The Extremophiles: Adaptation Mechanisms and Biotechnological Applications
Simple Summary
Abstract
1. Introduction
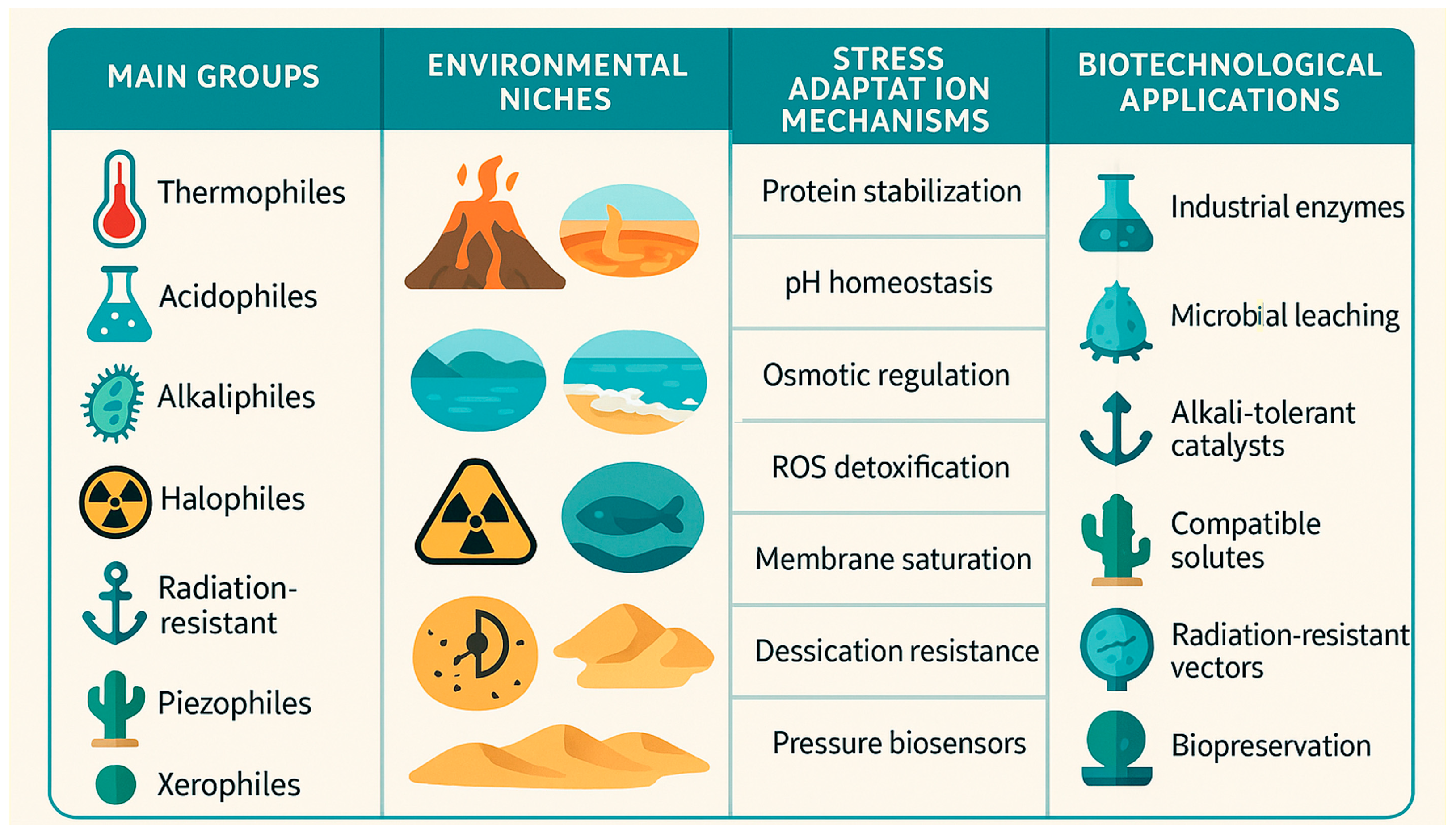
2. Extreme Conditions Triggering the Adaptation
3. Extreme Habitat as Key Decision Maker
4. Future Directions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rothschild, L.J.; Mancinelli, R.L. Life in Extreme Environments. Nature 2001, 409, 1092–1101. [Google Scholar] [CrossRef] [PubMed]
- Cockell, C.S. Vacant Habitats in the Universe. Trends Ecol. Evol. 2011, 26, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Pikuta, E.V.; Hoover, R.B.; Tang, J. Microbial Extremophiles at the Limits of Life. Crit. Rev. Microbiol. 2007, 33, 183–209. [Google Scholar] [CrossRef] [PubMed]
- Cavicchioli, R.; Charlton, T.; Ertan, H.; Mohd Omar, S.; Siddiqui, K.S.; Williams, T.J. Biotechnological Uses of Enzymes from Psychrophiles. Microb. Biotechnol. 2011, 4, 449–460. [Google Scholar] [CrossRef]
- Martínez-Espinosa, R.M. Microorganisms and Their Metabolic Capabilities in the Context of the Biogeochemical Nitrogen Cycle at Extreme Environments. Int. J. Mol. Sci. 2020, 21, 4228. [Google Scholar] [CrossRef]
- Merino, N.; Aronson, H.S.; Bojanova, D.P.; Feyhl-Buska, J.; Wong, M.L.; Zhang, S.; Giovannelli, D. Living at the Extremes: Extremophiles and the Limits of Life in a Planetary Context. Front. Microbiol. 2019, 10, 780. [Google Scholar] [CrossRef]
- Mastascusa, V.; Romano, I.; Di Donato, P.; Poli, A.; Della Corte, V.; Rotundi, A.; Bussoletti, E.; Quarto, M.; Pugliese, M.; Nicolaus, B. Extremophiles Survival to Simulated Space Conditions: An Astrobiology Model Study. Orig. Life Evol. Biosph. 2014, 44, 231–237. [Google Scholar] [CrossRef]
- Kochhar, N.; Shrivastava, S.; Ghosh, A.; Rawat, V.S.; Sodhi, K.K.; Kumar, M. Perspectives on the Microorganism of Extreme Environments and Their Applications. Curr. Res. Microb. Sci. 2022, 3, 100134. [Google Scholar] [CrossRef]
- Dalmaso, G.Z.; Ferreira, D.; Vermelho, A.B. Marine Extremophiles: A Source of Hydrolases for Biotechnological Applications. Mar. Drugs 2015, 13, 1925–1965. [Google Scholar] [CrossRef]
- Colman, D.R.; Poudel, S.; Hamilton, T.L.; Havig, J.R.; Selensky, M.J.; Shock, E.L.; Boyd, E.S. Geobiological Feedbacks and the Evolution of Thermoacidophiles. ISME J. 2018, 12, 225–236. [Google Scholar] [CrossRef]
- Mesbah, N.M. Industrial Biotechnology Based on Enzymes From Extreme Environments. Front. Bioeng. Biotechnol. 2022, 10, 870083. [Google Scholar] [CrossRef] [PubMed]
- Ishino, S.; Ishino, Y. DNA Polymerases as Useful Reagents for Biotechnology—The History of Developmental Research in the Field. Front. Microbiol. 2014, 5, 465. [Google Scholar] [CrossRef]
- Rampelotto, P.H. Extremophiles and Extreme Environments. Life 2013, 3, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Coker, J.A. Recent Advances in Understanding Extremophiles. F1000Research 2019, 8, 1917. [Google Scholar] [CrossRef] [PubMed]
- Dimitriu, T.; Szczelkun, M.D.; Westra, E.R. Evolutionary Ecology and Interplay of Prokaryotic Innate and Adaptive Immune Systems. Curr. Biol. 2020, 30, R1189–R1202. [Google Scholar] [CrossRef]
- Singh, P.; Jain, K.; Desai, C.; Tiwari, O.; Madamvar, D. Microbial Community Dynamics of Extremophiles/Extreme Environment. In Microbial Diversity in the Genomic Era; Academic Press: Cambridge, MA, USA, 2019; pp. 323–332. ISBN 978-0-12-814849-5. [Google Scholar]
- Grant, P.R.; Grant, B.R.; Huey, R.B.; Johnson, M.T.J.; Knoll, A.H.; Schmitt, J. Evolution Caused by Extreme Events. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372, 20160146. [Google Scholar] [CrossRef]
- Chevin, L.-M.; Hoffmann, A.A. Evolution of Phenotypic Plasticity in Extreme Environments. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372, 20160138. [Google Scholar] [CrossRef]
- Peng, Z.; Liu, G.; Huang, K. Cold Adaptation Mechanisms of a Snow Alga Chlamydomonas Nivalis During Temperature Fluctuations. Front. Microbiol. 2020, 11, 611080. [Google Scholar] [CrossRef]
- Perfumo, A.; Freiherr von Sass, G.J.; Nordmann, E.-L.; Budisa, N.; Wagner, D. Discovery and Characterization of a New Cold-Active Protease From an Extremophilic Bacterium via Comparative Genome Analysis and in Vitro Expression. Front. Microbiol. 2020, 11, 881. [Google Scholar] [CrossRef]
- Berlemont, R.; Gerday, C. 1.16—Extremophiles. In Comprehensive Biotechnology, 3rd ed.; Moo-Young, M.B.T.-C.B., Ed.; Pergamon: Oxford, UK, 2011; pp. 203–216. ISBN 978-0-444-64047-5. [Google Scholar]
- Maheshwari, R.; Bharadwaj, G.; Bhat, M.K. Thermophilic Fungi: Their Physiology and Enzymes. Microbiol. Mol. Biol. Rev. 2000, 64, 461–488. [Google Scholar] [CrossRef]
- Santos, F.; Antón, J. Extremophiles: Hypersaline Environments. In Encyclopedia of Microbiology, 4th ed.; Schmidt, T.M., Ed.; Academic Press: Oxford, UK, 2019; pp. 270–275. ISBN 978-0-12-811737-8. [Google Scholar]
- Blaser, M.J.; Cardon, Z.G.; Cho, M.K.; Dangl, J.L.; Donohue, T.J.; Green, J.L.; Knight, R.; Maxon, M.E.; Northen, T.R.; Pollard, K.S.; et al. Toward a Predictive Understanding of Earth’s Microbiomes to Address 21st Century Challenges. mBio 2016, 7, e00714–e00716. [Google Scholar] [CrossRef] [PubMed]
- Furhan, J. Adaptation, Production, and Biotechnological Potential of Cold-Adapted Proteases from Psychrophiles and Psychrotrophs: Recent Overview. J. Genet. Eng. Biotechnol. 2020, 18, 36. [Google Scholar] [CrossRef] [PubMed]
- Gunjal, A.; Waghmode, M.; Annasaheb Magar Mahavidyalaya, S.; Patil, N.; Aparna, G.; Meghmala, W.; Neha, P. Role of Extremozymes in Bioremediation. Res. J. Biotechnol. 2021, 16. Available online: https://pdeaamcollege.edu.in/ResearchUpload/41_37%20%20Role%20of%20extremozymes%20in%20bioremediation-%20Patil%20Waghmaode.pdf (accessed on 8 April 2025).
- Tesei, D.; Quartinello, F.; Guebitz, G.M.; Ribitsch, D.; Nöbauer, K.; Razzazi-Fazeli, E.; Sterflinger, K. Shotgun Proteomics Reveals Putative Polyesterases in the Secretome of the Rock-Inhabiting Fungus Knufia Chersonesos. Sci. Rep. 2020, 10, 9770. [Google Scholar] [CrossRef]
- Atanasova, N.; Stoitsova, S.; Paunova-krasteva, T.; Kambourova, M. Plastic Degradation by Extremophilic Bacteria. Int. J. Mol. Sci. 2021, 22, 5610. [Google Scholar] [CrossRef]
- Gumulya, Y.; Boxall, N.J.; Khaleque, H.N.; Santala, V.; Carlson, R.P.; Kaksonen, A.H. In a Quest for Engineering Acidophiles for Biomining Applications: Challenges and Opportunities. Genes 2018, 9, 116. [Google Scholar] [CrossRef]
- Jerez, C. Biomining of Metals: How to Access and Exploit Natural Resource Sustainably. Microb. Biotechnol. 2017, 10, 1191–1193. [Google Scholar] [CrossRef]
- Anwar, U.B.; Zwar, I.P.; de Souza, A.O. Biomolecules Produced by Extremophiles Microorganisms and Recent Discoveries. In New and Future Developments in Microbial Biotechnology and Bioengineering: Microbial Biomolecules: Properties, Relevance, and Their Translational Applications; Elsevier: Amsterdam, The Netherlands, 2020; pp. 247–270. [Google Scholar] [CrossRef]
- Stetter, K.O. Extremophiles and Their Adaptation to Hot Environments. FEBS Lett. 1999, 452, 22–25. [Google Scholar] [CrossRef]
- Chien, A.; Edgar, D.B.; Trela, J.M. Deoxyribonucleic Acid Polymerase from the Extreme Thermophile Thermus Aquaticus. J. Bacteriol. 1976, 127, 1550–1557. [Google Scholar] [CrossRef]
- Takagi, M.; Nishioka, M.; Kakihara, H.; Kitabayashi, M.; Inoue, H.; Kawakami, B.; Oka, M.; Imanaka, T. Characterization of DNA Polymerase from Pyrococcus Sp. Strain KOD1 and Its Application to PCR. Appl. Environ. Microbiol. 1997, 63, 4504–4510. [Google Scholar] [CrossRef]
- Gifford, D.R.; Bhattacharyya, A.; Geim, A.; Marshall, E.; Krašovec, R.; Knight, C.G. Environmental and Genetic Influence on the Rate and Spectrum of Spontaneous Mutations in Escherichia Coli. Microbiology 2024, 170, 001452. [Google Scholar] [CrossRef] [PubMed]
- Marie, V.; Lin, J. Cannibalistic Viruses in the Aquatic Environment: Role of Virophages in Manipulating Microbial Communities. Int. J. Environ. Sci. Technol. 2016, 13, 2097–2104. [Google Scholar] [CrossRef]
- Kobras, C.M.; Falush, D. Adapting for Life in the Extreme. eLife 2019, 8, e48999. [Google Scholar] [CrossRef]
- Polz, M.F.; Alm, E.J.; Hanage, W.P. Horizontal Gene Transfer and the Evolution of Bacterial and Archaeal Population Structure. Trends Genet. 2013, 29, 170–175. [Google Scholar] [CrossRef]
- Daly, M.J. A New Perspective on Radiation Resistance Based on Deinococcus Radiodurans. Nat. Rev. Microbiol. 2009, 7, 237–245. [Google Scholar] [CrossRef]
- Martínez-Puente, D.H.; Pérez-Trujillo, J.J.; Zavala-Flores, L.M.; García-García, A.; Villanueva-Olivo, A.; Rodríguez-Rocha, H.; Valdés, J.; Saucedo-Cárdenas, O.; Montes de Oca-Luna, R.; Loera-Arias, M.d.J. Plasmid DNA for Therapeutic Applications in Cancer. Pharmaceutics 2022, 14, 1861. [Google Scholar] [CrossRef] [PubMed]
- Sguazzi, G.; Fasani, G.; Renò, F.; Gino, S. Biobanks: Archives or Resources? Their Secondary Use for Forensic Purposes—A Systematic Review. Forensic Sci. 2024, 4, 42–61. [Google Scholar] [CrossRef]
- Uherek, C.; Wels, W. DNA-Carrier Proteins for Targeted Gene Delivery. Adv. Drug Deliv. Rev. 2000, 44, 153–166. [Google Scholar] [CrossRef]
- Benner, S.A.; Sismour, A.M. Synthetic Biology. Nat. Rev. Genet. 2005, 6, 533–543. [Google Scholar] [CrossRef]
- Foo, J.L.; Ching, C.B.; Chang, M.W.; Leong, S.S.J. The Imminent Role of Protein Engineering in Synthetic Biology. Biotechnol. Adv. 2012, 30, 541–549. [Google Scholar] [CrossRef]
- Kour, D.; Rana, K.L.; Kaur, T.; Singh, B.; Chauhan, V.S.; Kumar, A.; Rastegari, A.A.; Yadav, N.; Yadav, A.N.; Gupta, V.K. Extremophiles for Hydrolytic Enzymes Productions: Biodiversity and Potential Biotechnological Applications. In Bioprocessing for Biomolecules Production; John Wiley & Sons: Hoboken, NJ, USA, 2019; pp. 321–372. ISBN 9781119434436. [Google Scholar]
- Seckbach, J.; Chela-Flores, J. Extremophiles and Chemotrophs as Contributors to Astrobiological Signatures on Europa: A Review of Biomarkers of Sulfate-Reducers and Other Microorganisms. In Proceedings of the SPIE Optical Engineering + Applications, San Diego, CA, USA, 26–30 August 2007. [Google Scholar]
- Mangold, S.; Valdés, J.H.; Holmes, D.S.; Dopson, M. Sulfur Metabolism in the Extreme Acidophile Acidithiobacillus caldus. Front. Microbiol. 2011, 2, 17. [Google Scholar] [CrossRef] [PubMed]
- Marlow, J.J.; Steele, J.A.; Case, D.H.; Connon, S.A.; Levin, L.A.; Orphan, V.J. Microbial Abundance and Diversity Patterns Associated with Sediments and Carbonates from the Methane Seep Environments of Hydrate Ridge, OR. Front. Mar. Sci. 2014, 1, 44. [Google Scholar] [CrossRef]
- Witt, M.; Pozzi, R.; Diesch, S.; Hädicke, O.; Grammel, H. New Light on Ancient Enzymes—In Vitro CO2 Fixation by Pyruvate Synthase of Desulfovibrio Africanus and Sulfolobus acidocaldarius. FEBS J. 2019, 286, 4494–4508. [Google Scholar] [CrossRef]
- Narayanan, M.; Ali, S.S.; El-sheekh, M. A Comprehensive Review on the Potential of Microbial Enzymes in Multipollutant Bioremediation: Mechanisms, Challenges, and Future Prospects. J. Environ. Manag. 2023, 334, 117532. [Google Scholar] [CrossRef]
- Haque, S.; Singh, R.; Harakeh, S.M.; Teklemariam, A.D.; Alharthy, S.A.; Tripathi, S.C.; Singh, R.P.; Hassan, A.A.; Srivastava, N.; Gupta, V.K. Enzymes Based Biocatalysis for the Treatment of Organic Pollutants and Bioenergy Production. Curr. Opin. Green Sustain. Chem. 2022, 38, 100709. [Google Scholar] [CrossRef]
- May, S.W. Applications of Oxidoreductases. Curr. Opin. Biotechnol. 1999, 10, 370–375. [Google Scholar] [CrossRef] [PubMed]
- Burns, R.C.; Hardy, R.W. Nitrogen Fixation in Bacteria and Higher Plants. In Molecular Biology Biochemistry Biophysics; Springer: Berlin/Heidelberg, Germany, 1975; pp. 1–189. [Google Scholar] [CrossRef]
- Neemisha; Kumar, A.; Sharma, P.; Kaur, A.; Sharma, S.; Jain, R. Harnessing Rhizobacteria to Fulfil Inter-Linked Nutrient Dependency on Soil and Alleviate Stresses in Plants. J. Appl. Microbiol. 2022, 133, 2694–2716. [Google Scholar] [CrossRef]
- Timofeeva, A.M.; Galyamova, M.R.; Sedykh, S.E. Plant Growth-Promoting Soil Bacteria: Nitrogen Fixation, Phosphate Solubilization, Siderophore Production, and Other Biological Activities. Plants 2023, 12, 4074. [Google Scholar] [CrossRef]
- Steimbrüch, B.A.; Sartorio, M.G.; Cortez, N.; Albanesi, D.; Lisa, M.; Repizo, G.D. The Distinctive Roles Played by the Superoxide Dismutases of the Extremophile Acinetobactersp. Ver3. Sci. Rep. 2022, 12, 4321. [Google Scholar] [CrossRef]
- Tanwir, K.; Javed, M.T.; Shahid, M.; Akram, M.S.; Ali, Q. Chapter 32—Antioxidant Defense Systems in Bioremediation of Organic Pollutants. In Handbook of Bioremediation; Hasanuzzaman, M., Prasad, M.N.V., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 505–521. ISBN 978-0-12-819382-2. [Google Scholar]
- Cabej, N.R. 18—Species and Allopatric Speciation. In Epigenetic Principles of Evolution; Cabej, N.R., Ed.; Elsevier: London, UK, 2012; pp. 707–723. ISBN 978-0-12-415831-3. [Google Scholar]
- Laksanalamai, P.; Robb, F.T. Small Heat Shock Proteins from Extremophiles: A Review. Extremophiles 2004, 8, 1–11. [Google Scholar] [CrossRef]
- Assenberg, R.; Wan, P.T.; Geisse, S.; Mayr, L.M. Advances in Recombinant Protein Expression for Use in Pharmaceutical Research. Curr. Opin. Struct. Biol. 2013, 23, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Saibil, H. Chaperone Machines for Protein Folding, Unfolding and Disaggregation. Nat. Rev. Mol. Cell. Biol. 2013, 14, 630–642. [Google Scholar] [CrossRef] [PubMed]
- de Champdoré, M.; Staiano, M.; Rossi, M.; D’Auria, S. Proteins from Extremophiles as Stable Tools for Advanced Biotechnological Applications of High Social Interest. J. R. Soc. Interface 2007, 4, 183–191. [Google Scholar] [CrossRef]
- Weber, A.P.M.; Horst, R.J.; Barbier, G.G.; Oesterhelt, C. Metabolism and Metabolomics of Eukaryotes Living under Extreme Conditions. Int. Rev. Cytol. 2007, 256, 1–34. [Google Scholar] [CrossRef]
- Rappaport, H.B.; Oliverio, A.M. Extreme Environments Offer an Unprecedented Opportunity to Understand Microbial Eukaryotic Ecology, Evolution, and Genome Biology. Nat. Commun. 2023, 14, 4959. [Google Scholar] [CrossRef]
- Aguilera, A. Eukaryotic Organisms in Extreme Acidic Environments, the Río Tinto Case. Life 2013, 3, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Nancucheo, I.; Barrie Johnson, D. Acidophilic Algae Isolated from Mine-Impacted Environments and Their Roles in Sustaining Heterotrophic Acidophiles. Front. Microbiol. 2012, 3, 325. [Google Scholar] [CrossRef]
- Lin, S. Algae. By Linda E Graham and, Lee W Wilcox. Upper Saddle River (New Jersey): Prentice Hall. $74.67. Xvi + 700 p; Ill.; Taxonomic and Subject Indexes. ISBN: 0-13-660333-5. 2000. Q. Rev. Biol. 2002, 77, 70–71. [Google Scholar] [CrossRef]
- McFadden, G.I. Origin and Evolution of Plastids and Photosynthesis in Eukaryotes. Cold Spring Harb. Perspect. Biol. 2014, 6, a016105. [Google Scholar] [CrossRef]
- Elias, M.; Archibald, J.M. Sizing up the Genomic Footprint of Endosymbiosis. Bioessays 2009, 31, 1273–1279. [Google Scholar] [CrossRef]
- Ramos, A.A.; Polle, J.; Tran, D.; Cushman, J.C.; Jin, E.S.; Varela, J.C. The Unicellular Green Alga Dunaliella Salina Teod. as a Model for Abiotic Stress Tolerance: Genetic Advances and Future Perspectives. Algae 2011, 26, 3–20. [Google Scholar] [CrossRef]
- Khan, M.I.; Shin, J.H.; Kim, J.D. The Promising Future of Microalgae: Current Status, Challenges, and Optimization of a Sustainable and Renewable Industry for Biofuels, Feed, and Other Products. Microb. Cell Fact. 2018, 17, 36. [Google Scholar] [CrossRef]
- Downing, J.M.; Lock, S.C.L.; Iovinella, M.; Davey, J.W.; Mackinder, L.C.M.; Chong, J.P.J.; Ashton, P.D.; Feichtinger, G.A.; James, S.; Jeffares, D.C.; et al. Comparisons between Complete Genomes of the Eukaryotic Extremophile Galdieria Sulphuraria Reveals Complex Nuclear Chromosomal Structures. bioRxiv 2022. [Google Scholar] [CrossRef]
- Clapcott, J.E.; Goodwin, E.O.; Harding, J.S. Identifying Catchment-Scale Predictors of Coal Mining Impacts on New Zealand Stream Communities. Environ. Manag. 2016, 57, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Hogsden, K.L.; Harding, J.S. Consequences of Acid Mine Drainage for the Structure and Function of Benthic Stream Communities: A Review. Freshw. Sci. 2011, 31, 108–120. [Google Scholar] [CrossRef]
- Stefanidou, N.; Genitsaris, S.; Lopez-bautista, J.M.; Sommer, U.; Moustaka-Gouni, M. Unicellular Eukaryotic Community Response to Temperature and Salinity Variation in Mesocosm Experiments. Front. Microbiol. 2018, 9, 2444. [Google Scholar] [CrossRef]
- Moustaka-Gouni, M.; Kormas, K.A.; Scotti, M.; Vardaka, E.; Sommer, U. Warming and Acidification Effects on Planktonic Heterotrophic Pico- and Nanoflagellates in a Mesocosm Experiment. Protist 2016, 167, 389–410. [Google Scholar] [CrossRef]
- Loreau, M.; Naeem, S.; Inchausti, P.; Bengtsson, J.; Grime, J.P.; Hector, A.; Hooper, D.U.; Huston, M.A.; Raffaelli, D.; Schmid, B.; et al. Biodiversity and Ecosystem Functioning: Current Knowledge and Future Challenges. Science 2001, 294, 804–808. [Google Scholar] [CrossRef]
- Rajapakse, K.; Drobne, D.; Kastelec, D.; Kogej, K.; Makovec, D.; Gallampois, C.; Amelina, H.; Danielsson, G.; Fanedl, L.; Marinsek-Logar, R.; et al. Proteomic Analyses of Early Response of Unicellular Eukaryotic Microorganism Tetrahymena Thermophila Exposed to TiO2 Particles. Nanotoxicology 2016, 10, 542–556. [Google Scholar] [CrossRef]
- Yang, W.W.; Wang, Y.; Huang, B.; Wang, N.X.; Wei, Z.B.; Luo, J.; Miao, A.J.; Yang, L.Y. TiO2 Nanoparticles Act as a Carrier of Cd Bioaccumulation in the Ciliate Tetrahymena Thermophila. Environ. Sci. Technol. 2014, 48, 7568–7575. [Google Scholar] [CrossRef]
- Merola, A.; Rosa, C.; Paolo De, L.; Raffaele, G.; Aldo, M.; Taddei, R. Revision of Cyanidium Caldarium. Three Species of Acidophilic Algae. G. Bot. Ital. 1981, 115, 189–195. [Google Scholar] [CrossRef]
- Matsuzaki, M.; Misumi, O.; Shin-i, T.; Maruyama, S.; Takahara, M.; Miyagishima, S.; Mori, T.; Nishida, K.; Yagisawa, F.; Nishida, K.; et al. Genome Sequence of the Ultrasmall Unicellular Red Alga Cyanidioschyzon Merolae 10D. Nature 2004, 428, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Nozaki, H.; Takano, H.; Misumi, O.; Terasawa, K.; Matsuzaki, M.; Maruyama, S.; Nishida, K.; Yagisawa, F.; Yoshida, Y.; Fujiwara, T.; et al. A 100%-Complete Sequence Reveals Unusually Simple Genomic Features in the Hot-Spring Red Alga Cyanidioschyzon Merolae. BMC Biol. 2007, 5, 28. [Google Scholar] [CrossRef]
- Hirooka, S.; Tomita, R.; Fujiwara, T.; Ohnuma, M.; Kuroiwa, H.; Kuroiwa, T.; Miyagishima, S. Efficient Open Cultivation of Cyanidialean Red Algae in Acidified Seawater. Sci. Rep. 2020, 10, 13794. [Google Scholar] [CrossRef]
- Castellanos-Huerta, I.; Gómez-Verduzco, G.; Tellez-Isaias, G.; Ayora-Talavera, G.; Bañuelos-Hernández, B.; Petrone-García, V.M.; Fernández-Siurob, I.; Garcia-Casillas, L.A.; Velázquez-Juárez, G. Dunaliella Salina as a Potential Biofactory for Antigens and Vehicle for Mucosal Application. Processes 2022, 10, 1776. [Google Scholar] [CrossRef]
- Raja, R.; Hemaiswarya, S.; Rengasamy, R. Exploitation of Dunaliella for Beta-Carotene Production. Appl. Microbiol. Biotechnol. 2007, 74, 517–523. [Google Scholar] [CrossRef]
- Ianutsevich, E.A.; Danilova, O.A.; Grum-Grzhimaylo, O.A.; Tereshina, V.M. The Role of Osmolytes and Membrane Lipids in the Adaptation of Acidophilic Fungi. Microorganisms 2023, 11, 1733. [Google Scholar] [CrossRef]
- Aguilera, A.; González-Toril, E. Eukaryotic Life in Extreme Environments: Acidophilic Fungi. In Fungi in Extreme Environments: Ecological Role and Biotechnological Significance; Tiquia-Arashiro, S.M., Grube, M., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 21–38. ISBN 978-3-030-19030-9. [Google Scholar]
- Moreno-Perlin, T.; Valdés-Muñoz, G.; Jiménez-Gómez, I.; Gunde-Cimerman, N.; Yarzábal Rodríguez, L.A.; Sánchez-Carbente, M.d.R.; Vargas-Fernández, A.; Gutiérrez-Cepeda, A.; Batista-García, R.A. Extremely Chaotolerant and Kosmotolerant Aspergillus Atacamensis—A Metabolically Versatile Fungus Suitable for Recalcitrant Biosolid Treatment. Front. Microbiol. 2023, 14, 1191312. [Google Scholar] [CrossRef]
- Fernández-López, M.G.; Batista-García, R.A.; Aréchiga-Carvajal, E.T. Alkaliphilic/Alkali-Tolerant Fungi: Molecular, Biochemical, and Biotechnological Aspects. J. Fungi 2023, 9, 652. [Google Scholar] [CrossRef]
- Jančič, S.; Zalar, P.; Kocev, D.; Schroers, H.-J.; Džeroski, S.; Gunde-Cimerman, N. Halophily Reloaded: New Insights into the Extremophilic Life-Style of Wallemia with the Description of Wallemia Hederae Sp. Nov. Fungal Divers. 2016, 76, 97–118. [Google Scholar] [CrossRef]
- Goldstein, B. Tardigrades and Their Emergence as Model Organisms. Curr. Top. Dev. Biol. 2022, 147, 173–198. [Google Scholar] [PubMed]
- Goldstein, B.; Blaxter, M. Tardigrades. Curr. Biol. 2002, 12, R475. [Google Scholar] [CrossRef]
- Hesgrove, C.; Boothby, T.C. The Biology of Tardigrade Disordered Proteins in Extreme Stress Tolerance. Cell Commun. Signal. 2020, 18, 178. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, A.; Tanaka, S.; Yamaguchi, S.; Kuwahara, H.; Takamura, C.; Imajoh-Ohmi, S.; Horikawa, D.D.; Toyoda, A.; Katayama, T.; Arakawa, K.; et al. Two Novel Heat-Soluble Protein Families Abundantly Expressed in an Anhydrobiotic Tardigrade. PLoS ONE 2012, 7, e44209. [Google Scholar] [CrossRef] [PubMed]
- Makarov, K.V.; Sundukov, Y.u.N.; Matalin, A.V. Ground Beetles (Coleoptera, Carabidae) in Fumarole Fields of Kunashir Island, Kuril Archipelago, Russia. Acta Zool. Acad. Sci. Hung. 2020, 6, 97–146. [Google Scholar] [CrossRef]
- Chng, Y.R.; Ong, J.L.Y.; Ching, B.; Chen, X.L.; Hiong, K.C.; Wong, W.P.; Chew, S.F.; Lam, S.H.; Ip, Y.K. Molecular Characterization of Aquaporin 1 and Aquaporin 3 from the Gills of the African Lungfish, Protopterus Annectens, and Changes in Their Branchial MRNA Expression Levels and Protein Abundance during Three Phases of Aestivation. Front. Physiol. 2016, 7, 532. [Google Scholar] [CrossRef]
- Meir, J.U.; Ponganis, P.J. High-Affinity Hemoglobin and Blood Oxygen Saturation in Diving Emperor Penguins. J. Exp. Biol. 2009, 212, 3330–3338. [Google Scholar] [CrossRef]
- Golikov, A.V.; Ceia, F.R.; Sabirov, R.M.; Ablett, J.D.; Gleadall, I.G.; Gudmundsson, G.H.; Hoving, H.J.T.; Judkins, H.; Palsson, J.; Reid, A.L.; et al. The First Global Deep-Sea Stable Isotope Assessment Reveals the Unique Trophic Ecology of Vampire Squid Vampyroteuthis infernalis (Cephalopoda). Sci. Rep. 2019, 9, 19099. [Google Scholar] [CrossRef]
| Type of Extreme Condition | Description | Biological Consequences |
|---|---|---|
| Alkaline or acidic environment | Natural habitats with a pH above 9, like alkaline/soda lakes, limestone caves, and some hot springs, or with a pH under 5, such as volcanic lakes, acidic wetlands, streams, and soil and mine drainage, which are extremely acidic or basic either persistently, periodically, or for short periods. | Protein denaturation, cell membrane damage, enzyme inactivation, disruption of internal pH balance, and altered metabolic processes. |
| Cold | Habitats periodically or consistently below −17 °C either persistently, with regular frequency, or for short periods, like mountains, polar sites, and deep oceans. | Cell membrane damage, intracellular ice formation, dehydration, enzymatic inhibition and cellular damage, and cold adaptation. |
| Hot | Broadly defined, these habitats experience temperatures exceeding 40 °C either constantly, periodically, or for protracted periods. Examples include volcanic regions and geothermal streams. | Dehydration, cell membrane damage, protein denaturation, DNA denaturation, enzyme deactivation, and disruption of biological processes. |
| Hypersaline | Environments with salt concentrations greater than that of seawater, i.e., >3.5%, including salt lakes and mines. | Osmotic stress, shrinkage of the cell, desiccation, and enzyme inactivation. |
| High pressure | Habitats exposed to extreme hydrostatic pressure, such as ocean depths beyond 2000 m and deep lakes. | Cellular compression, enzyme inactivation, membrane disruption, possible DNA and protein denaturation, and cellular adaptation. |
| Radiation | Environments with background radiation levels exceeding the natural average annual exposure of approximatively 2.4 mSv (240 mrem). | DNA damage, cell death, carcinogenesis, cell dysfunction, and cell cycle arrest. |
| Absence of water | Habitats lacking free water whether persistently, periodically, or for short periods, including hot and cold desert environments, and some endolithic habitats. | Dehydration, protein denaturation, cell dysfunction, impaired cellular communication and functions, metabolic inactivity, growth arrest, and death. |
| Absence of light | Regions inaccessible to sunlight, like deep ocean environments and caves. | Reduced energy production, retarded biological rhythms, dependency on alternative energy source, reduced biodiversity, and increased adaptation and specialization. |
| Absence of oxygen | Habitats lacking free oxygen—whether persistently, periodically, or for protracted periods, including habitats within deeper sediments. | Oxygen deprivation, cellular damage and death. Development of semi- or full anaerobe metabolism. |
| Absence of nutrients | Areas on Earth that are nutrient-poor, such as the open ocean, deserts, and high-altitude regions. | Nutrient deficiency and energy depletion, organ dysfunction, and damages leading to death or growth arrest. |
| Human-made extreme environment | Anthropogenically affected habitats, including waste depots, mine tailings, oil-contaminated habitats, and areas polluted by heavy metals or organic compounds. | Severe cellular and tissue damage. |
| Type of Extremophile | Type of Environment | Taxonomic Families of Extremophiles |
|---|---|---|
| Psychrophile | Cold environment | Mostly bacteria, archaea, and eukaryotes (algae) |
| Thermophile and acidophilic thermophile | Hot or hot acidic environment | Mostly bacteria, archaea, and rarely eukaryotes (fungi and algae) |
| Halophile or osmophile | High salt/high sugar | Mostly bacteria, archaea, and rarely eukaryotes (fungi and algae) |
| Acidophile | Acidic environment | Mostly bacteria, archaea, and eukaryotes (algae) |
| Alkaliphile | Alkaline environment | Mostly bacteria, archaea, and eukaryotes (black fungi and algae) |
| Barophile or Piezophile | High-pressure environment | Mostly bacteria, archaea, and rarely eukaryotes (single cell protists, deep-sea fish, and invertebrates) |
| Xerophile | Dry environment | Mostly bacteria, archaea, and eukaryotes (fungi) |
| Radiotolerant | High level of radiating environment | Mostly bacteria, archaea, and eukaryotes (fungi) |
| Endolithic extremophile | Rocky environment | Mostly bacteria, archaea, and eukaryotes (black fungi, lichens, and algae) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marzban, G.; Tesei, D. The Extremophiles: Adaptation Mechanisms and Biotechnological Applications. Biology 2025, 14, 412. https://doi.org/10.3390/biology14040412
Marzban G, Tesei D. The Extremophiles: Adaptation Mechanisms and Biotechnological Applications. Biology. 2025; 14(4):412. https://doi.org/10.3390/biology14040412
Chicago/Turabian StyleMarzban, Gorji, and Donatella Tesei. 2025. "The Extremophiles: Adaptation Mechanisms and Biotechnological Applications" Biology 14, no. 4: 412. https://doi.org/10.3390/biology14040412
APA StyleMarzban, G., & Tesei, D. (2025). The Extremophiles: Adaptation Mechanisms and Biotechnological Applications. Biology, 14(4), 412. https://doi.org/10.3390/biology14040412








