. Res. Dev. 2015, 6, 328. [Google Scholar] [CrossRef]
- Tamele, I.J.; Loureiro, P.V. Lead, Mercury and Cadmium in Fish and Shellfish from the Indian Ocean and Red Sea (African Countries): Public Health Challenges. J. Mar. Sci. Eng. 2020, 8, 344. [Google Scholar] [CrossRef]
- Smith, S.B.; Gill, C.A.; Lunt, D.K.; Brooks, M.A. Regulation of fat and fatty acid composition in beef cattle. Asian-Australas. J. Anim. Sci. 2009, 22, 1225–1233. [Google Scholar] [CrossRef]
- Thompson, L.V. Oxidative stress, mitochondria and mtDNA-mutator mice. Exp. Gerontol. 2006, 41, 1220–1222. [Google Scholar] [CrossRef][Green Version]
- Colombo, R.; Olmo, E. Biologia; Cellula e Tessuti; Edi Ermes: Milan, Italy, 2013. [Google Scholar]
- Lee, H.-C.; Wei, Y.-H. Mitochondrial role in life and death of the cell. J. Biomed. Sci. 2000, 7, 2–15. [Google Scholar] [CrossRef]
- Yapa, N.M.B.; Lisnyak, V.; Reljic, B.; Ryan, M.T. Mitochondrial dynamics in health and disease. FEBS Lett. 2021, 595, 1184–1204. [Google Scholar] [CrossRef] [PubMed]
- Castellani, C.A.; Longchamps, R.J.; Sun, J.; Guallar, E.; Arking, D.E. Thinking outside the nucleus: Mitochondrial DNA copy number in health and disease. Mitochondrion 2020, 53, 214–223. [Google Scholar] [CrossRef]
- Filograna, R.; Mennuni, M.; Alsina, D.; Larsson, N.G. Mitochondrial DNA Copy Number in Human Disease: The More the Better? FEBS Lett. 2021, 595, 976–1002. [Google Scholar] [CrossRef]
- Radzak, S.M.A.; Khair, S.Z.N.M.; Ahmad, F.; Patar, A.; Idris, Z.; Yusoff, A.A.M. Insights regarding mitochondrial DNA copy number alterations in human cancer (Review). Int. J. Mol. Med. 2022, 50, 104. [Google Scholar] [CrossRef] [PubMed]
- Avilés-Ramírez, C.; Moreno-Godínez, M.E.; Bonner, M.R.; Parra-Rojas, I.; Flores-Alfaro, E.; Ramírez, M.; Huerta-Beristain, G.; Ramírez-Vargas, M.A. Effects of exposure to environmental pollutants on mitochondrial DNA copy number: A meta-analysis. Environ. Sci. Pollut. Res. 2022, 29, 43588–43606. [Google Scholar] [CrossRef]
- He, W.J.; Li, C.; Huang, Z.; Geng, S.; Rao, V.S.; Kelly, T.N.; Hamm, L.L.; Grams, M.E.; Arking, D.E.; Appel, L.J.; et al. Association of Mitochondrial DNA Copy Number with Risk of Progression of Kidney Disease. Clin. J. Am. Soc. Nephrol. 2022, 17, 966–975. [Google Scholar] [CrossRef] [PubMed]
- Picca, A.; Guerra, F.; Calvani, R.; Coelho-Júnior, H.J.; Leeuwenburgh, C.; Bucci, C.; Marzetti, E. The contribution of mitochondrial DNA alterations to aging, cancer, and neurodegeneration. Exp. Gerontol. 2023, 178, 112203. [Google Scholar] [CrossRef] [PubMed]
- Alegría-Torres, J.A. The mitochondrial DNA copy number used as biomarker. Int. J. Mol. Biol. 2018, 3, 117–119. [Google Scholar] [CrossRef][Green Version]
- Bess, A.S.; Crocker, T.L.; Ryde, I.T.; Meyer, J.N. Mitochondrial dynamics and autophagy aid in removal of persistent mitochondrial DNA damage in Caenorhabditis elegans. Nucleic Acids Res. 2012, 40, 7916–7931. [Google Scholar] [CrossRef]
- Reznik, E.; Miller, M.L.; Şenbabaoğlu, Y.; Riaz, N.; Sarungbam, J.; Tickoo, S.K.; Al-Ahmadie, H.A.; Lee, W.; Seshan, V.E.; Hakimi, A.A.; et al. Mitochondrial DNA copy number variation across human cancers. Elife 2016, 5, e10769. [Google Scholar] [CrossRef]
- Xu, Y.; Li, H.; Hedmer, M.; Hossain, M.B.; Tinnerberg, H.; Broberg, K.; Albin, M. Occupational exposure to particles and mitochondrial DNA-relevance for blood pressure. Environ. Health 2017, 16, 22. [Google Scholar] [CrossRef]
- Mutlu, A.G. Increase in Mitochondrial DNA Copy Number in Response to Ochratoxin a and Methanol-Induced Mitochondrial DNA Damage in Drosophila. Bull. Environ. Contam. Toxicol. 2012, 89, 1129–1132. [Google Scholar] [CrossRef]
- Sanders, L.H.; Howlett, E.H.; McCoy, J.; Greenamyre, J.T. Mitochondrial DNA Damage as a Peripheral Biomarker for Mitochondrial Toxin Exposure in Rats. Toxicol. Sci. 2014, 142, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.H.; Zhang, P.; Ma, H.K.; Xu, W.Y.; Sun, J.Q.; Yan, B.L.; Zhang, Q.Q.; Gao, H. Effect of temperature and salinity on mtDNA copy number of the ridgetail white prawn, Palaemon carinicauda Holthuis, 1950 (Decapoda, Palaemonidae). Crustaceana 2018, 91, 1061–1072. [Google Scholar] [CrossRef]
- Calogero, G.S.; Giuga, M.; D’Urso, V.; Ferrito, V.; Pappalardo, A.M. First Report of Mitochondrial DNA Copy Number Variation in Opsius heydeni (Insecta, Hemiptera, Cicadellidae) from Polluted and Control Sites. Animals 2023, 13, 1793. [Google Scholar] [CrossRef]
- Frapiccini, E.; Panfili, M.; Guicciardi, S.; Santojanni, A.; Marini, M.; Truzzi, C.; Annibaldi, A. Effects of biological factors and seasonality on the level of polycyclic aromatic hydrocarbons in red mullet (Mullus barbatus). Environ. Pollut. 2020, 258, 113742. [Google Scholar] [CrossRef]
- Kontas, A.; Alyuruk, H.; Bilgin, M.; Uluturhan, E.; Ünlüoğlu, A.; Darilmaz, E.; Altay, O. Metal Bioaccumulation and Potential Health Risk Assessment in Different Tissues of Three Commercial Fish Species (Merluccius merluccius, Mullus barbatus, and Pagellus erythrinus) from Edremit Bay (Aegean Sea), Turkey. Biol. Trace Elem. Res. 2021, 200, 868–880. [Google Scholar] [CrossRef]
- Girolametti, F.; Frapiccini, E.; Annibaldi, A.; Illuminati, S.; Panfili, M.; Marini, M.; Santojanni, A.; Truzzi, C. Total Mercury (THg) content in red mullet (Mullus barbatus) from Adriatic Sea (Central Mediterranean Sea): Relation to biological parameters, sampling area and human health risk assessment. Appl. Sci. 2022, 12, 10083. [Google Scholar] [CrossRef]
- Squillante, J.; Scivicco, M.; Ariano, A.; Nolasco, A.; Esposito, F.; Cacciola, N.A.; Severino, L.; Cirillo, T. Occurence of phtalate esters and preliminary data on microplastics in fish from the Tyrrhenian Sea (Italy) and impact on human health. Environ. Pollut. 2023, 316, 120664. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, M.; Romano, E.; Ausili, A.; Fattorini, D.; Gorbi, S.; Maggi, C.; Salmeri, A.; Manta, D.S.; Sesta, G.; Sprovieri, M.; et al. 10-year time course of Hg and organic compounds in Augusta Bay: Bioavailability and biological effects in marine organisms. Front. Public Health 2022, 10, 968296. [Google Scholar] [CrossRef]
- Bonsignore, M.; Salvagio Manta, D.; Oliveri, E.; Sprovieri, M.; Basilone, G.; Bonanno, A.; Falco, F.; Traina, A.; Mazzola, S. Mercury in fishes from Augusta Bay (southern Italy): Risk assessment and health implication. Food Chem. Toxicol. 2013, 56, 184–194. [Google Scholar] [CrossRef]
- Copat, C.; Arena, G.; Fiore, M.; Ledda, C.; Fallico, R.; Sciacca, S. Heavy metals concentration in fish and shellfish from eastern Mediterranean Sea: Consumpion advisories. Food Chem. Toxicol. 2013, 53, 33–37. [Google Scholar] [CrossRef]
- Bonsignore, M.; Andolfi, N.; Barra, M.; Madeddu, A.; Tisano, F.; Ingallinella, V.; Castorina, M.; Sprovieri, M. Assessment of mercury exposure in human populations: A status report from Augusta Bay (southern Italy). Environ. Res. 2016, 150, 592–599. [Google Scholar] [CrossRef]
- Traina, A.; Bono, G.; Bonsignore, M.; Falco, F.; Giuga, M.; Quinci, E.M.; Vitale, S.; Sprovieri, M. Heavy metals concentrations in some commercially key species from Sicilian coasts (Mediterranean Sea): Potential human health risk estimation. Ecotoxicol. Environ. Saf. 2019, 168, 466–478. [Google Scholar] [CrossRef]
- ARPA. Convenzione per l’aggiornamento del Quadro Conoscitivo Sullo Stato di Qualità Delle Acque Sotterranee, Superficiali, Interne, Marino-Costiere ai Fini Della Revisione del Piano di Gestione del Distretto Idrografico Della Regione Sicilia; ARPA Sicilia: Palermo, Italy, 2018. [Google Scholar]
- Sprovieri, M.; Oliveri, E.; Di Leonardo, R.; Romano, E.; Ausili, A.; Gabellini, M.; Barra, M.; Tranchida, G.; Bellanca, A.; Neri, R.; et al. The key role played by the Augusta basin (southern Italy) in the mercury contamination of the Mediterranean Sea. J. Environ. Monit. 2011, 13, 1753–1760. [Google Scholar] [CrossRef] [PubMed]
- Manuguerra, S.; Giuga, M.; Bonsignore, M.; Arena, R.; Traina, A.; Bellante, A.; Santulli, A.; Messina, C.M.; Sprovieri, M. Bioaccumulation, Markers of Stress, Welfare and Quality in Fish From an Impacted Area: The side Effects of the Contaminants in the Marine Food System. Fish Shellfish Immunol. 2024; submitted. [Google Scholar]
- Ausili, A.; Gabellini, M.; Cammarata, G.; Fattorini, D.; Benedetti, M.; Pisanelli, B.; Gorbi, S.; Regoli, F. Ecotoxicological and human health risk in a petrochemical district of southern Italy. Mar. Environ. Res. 2008, 66, 215–217. [Google Scholar] [CrossRef] [PubMed]
- Messina, C.M.; Manuguerra, S.; Arena, R.; Espinosa-Ruiz, C.; Curcuraci, E.; Esteban, M.A.; Santulli, A. Contaminant-induced oxidative stress underlies biochemical, molecular and fatty acid profile changes, in gilthead seabream (Sparus aurata L.). Res. Vet. Sci. 2023, 159, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, P.J.P.; Bauchot, M.L.; Hureau, J.C.; Nielsen, J.; Tortonese, E. Fishes of the North-Eastern Atlantic and the Mediterranean; UNESCO: Bungay, UK, 1984. [Google Scholar]
- Environmnetal Protection Agency. Method 7473: Mercury in Solids and Solutions by Thermal 420 Decomposition, Amalgamation, and Atomic Absorption Spectrophotometry; US EPA: Washington, DC, USA, 2007. [Google Scholar]
- Dwiyitno, D.; Hoffman, S.; Parmentier, K.; Van Keer, C. Universal primer design for crustacean and bivalve-mollusc authenticity based on cytochrome-b gene. Biodiversitas 2022, 23, 17–24. [Google Scholar] [CrossRef]
- Messina, C.M.; Arena, R.; Manuguerra, S.; La Barbera, L.; Curcuraci, E.; Renda, G.; Santulli, A. Valorization of Side Stream Products from Sea Cage Fattened Bluefin Tuna (Thunnus thynnus): Production and In Vitro Bioactivity Evaluation of Enriched ω-3 Polyunsaturated Fatty Acids. Mar. Drugs 2022, 20, 309. [Google Scholar] [CrossRef]
- Hartmann, N.; Reichwald, K.; Wittig, I.; Dröse, S.; Schmeisser, S.; Lück, C.; Hahn, C.; Graf, M.; Gausmann, U.; Terzibasi, E.; et al. Mitochondrial DNA Copy Number and Function Decrease with Age in the Short-Lived Fish Nothobranchius furzeri. Aging Cell 2011, 10, 824–831. [Google Scholar] [CrossRef]
- Rooney, J.P.; Ryde, I.T.; Sanders, L.H.; Howlett, E.H.; Colton, M.D.; Germ, K.E.; Mayer, G.D.; Greenamyre, J.T.; Meyer, J.N. PCR based determination of mitochondrial DNA copy number in multiple species. Methods Mol. Biol. 2015, 1241, 23–38. [Google Scholar] [CrossRef]
- Giuga, M. Impiego di Indicatori Chimici, Biochimici e Molecolari per la Valutazione Degli Effetti Indotti da Contaminanti Am-bientali in Organismi Marini: Dall’early Warning Alla Previsione Degli Effetti a Lungo Termine Sulle Performance. Ph.D. Thesis, University of Catania, Catania, Italy, 2023. [Google Scholar]
- Di Bella, C.; Traina, A.; Giosuè, C.; Carpintieri, D.; Lo Dico, G.M.; Bellante, A.; Del Core, M.; Falco, F.; Gherardi, S.; Uccello, M.M.; et al. Heavy metals and PAHs in meat, milk and seafood from Augusta area (southern Italy): Contamination levels, dietary intake and human exposure assessment. Front. Public Health 2020, 8, 273. [Google Scholar] [CrossRef]
- Relini, G.; Bertrand, J.; Zamboni, A. Sintesi delle conoscenze sulle risorse da pesca dei fondi del Mediterraneo centrale (Italia e Corsica). Biol. Mar. Mediterr. 1999, 6, 1–868. [Google Scholar]
- Osteryoung, K.W.; Nunnari, J. The Division of Endosymbiotic Organelles. Science 2003, 302, 1698–1704. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.C. Mitochondrial Fusion and Fission in Mammals. Ann. Rev. Cell Dev. Biol. 2006, 22, 79–99. [Google Scholar] [CrossRef] [PubMed]
- Karbowski, M.; Youle, R.J. Dynamics of Mitochondrial Morphology in Healthy Cells and during Apoptosis. Cell Death Differ. 2003, 10, 870–880. [Google Scholar] [CrossRef]
- Roubicek, D.A.; de Souza-Pinto, N.C. Mitochondria and Mitochondrial DNA as Relevant Targets for Environmental Contaminants. Toxicology 2017, 391, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.N.; Czajka, A. Is Mitochondrial DNA Content a Potential Biomarker of Mitochondrial Dysfunction? Mitochondrion 2013, 13, 481–492. [Google Scholar] [CrossRef]
- Youle, R.J.; van der Bliek, A. Mitochondrial Fission, Fusion, and Stress. Science 2012, 337, 1062–1065. [Google Scholar] [CrossRef]
- Jones, M.D.; Naylor, K. Simple to Complex: The Role of Actin and Microtubules in Mitochondrial Dynamics in Amoeba, Yeast, and Mammalian Cells. Int. J. Mol. Sci. 2022, 23, 9402. [Google Scholar] [CrossRef]
- Hu, Z.; Xu, L.; Song, H.; Feng, J.; Zhou, C.; Yang, M.J.; Shi, P.; Li, Y.R.; Guo, Y.J.; Li, H.Z.; et al. Effect of heat and hypoxia stress on mitochondrion and energy metabolism in the gill of hard clam. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2023, 266, 109556. [Google Scholar] [CrossRef]
- Maremonti, E.; Brede, D.A.; Olsen, A.-K.; Eide, D.M.; Berg, E.S. Ionizing radiation, genotoxic stress, and mitochondrial DNA copy-number variation in Caenorhabditis elegans: Droplet digital PCR analysis. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2020, 858–860, 503277. [Google Scholar] [CrossRef] [PubMed]
- Malakhova, L.; Bezlepkin, V.G.; Antipova, V.; Ushakova, T.; Fomenko, L.; Sirota, N.; Gaziev, A.I. The increase in mitochondrial DNA copy number in the tissues of γ-irradiated mice. Cell. Mol. Biol. Lett. 2005, 10, 721. [Google Scholar]
- Nugent, S.; Mothersill, C.E.; Seymour, C.; McClean, B.; Lyng, F.M.; Murphy, J.E.J. Altered mitochondrial function and genome frequency post exposure to γ-radiation and bystander factors. Int. J. Radiat. Biol. 2010, 86, 829–841. [Google Scholar] [CrossRef]
- Kam, W.; Lake, V.; Banos, C.; Davies, J.; Banati, R. Apparent polyploidization after gamma irradiation: Pitfalls in the use of quantitative polymerase chain reaction (qPCR) for the estimation of mitochondrial and nuclear DNA gene copy numbers. Int. J. Mol. Sci. 2013, 14, 11544–11559. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-Y.; Kim, H.-R.; Kang, M.-G.; Trang, N.T.D.; Baek, H.-J.; Moon, J.-D.; Shin, J.-H.; Suh, S.-P.; Ryang, D.-W.; Kook, H.; et al. Profiling of Biomarkers for the Exposure of Polycyclic Aromatic Hydrocarbons: Lamin-A/C Isoform 3, Poly[ADP-Ribose] Polymerase 1, and Mitochondria Copy Number Are Identified as Universal Biomarkers. BioMed Res. Int. 2014, 2014, 605135. [Google Scholar] [CrossRef]
- Das, A.; Bank, S.; Chatterjee, S.; Paul, N.; Sarkar, K.; Chatterjee, A.; Chakraborty, S.; Banerjee, C.; Majumdar, A.; Das, M.; et al. Bifenthrin disrupts cytochrome c oxidase activity and reduces mitochondrial DNA copy number through oxidative damage in pool barb (Puntius sophore). Chemosphere 2023, 332, 138848. [Google Scholar] [CrossRef]
- Martín-del-Campo, R.; Bárcenas-Ibarra, A.; Lund, G.; Rodríguez-Rìos, D.; Yong-Villalobos, L.; García-Hernandez, J.; García-Gasca, A. Mercury Concentration, DNA Methylation, and Mitochondrial DNA Damage in Olive Ridley Sea Turtle Embryos With Schistosomus Reflexus Syndrome. Vet. Pathol. 2019, 56, 940–949. [Google Scholar] [CrossRef]
- Bowman, A.; Martinez-Levasseur, L.M.; Acevedo-Whitehouse, K.; Gendron, D.; Birch-Machin, M.A. The simultaneous detection of mitochondrial DNA damage from sun-exposed skin of three whale species and its association with UV-induced microscopic lesions and apoptosis. Mitochondrion 2013, 13, 342–349. [Google Scholar] [CrossRef]
- Fan, X.; Zhang, D.; Hou, T.; Zhang, Q.; Tao, L.; Bian, C.; Wang, Z. Mitochondrial DNA stress-mediated health risk to Dibutyl Phthalate contamination on Zebrafish (Danio rerio) at early life stage. Environ. Sci. Technol. 2024, 58, 7731–7742. [Google Scholar] [CrossRef]
- Baek, J.H.; Son, H.; Jeong, Y.H.; Park, S.W.; Kim, H.J. Chronological Aging Standard Curves of Telomere Length and Mitochondrial DNA Copy Number in Twelve Tissues of C57BL/6 Male Mouse. Cells 2019, 8, 247. [Google Scholar] [CrossRef]
- Stier, A.; Hsu, B.-Y.; Marciau, C.; Doligez, B.; Gustafsson, L.; Bize, P.; Ruuskanen, S.; Stier, A.; Hsu, B.-Y.; Marciau, C.; et al. Born to be young? Prenatal thyroid hormones increase early-life telomere length in wild collared flycatchers. Biol. Lett. 2020, 16, 20200364. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Liu, Y.; An, Z.; Huang, D.; Qi, Y.; Zhang, Y. Mediating effect of ROS on mtDNA damage and low ATP content induced by arsenic trioxide in mouse oocytes. Toxicol. In Vitro 2011, 25, 979–984. [Google Scholar] [CrossRef]
- Xia, T.; Korge, P.; Weiss, J.N.; Li, N.; Venkatesen, M.I.; Sioutas, C.; Nel, A. Quinones and Aromatic Chemical Compounds in Particulate Matter Induce Mitochondrial Dysfunction: Implications for Ultrafine Particle Toxicity. Environ. Health Perspect. 2004, 112, 1347–1358. [Google Scholar] [CrossRef] [PubMed]
- Fetterman, J.L.; Sammy, M.J.; Ballinger, S.W. Mitochondrial toxicity of tobacco smoke and air pollution. Toxicology 2017, 391, 18–33. [Google Scholar] [CrossRef] [PubMed]
- Abdullaev, S.; Bulanova, T.; Timoshenko, G.; Gaziev, A.I. Increase of mtDNA number and its mutant copies in rat brain after exposure to 150 MeV protons. Mol. Biol. Rep. 2020, 47, 4815–4820. [Google Scholar] [CrossRef]
- Syromyatnikov, M.Y.; Gureev, A.P.; Mikhaylov, E.V.; Parshin, P.A.; Popov, V.N. Pesticides Effect on the Level of MtDNA Damage in Bumblebees Heads (Bombus terrestris L.). Period. Tche Quim. 2020, 17, 395–402. [Google Scholar] [CrossRef]


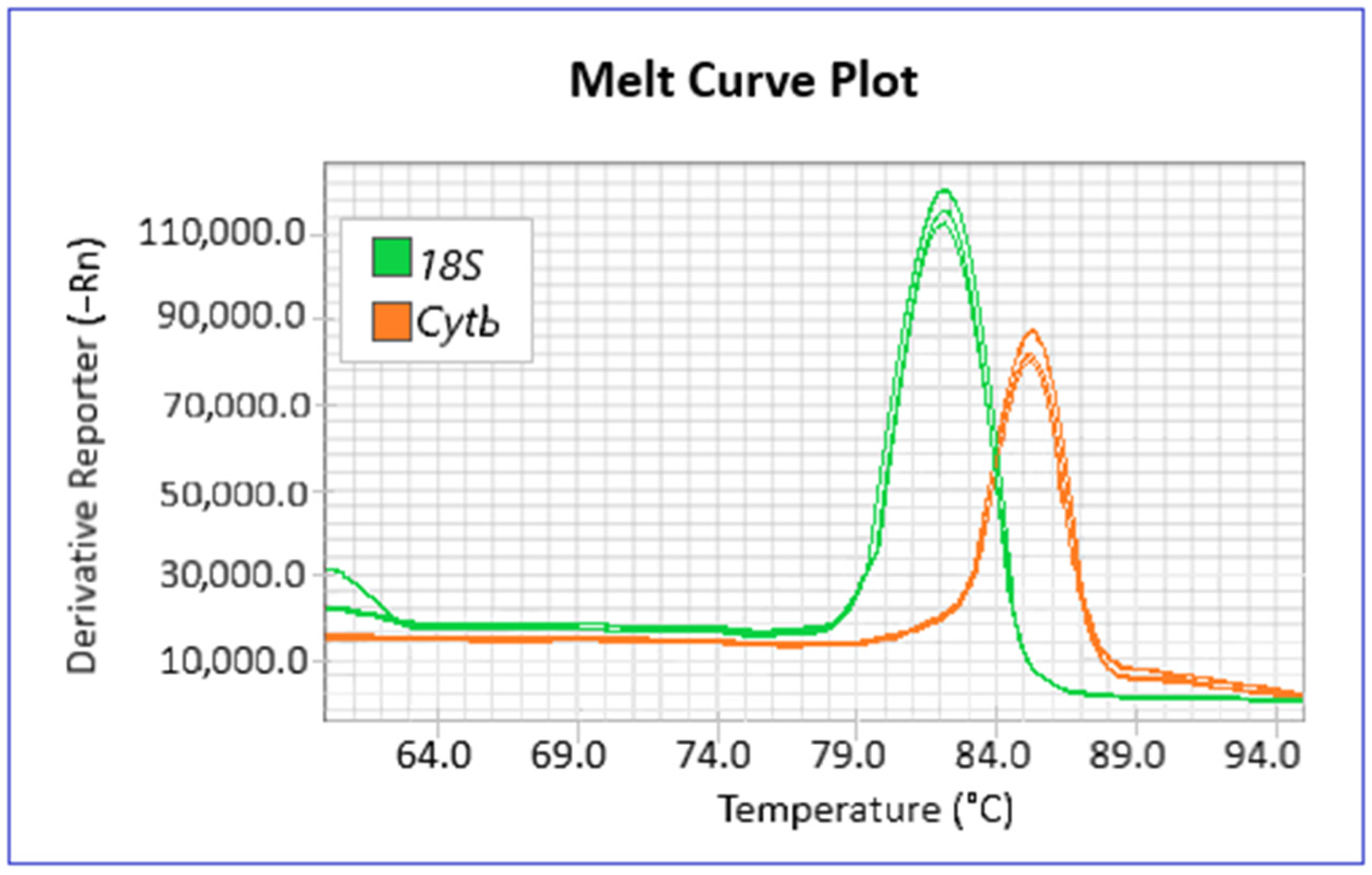
| Species | Marine Habitat | Sampling | Total Length (Mean ± s.d. in cm) | Weight (Mean ± s.d. in g) | Hg w.w. (Mean in μg g−1) | |||
|---|---|---|---|---|---|---|---|---|
| N° | Area | Station | Fishing Gear | |||||
| Mullus barbatus | benthic | 6 | SNI | F 1-2-3 | trammel net | 19 ± 2.7 | 80.4 ± 31.5 | 0.84 ± 0.20 |
| 9 | CTR | F 8 | trammel net | 16.6 ± 1.5 | 48.7 ± 12.9 | 0.30 ± 0.12 | ||
| Pagellus erythrinus | demersal | 10 | SNI | F 1-2-3 | trammel net | 19.4 ± 4.1 | 109.6 ± 64.7 | 0.61 ± 0.24 |
| 8 | CTR | F 4-5-6-7 | trammel net | 18.2 ± 2 | 108.2 ± 48.7 | 0.25 ± 0.10 | ||
| Diplodus annularis | demersal | 10 | SNI | F 1-2-3 | trammel net | 12.7 ± 0.7 | 35.8 ± 5.6 | 0.84 ± 0.24 |
| 10 | CTR | F 4-5-6-7 | trawl net | 13.1 ± 1.5 | 40.7 ± 13 | 0.34 ± 0.11 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giuga, M.; Ferrito, V.; Calogero, G.S.; Traina, A.; Bonsignore, M.; Sprovieri, M.; Pappalardo, A.M. Differential Cellular Response to Mercury in Non-Farmed Fish Species Based on Mitochondrial DNA Copy Number Variation Analysis. Biology 2024, 13, 691. https://doi.org/10.3390/biology13090691
Giuga M, Ferrito V, Calogero GS, Traina A, Bonsignore M, Sprovieri M, Pappalardo AM. Differential Cellular Response to Mercury in Non-Farmed Fish Species Based on Mitochondrial DNA Copy Number Variation Analysis. Biology. 2024; 13(9):691. https://doi.org/10.3390/biology13090691
Chicago/Turabian StyleGiuga, Marta, Venera Ferrito, Giada Santa Calogero, Anna Traina, Maria Bonsignore, Mario Sprovieri, and Anna Maria Pappalardo. 2024. "Differential Cellular Response to Mercury in Non-Farmed Fish Species Based on Mitochondrial DNA Copy Number Variation Analysis" Biology 13, no. 9: 691. https://doi.org/10.3390/biology13090691
APA StyleGiuga, M., Ferrito, V., Calogero, G. S., Traina, A., Bonsignore, M., Sprovieri, M., & Pappalardo, A. M. (2024). Differential Cellular Response to Mercury in Non-Farmed Fish Species Based on Mitochondrial DNA Copy Number Variation Analysis. Biology, 13(9), 691. https://doi.org/10.3390/biology13090691







