FAM98 Family Proteins Play Distinct Roles in Osteoclastogenesis and Bone Resorption
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Bone Marrow Monocyte and Osteoclast Cultures
2.3. Tartrate-Resistant Acid Phosphatase (TRAP) Staining
2.4. Real-Time Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
2.5. Lentiviral Transduction
2.6. Immunoblotting
2.7. Immunofluorescence
2.8. Resorption Pit Staining
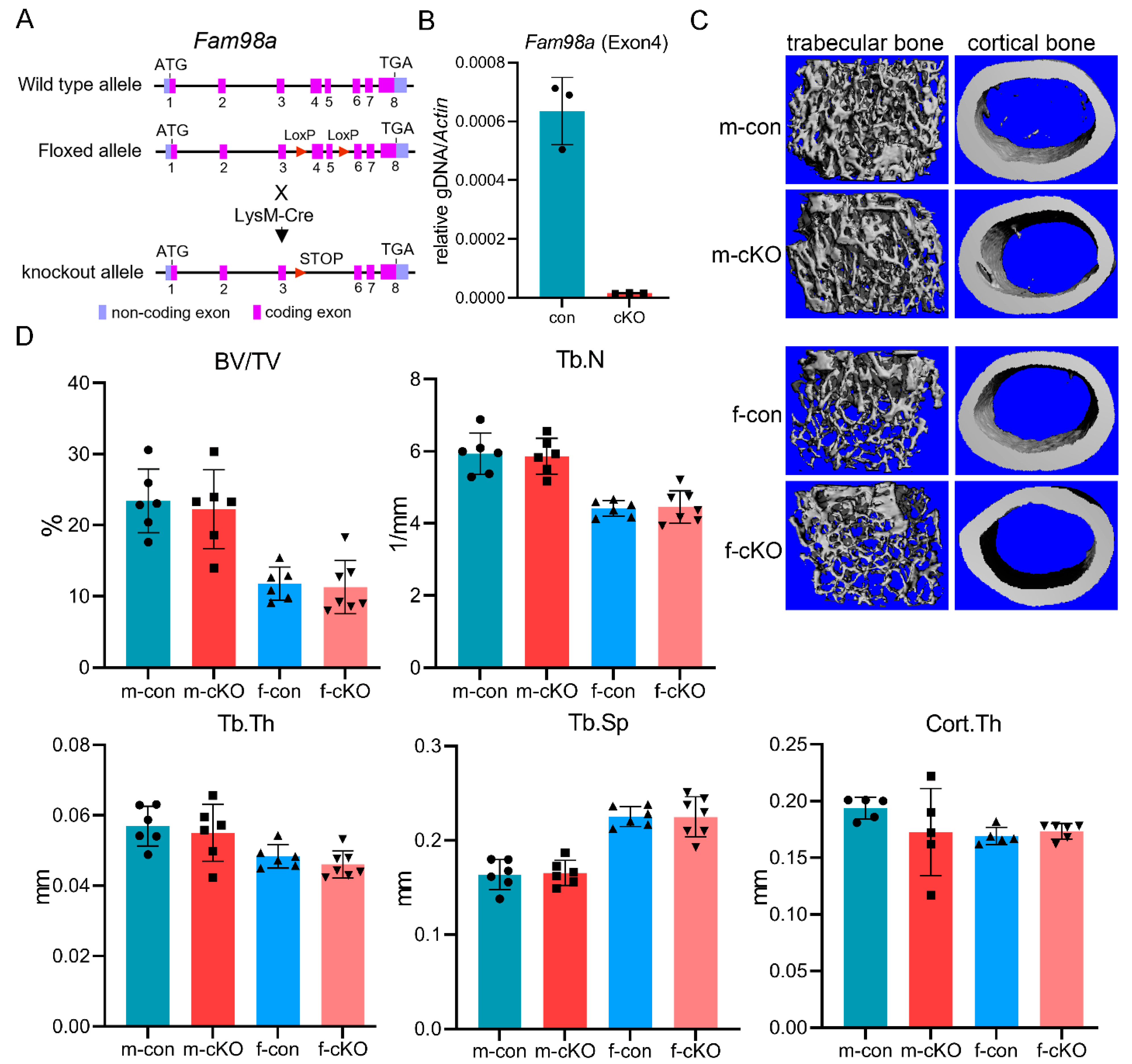
2.9. Generation of Fam98a-Flox and Fam98a-Flox;LysM-Cre Mice
2.10. Micro-CT
3. Results
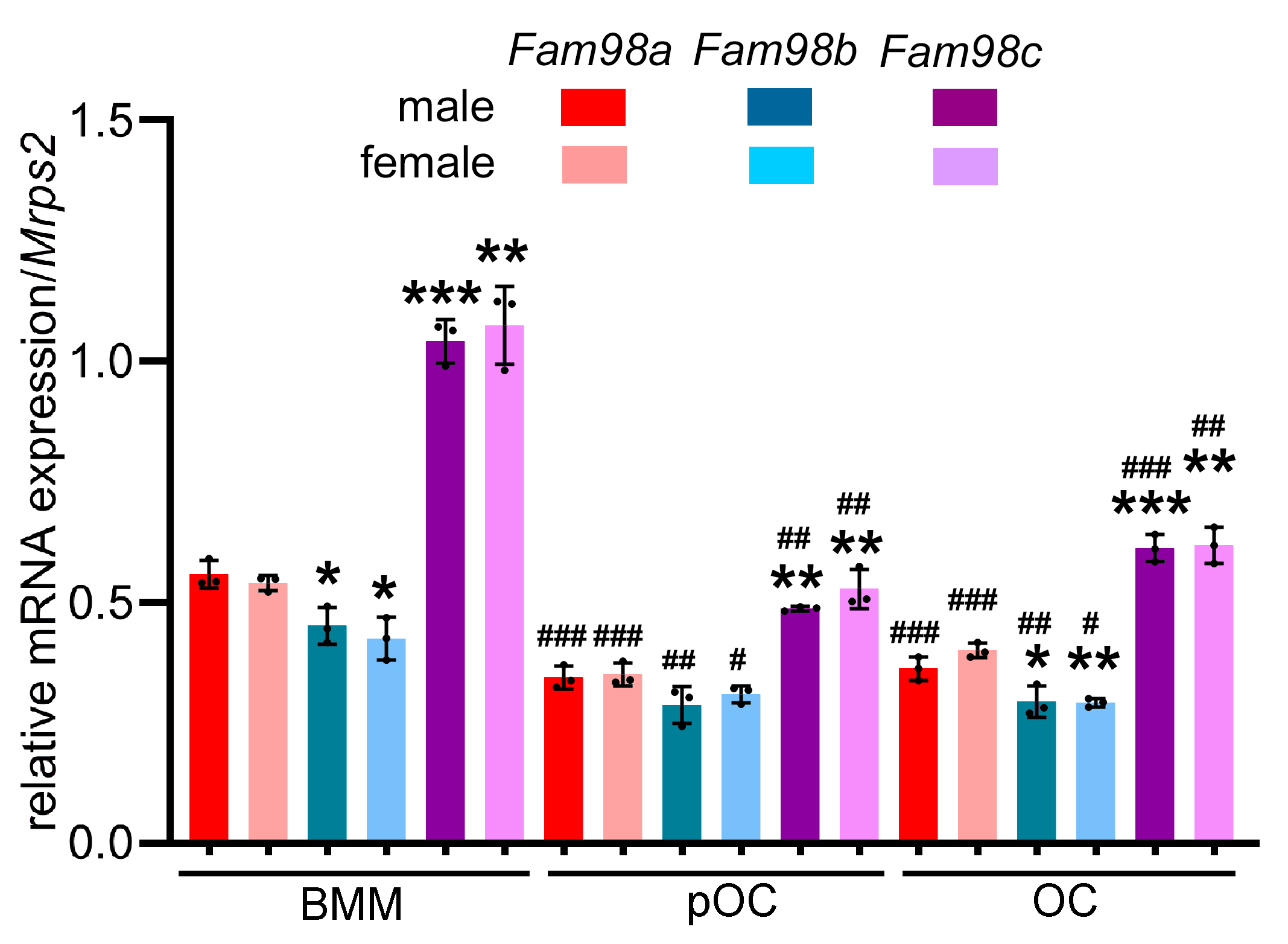
3.1. All Three Murine Fam98 Family Genes Are Expressed in Precursor and Mature Osteoclasts
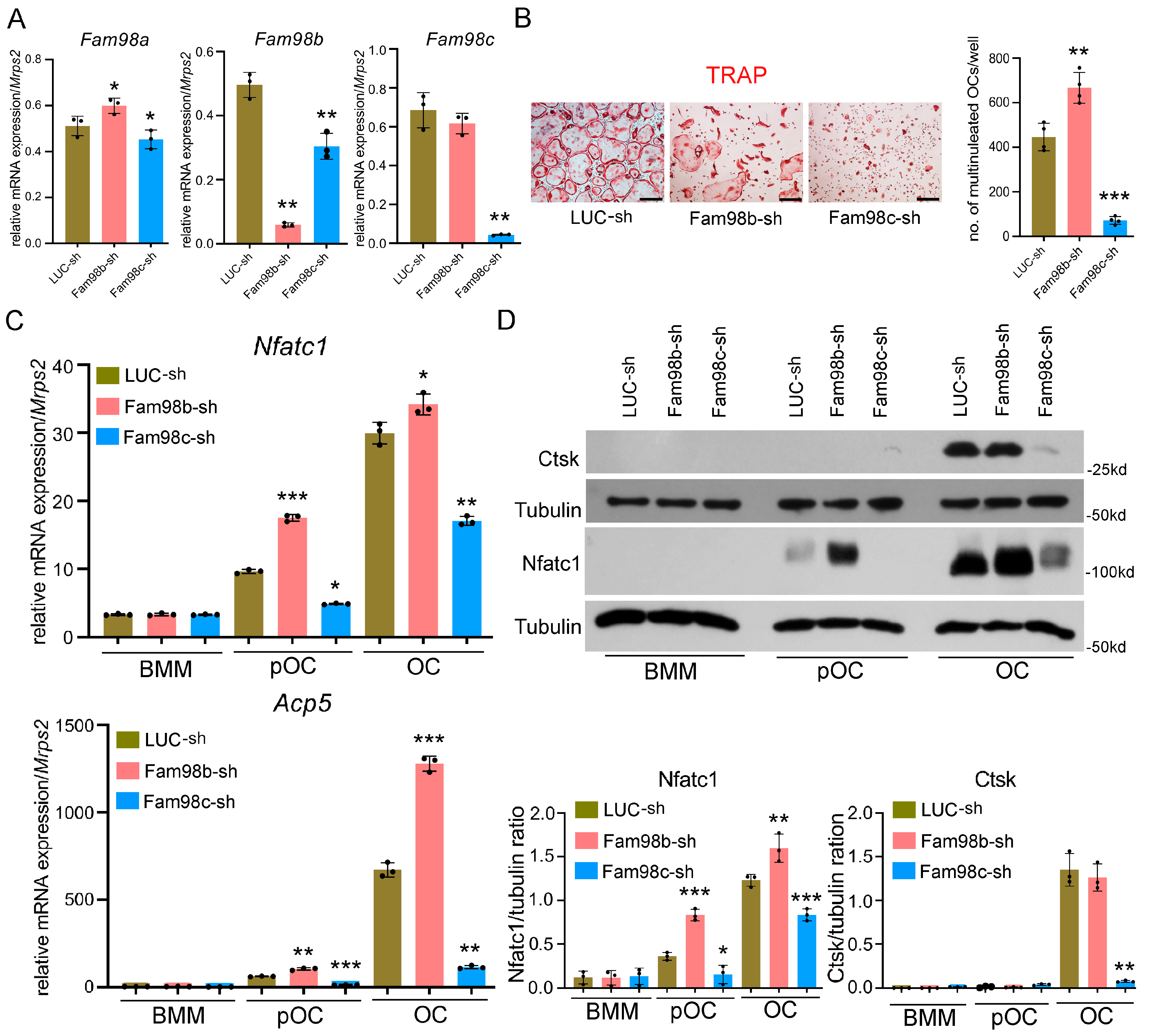
3.2. Knockdown of Fam98c but Not Fam98b Expression Attenuates Osteoclastogenesis In Vitro
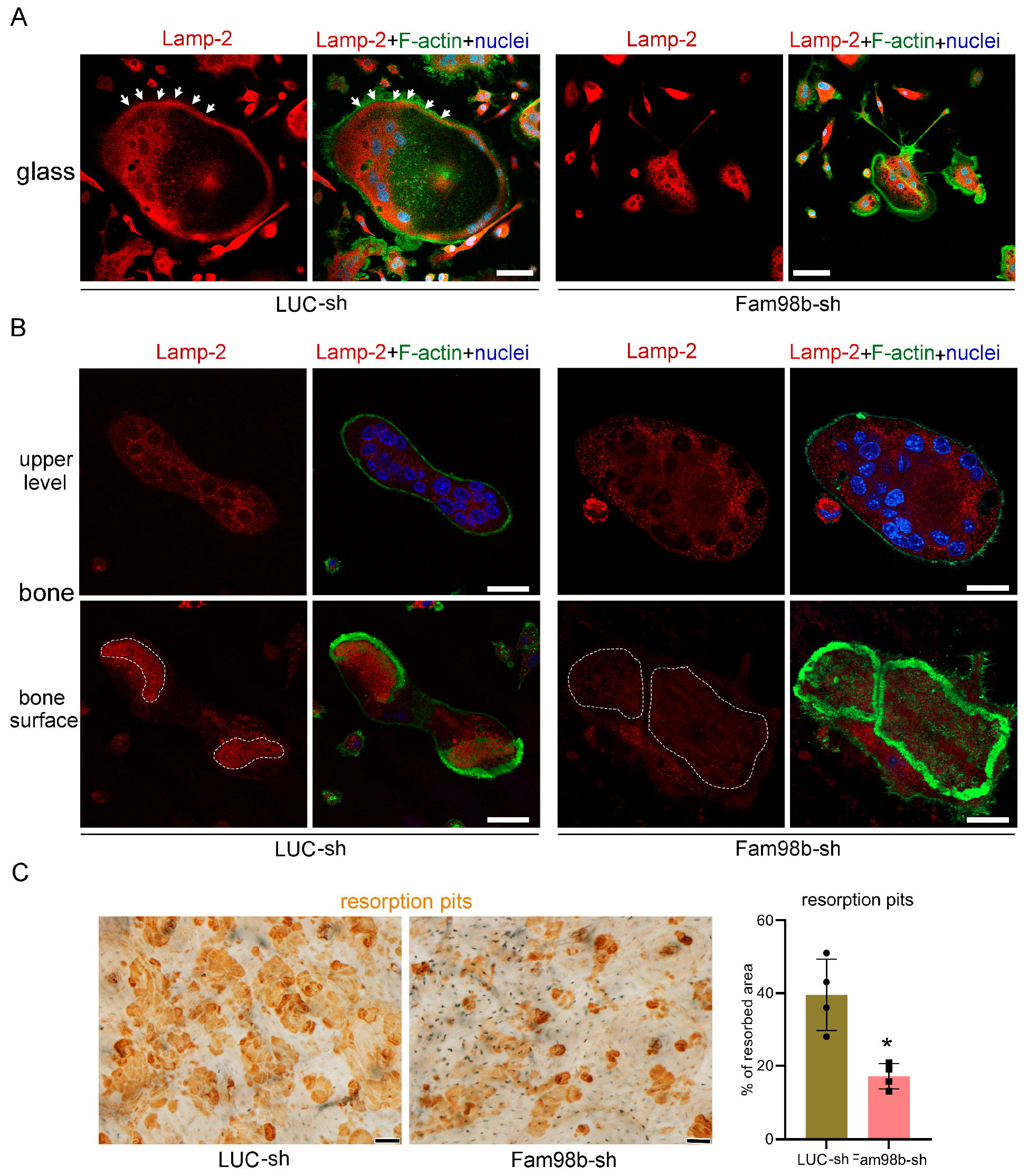
3.3. Decreased Expression of Fam98b in Osteoclasts Inhibits Lysosome Trafficking and Bone Resorption In Vitro
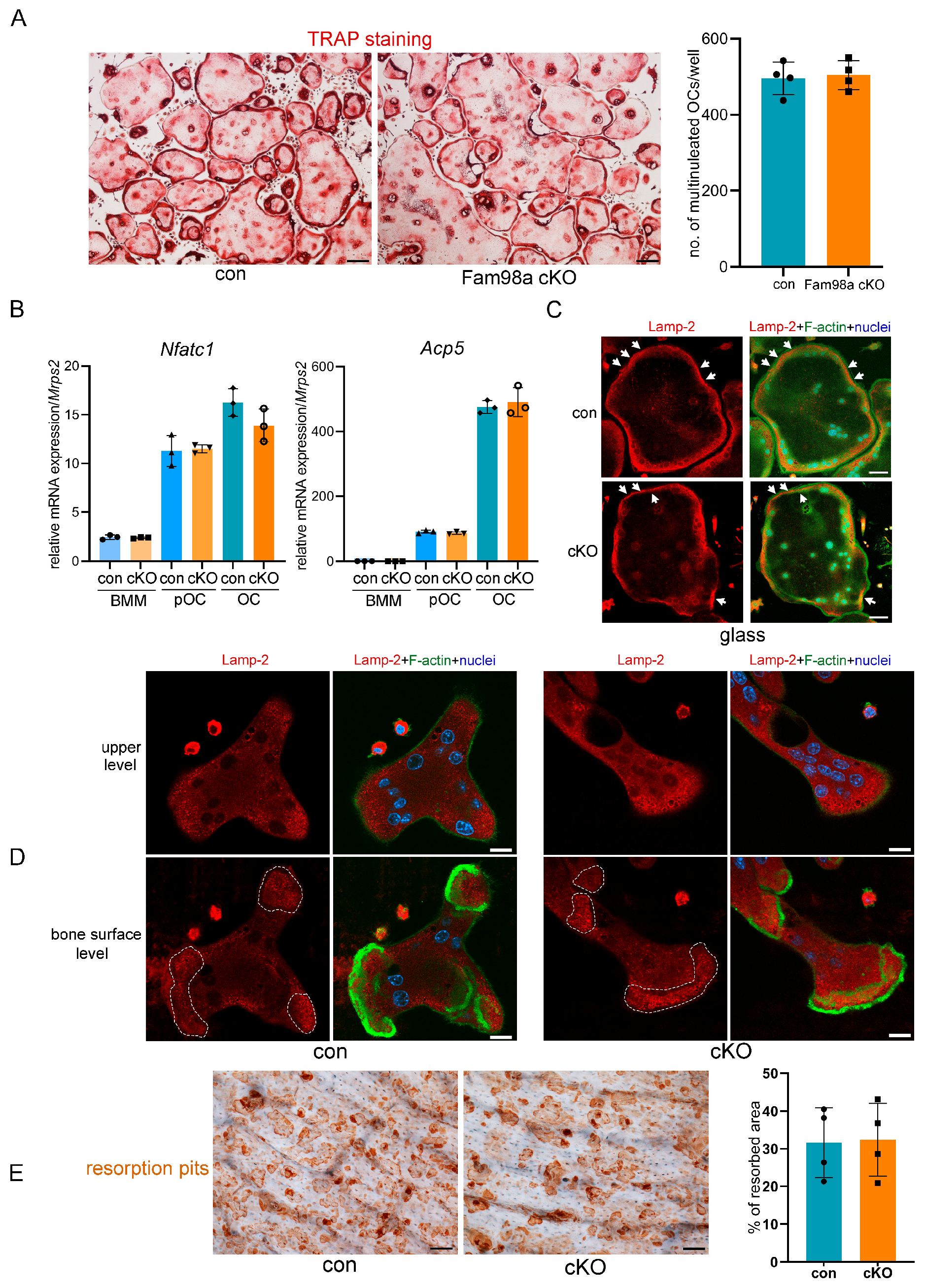
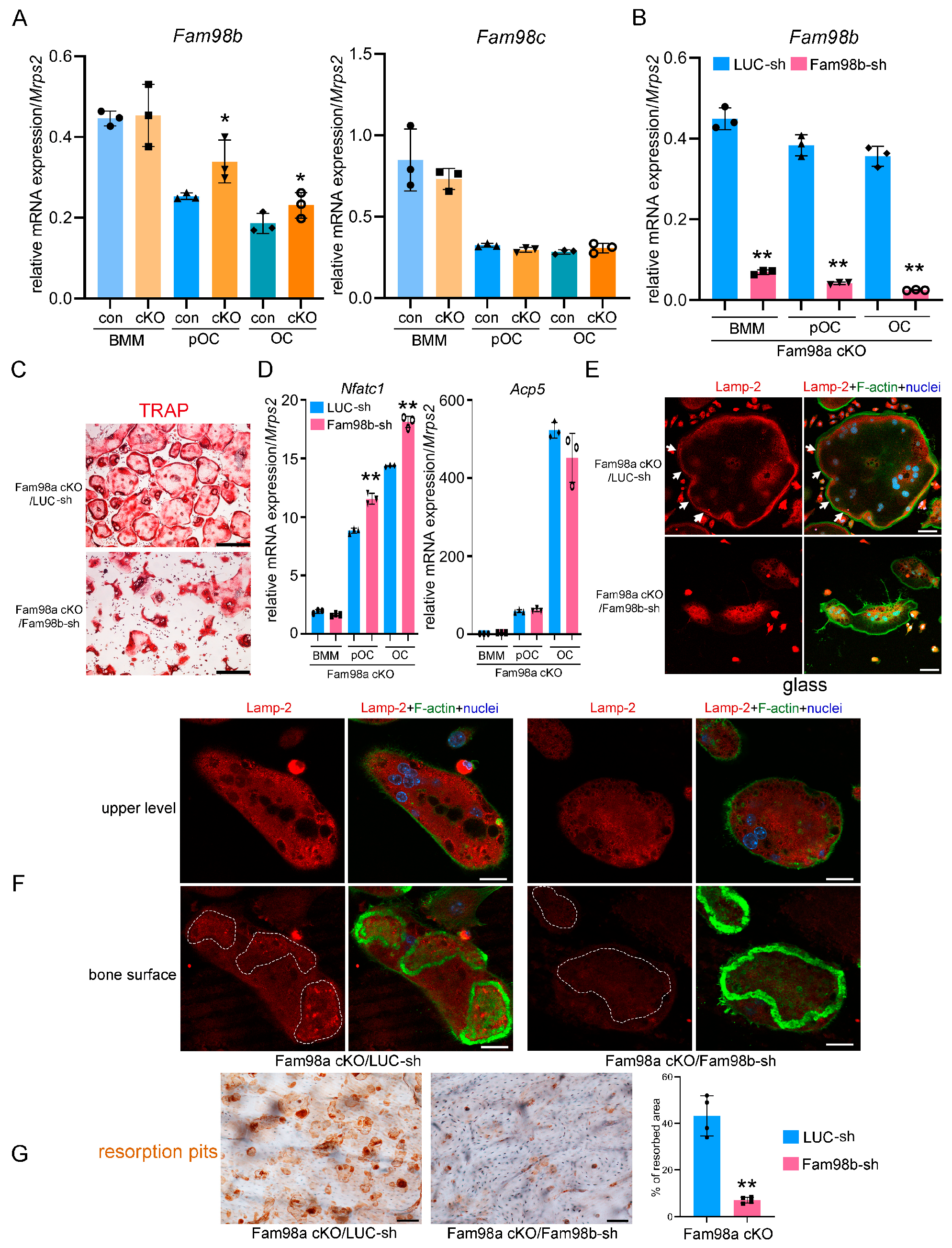
3.4. Fam98a and Fam98b Compensate in Regulation of Lysosome Secretion and Bone Resorption in Osteoclasts
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Teitelbaum, S.L. Bone resorption by osteoclasts. Science 2000, 289, 1504–1508. [Google Scholar] [CrossRef] [PubMed]
- Boyle, W.J.; Simonet, W.S.; Lacey, D.L. Osteoclast differentiation and activation. Nature 2003, 423, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Palagano, E.; Menale, C.; Sobacchi, C.; Villa, A. Genetics of Osteopetrosis. Curr. Osteoporos. Rep. 2018, 16, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Huybrechts, Y.; Van Hul, W. Osteopetrosis associated with PLEKHM1 and SNX10 genes, both involved in osteoclast vesicular trafficking. Bone 2022, 164, 116520. [Google Scholar] [CrossRef]
- Van Wesenbeeck, L.; Odgren, P.R.; Coxon, F.P.; Frattini, A.; Moens, P.; Perdu, B.; MacKay, C.A.; Van Hul, E.; Timmermans, J.P.; Vanhoenacker, F.; et al. Involvement of PLEKHM1 in osteoclastic vesicular transport and osteopetrosis in incisors absent rats and humans. J. Clin. Investig. 2007, 117, 919–930. [Google Scholar] [CrossRef]
- Fujiwara, T.; Ye, S.; Castro-Gomes, T.; Winchell, C.G.; Andrews, N.W.; Voth, D.E.; Varughese, K.I.; Mackintosh, S.G.; Feng, Y.; Pavlos, N.; et al. PLEKHM1/DEF8/RAB7 complex regulates lysosome positioning and bone homeostasis. JCI Insight 2016, 1, e86330. [Google Scholar] [CrossRef]
- Soldati, T.; Rancaño, C.; Geissler, H.; Pfeffer, S.R. Rab7 and Rab9 are recruited onto late endosomes by biochemically distinguishable processes. J. Biol. Chem. 1995, 270, 25541–25548. [Google Scholar] [CrossRef]
- Méresse, S.; Gorvel, J.P.; Chavrier, P. The rab7 GTPase resides on a vesicular compartment connected to lysosomes. J. Cell Sci. 1995, 108 Pt 11, 3349–3358. [Google Scholar] [CrossRef]
- Vitelli, R.; Santillo, M.; Lattero, D.; Chiariello, M.; Bifulco, M.; Bruni, C.B.; Bucci, C. Role of the small GTPase Rab7 in the late endocytic pathway. J. Biol. Chem. 1997, 272, 4391–4397. [Google Scholar] [CrossRef]
- Mukhopadhyay, A.; Barbieri, A.M.; Funato, K.; Roberts, R.; Stahl, P.D. Sequential actions of Rab5 and Rab7 regulate endocytosis in the Xenopus oocyte. J. Cell Biol. 1997, 136, 1227–1237. [Google Scholar] [CrossRef]
- Zhao, H.; Laitala-Leinonen, T.; Parikka, V.; Väänänen, H.K. Downregulation of small GTPase Rab7 impairs osteoclast polarization and bone resorption. J. Biol. Chem. 2001, 276, 39295–39302. [Google Scholar] [CrossRef] [PubMed]
- Das, B.K.; Minocha, T.; Kunika, M.D.; Kannan, A.; Gao, L.; Mohan, S.; Xing, W.; Varughese, K.I.; Zhao, H. Molecular and functional mapping of Plekhm1-Rab7 interaction in osteoclasts. JBMR Plus 2024, 8, ziae034. [Google Scholar] [CrossRef]
- Akter, K.A.; Mansour, M.A.; Hyodo, T.; Ito, S.; Hamaguchi, M.; Senga, T. FAM98A is a novel substrate of PRMT1 required for tumor cell migration, invasion, and colony formation. Tumour Biol. 2016, 37, 4531–4539. [Google Scholar] [CrossRef]
- Ozeki, K.; Sugiyama, M.; Akter, K.A.; Nishiwaki, K.; Asano-Inami, E.; Senga, T. FAM98A is localized to stress granules and associates with multiple stress granule-localized proteins. Mol. Cell. Biochem. 2019, 451, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Pérez-González, A.; Pazo, A.; Navajas, R.; Ciordia, S.; Rodriguez-Frandsen, A.; Nieto, A. hCLE/C14orf166 associates with DDX1-HSPC117-FAM98B in a novel transcription-dependent shuttling RNA-transporting complex. PLoS ONE 2014, 9, e90957. [Google Scholar] [CrossRef] [PubMed]
- Popow, J.; Jurkin, J.; Schleiffer, A.; Martinez, J. Analysis of orthologous groups reveals archease and DDX1 as tRNA splicing factors. Nature 2014, 511, 104–107. [Google Scholar] [CrossRef]
- Pazo, A.; Pérez-González, A.; Oliveros, J.C.; Huarte, M.; Chavez, J.P.; Nieto, A. hCLE/RTRAF-HSPC117-DDX1-FAM98B: A New Cap-Binding Complex That Activates mRNA Translation. Front. Physiol. 2019, 10, 92. [Google Scholar] [CrossRef]
- Kroupova, A.; Ackle, F.; Asanović, I.; Weitzer, S.; Boneberg, F.M.; Faini, M.; Leitner, A.; Chui, A.; Aebersold, R.; Martinez, J.; et al. Molecular architecture of the human tRNA ligase complex. eLife 2021, 10, e71656. [Google Scholar] [CrossRef]
- Shaheen, R.; Szymanska, K.; Basu, B.; Patel, N.; Ewida, N.; Faqeih, E.; Al Hashem, A.; Derar, N.; Alsharif, H.; Aldahmesh, M.A.; et al. Characterizing the morbid genome of ciliopathies. Genome Biol. 2016, 17, 242. [Google Scholar] [CrossRef]
- Takeshita, S.; Kaji, K.; Kudo, A. Identification and characterization of the new osteoclast progenitor with macrophage phenotypes being able to differentiate into mature osteoclasts. J. Bone Miner. Res. 2000, 15, 1477–1488. [Google Scholar] [CrossRef]
- Zhao, H.; Väänänen, H.K. Pharmacological sequestration of intracellular cholesterol in late endosomes disrupts ruffled border formation in osteoclasts. J. Bone Miner. Res. 2006, 21, 456–465. [Google Scholar] [CrossRef] [PubMed]
- Lewis, V.; Green, S.A.; Marsh, M.; Vihko, P.; Helenius, A.; Mellman, I. Glycoproteins of the lysosomal membrane. J. Cell Biol. 1985, 100, 1839–1847. [Google Scholar] [CrossRef] [PubMed]
- Das, B.K.; Wang, L.; Fujiwara, T.; Zhou, J.; Aykin-Burns, N.; Krager, K.J.; Lan, R.; Mackintosh, S.G.; Edmondson, R.; Jennings, M.L.; et al. Transferrin receptor 1-mediated iron uptake regulates bone mass in mice via osteoclast mitochondria and cytoskeleton. Elife 2022, 11, e73539. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Xu, Y.; Zhang, Y.; Duan, X. Molecular Mechanisms of Craniofacial and Dental Abnormalities in Osteopetrosis. Int. J. Mol. Sci. 2023, 24, 10412. [Google Scholar] [CrossRef] [PubMed]
- Schou, K.B.; Andersen, J.S.; Pedersen, L.B. A divergent calponin homology (NN-CH) domain defines a novel family: Implications for evolution of ciliary IFT complex B proteins. Bioinformatics 2014, 30, 899–902. [Google Scholar] [CrossRef]
- Akter, K.A.; Mansour, M.A.; Hyodo, T.; Senga, T. FAM98A associates with DDX1-C14orf166-FAM98B in a novel complex involved in colorectal cancer progression. Int. J. Biochem. Cell Biol. 2017, 84, 1–13. [Google Scholar] [CrossRef]
- Sutton, M.M.; Duffy, M.P.; Verbruggen, S.W.; Jacobs, C.R. Osteoclastogenesis Requires Primary Cilia Disassembly and Can Be Inhibited by Promoting Primary Cilia Formation Pharmacologically. Cells Tissues Organs 2024, 213, 235–244. [Google Scholar] [CrossRef]
- Yuan, X.; Cao, J.; He, X.; Serra, R.; Qu, J.; Cao, X.; Yang, S. Ciliary IFT80 balances canonical versus non-canonical hedgehog signalling for osteoblast differentiation. Nat. Commun. 2016, 7, 11024. [Google Scholar] [CrossRef]
- Moraes de Lima Perini, M.; Pugh, J.N.; Scott, E.M.; Bhula, K.; Chirgwin, A.; Reul, O.N.; Berbari, N.F.; Li, J. Primary cilia in osteoblasts and osteocytes are required for skeletal development and mechanotransduction. bioRxiv 2023. [Google Scholar] [CrossRef]

| Reagent Type | Designation | Source | Catalog Number | Information |
|---|---|---|---|---|
| Cell cultures | Alpha-MEM | MilliporeSigma, St. Louis, MO, USA | M0644-10 × 1 L | |
| High glucose DMEM | MilliporeSigma, St. Louis, MO, USA | D5648-10 × 1 L | ||
| 10× penicillin–streptomycin-L-glutamine | MilliporeSigma, St. Louis, MO, USA | G1146 | ||
| Fetal bovine serum (FBS) | R&D Systems, Minneapolis, MN, USA | |||
| 10× Red Blood Cell Lysis Buffer | Abcam, Cambridge, MA, USA | ab204733 | ||
| 10× Trypsin/EDTA | Thermo-Fisher Scientific, Waltham, MA, USA | 15400-054 | ||
| Puromycin | MilliporeSigma, St. Louis, MO, USA | P8833-10MG | 6 µg/mL | |
| Antibodies | Mouse monoclonal anti-Nfatc1 | Santa Cruz Biotechnology, Dallas, TX, USA | sc-7294 | WB 1:250 |
| Mouse monoclonal anti-Cathepsin K | MilliporeSigma, St. Louis, MO, USA | MAB3324 | WB 1:2000 | |
| Mouse monoclonal anti-Tubulin | MilliporeSigma, St. Louis, MO, USA | T9026 | WB 1:5000 | |
| Rat monoclonal anti-Lamp-2 | Developmental Studies Hybridoma Bank, Iowa City, IA, USA | GL2A7 | IF 1:200 | |
| HRP-goat anti-mouse | Cell Signaling Technology, Danvers, MA, USA | 7076 | WB 1:5000 | |
| TRITC AffiniPure donkey anti-rat | Jackson Immunoresearch, West Grove, PA, USA | 712-025-150 | IF 1:200 | |
| Primers | Murine Fam98a-exon 4 forward | IDT, Coralville, IA, USA | 5′-TTGTGTCAGTTAAAGGAAAC-3′ | |
| Murine Fam98a-exon 4 backward | IDT, Coralville, IA, USA | 5′-TGGGCTGGTCCCATCGGCTTC-3′ | ||
| Murine β-actin gDNA forward | IDT, Coralville, IA, USA | 5′-TTCGCCATGGATGACGATATC-3′ | ||
| Murine β-actin gDNA backward | IDT, Coralville, IA, USA | 5′-GAATACAGCCCGGGGAGCATC-3′ | ||
| Murine Fam98a-flox forward | IDT, Coralville, IA, USA | 5′-GAGCCCAGGTTGGCCTCAGATTCT-3′ | ||
| Murine Fam98a-flox backward | IDT, Coralville, IA, USA | 5′-ACTGACGCTGTCACACGTGACTCC-3′ | ||
| Murine Fam98a-FAM | Thermo-Fisher Scientific, Waltham, MA, USA | Mm01223835_m1 | qPCR primer | |
| Murine Fam98b-FAM | Thermo-Fisher Scientific, Waltham, MA, USA | Mm01277500_m1 | qPCR primer | |
| Murine Fam98c -FAM | Thermo-Fisher Scientific, Waltham, MA, USA | Mm00503820_m1 | qPCR primer | |
| Murine Nfatc1-FAM | Thermo-Fisher Scientific, Waltham, MA, USA | Mm00479445_m1 | qPCR primer | |
| Murine Acp5-FAM | Thermo-Fisher Scientific, Waltham, MA, USA | Mm00475698_m1 | qPCR primer | |
| Murine Mrps2 | Thermo-Fisher Scientific, Waltham, MA, USA | Mm00475529_m1 | qPCR primer | |
| Plasmids | pLKO.1 Fam98b-sh | MilliporeSigma, St. Louis, MO, USA | TRCN 0000190520 | lentiviral targeting vector |
| pLKO.1 Fam98c-sh | MilliporeSigma, St. Louis, MO, USA | TRCN 0000192515 | lentiviral targeting vector | |
| pLKO.1 LUC-sh | MilliporeSigma, St. Louis, MO, USA | SHC007 | lentiviral targeting vector | |
| pMD2.G | Addgene, Watertown, MA, USA | 12259 | lentiviral packaging vector | |
| pCMV-delta R8.2 | Addgene, Watertown, MA, USA | 12263 | lentiviral packaging vector | |
| Compounds and solutions | Protamine sulfate | MilliporeSigma, St. Louis, MO, USA | P4020-1G | lentiviral transduction |
| Opti-MEM | Thermo-Fisher Scientific, Waltham, MA, USA | 11058-021 | plasmid transfection | |
| TransIT-LT1 | Mirus Bio LLC, Madison, WI, USA | MIR2300 | plasmid transfection | |
| NaNO2 | MilliporeSigma, St. Louis, MO, USA | S2252 | TRAP staining | |
| NaK tartrate | MilliporeSigma, St. Louis, MO, USA | S6170 | TRAP staining | |
| Na acetate | MilliporeSigma, St. Louis, MO, USA | S2889 | TRAP staining | |
| Pararosaniline | MilliporeSigma, St. Louis, MO, USA | P3750 | TRAP staining | |
| Naphthol AS-BI | MilliporeSigma, St. Louis, MO, USA | 1802 | TRAP staining | |
| RIPA buffer | MilliporeSigma, St. Louis, MO, USA | R-0278 | cell lysis | |
| cOmplete, EDTA-free Protease Inhibitor Cocktail | MilliporeSigma, St. Louis, MO, USA | 4693159001 | cell lysis | |
| Enhanced chemiluminescent detection reagents (ECL) | MilliporeSigma, St. Louis, MO, USA | WBKLS0100 | WB | |
| Paraformaldehyde | MilliporeSigma, St. Louis, MO, USA | P6148 | cell fixation | |
| Saponin | MilliporeSigma, St. Louis, MO, USA | S-4521-10G | IF | |
| Bovine serum albumin | MilliporeSigma, St. Louis, MO, USA | BSAV-RO | IF | |
| Alexa Fluro-488 phalloidin | Thermo-Fisher Scientific, Waltham, MA, USA | A12379 | IF | |
| Peroxidase-conjugated WGA (wheat germ agglutinin) lectin | MilliporeSigma, St. Louis, MO, USA | L-7017 | pit staining | |
| 3,3′-diaminobenzidine (DAB) tablets | MilliporeSigma, St. Louis, MO, USA | D-5905 | pit staining | |
| 30% H2O2 | MilliporeSigma, St. Louis, MO, USA | 216763 | pit staining | |
| Kits | RNeasy mini kit | Qiagen, Germantown, MD, USA | 74104 | RNA purification |
| High-capacity cDNA RT kit | Thermo-Fisher Scientific, Waltham, MA, USA | 4368813 | cDNA RT | |
| TaqMan Gene Expression Master Mix | Thermo-Fisher Scientific, Waltham, MA, USA | 4369016 | qPCR reaction |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Minocha, T.; Das, B.K.; Kunika, M.D.; Kannan, A.; Gao, L.; Mohan, S.; Xing, W.; Varughese, K.I.; Zhao, H. FAM98 Family Proteins Play Distinct Roles in Osteoclastogenesis and Bone Resorption. Biology 2025, 14, 45. https://doi.org/10.3390/biology14010045
Wang L, Minocha T, Das BK, Kunika MD, Kannan A, Gao L, Mohan S, Xing W, Varughese KI, Zhao H. FAM98 Family Proteins Play Distinct Roles in Osteoclastogenesis and Bone Resorption. Biology. 2025; 14(1):45. https://doi.org/10.3390/biology14010045
Chicago/Turabian StyleWang, Lei, Tarun Minocha, Bhaba K. Das, Mikaela D. Kunika, Aarthi Kannan, Ling Gao, Subburaman Mohan, Weirong Xing, Kottayil I. Varughese, and Haibo Zhao. 2025. "FAM98 Family Proteins Play Distinct Roles in Osteoclastogenesis and Bone Resorption" Biology 14, no. 1: 45. https://doi.org/10.3390/biology14010045
APA StyleWang, L., Minocha, T., Das, B. K., Kunika, M. D., Kannan, A., Gao, L., Mohan, S., Xing, W., Varughese, K. I., & Zhao, H. (2025). FAM98 Family Proteins Play Distinct Roles in Osteoclastogenesis and Bone Resorption. Biology, 14(1), 45. https://doi.org/10.3390/biology14010045






