Genome-Wide Dissection of Novel QTLs and Genes Associated with Weed Competitiveness in Early-Backcross Selective Introgression-Breeding Populations of Rice (Oryza sativa L.)
Simple Summary
Abstract
1. Background
2. Methods
2.1. Plant Materials
2.2. Phenotypic Screening for Weed Competitiveness
2.3. Statistical Analysis
2.4. SNP Extraction and Physical Map Construction
2.5. QTL Mapping and Candidate Gene Extraction
3. Results
3.1. ESG Performance of Parental Lines and EB-SILs
3.2. ESV Performance of Parental Lines and EB-SILs
3.3. Correlation Analysis Among Measured Traits
3.4. Principal Component Analysis Among Measured Traits
3.5. SNP Markers Generated by (tGBS®) Sequences for QTL Mapping
3.6. Identification of QTLs for Weed Competitive Traits
3.7. Candidate Genes Associated with ESG and ESV Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Yu, S.; Ali, J.; Zhou, S.; Ren, G.; Xie, H.; Xu, J.; Yu, X.; Zhou, F.; Peng, S.; Ma, L.; et al. From Green Super Rice to Green Agriculture: Reaping the Promise of Functional Genomics Research. Mol. Plant 2022, 15, 9–26. [Google Scholar] [CrossRef] [PubMed]
- Ali, J.; Anumalla, M.; Murugaiyan, V.; Li, Z. Green Super Rice (GSR) Traits: Breeding and Genetics for Multiple Biotic and Abiotic Stress Tolerance in Rice. In Rice Improvement; Springer International Publishing: Cham, Switzerland, 2021; pp. 59–97. [Google Scholar] [CrossRef]
- Arouna, A.; Fatognon, I.A.; Saito, K.; Futakuchi, K. Moving toward Rice Self-Sufficiency in Sub-Saharan Africa by 2030: Lessons Learned from 10 Years of the Coalition for African Rice Development. World Dev. Perspect. 2021, 21, 100291. [Google Scholar] [CrossRef]
- Samal, P.; Babu, S.C.; Mondal, B.; Mishra, S.N. The Global Rice Agriculture towards 2050: An Inter-Continental Perspective. Outlook Agric. 2022, 51, 164–172. [Google Scholar] [CrossRef]
- McNally, K.L.; Henry, A. Tools for Using the International Rice Genebank to Breed for Climate-Resilient Varieties. PLoS Biol. 2023, 21, e3002215. [Google Scholar] [CrossRef]
- Marasini, S.; Joshi, T.; Amgain, L. Direct Seeded Rice Cultivation Method: A New Technology for Climate Change and Food Security. J. Agric. Environ. 2018, 17, 30–38. [Google Scholar] [CrossRef]
- Peramaiyan, P.; Srivastava, A.K.; Kumar, V.; Seelan, L.P.; Banik, N.C.; Khandai, S.; Parida, N.; Kumar, V.; Das, A.; Pattnaik, S.; et al. Crop Establishment and Diversification Strategies for Intensification of Rice-Based Cropping Systems in Rice-Fallow Areas in Odisha. Field Crops Res. 2023, 302, 109078. [Google Scholar] [CrossRef] [PubMed]
- Connor, M.; Cuong, O.Q.; Demont, M.; Sander, B.O.; Nelson, K. The Influence of Climate Change Knowledge on Consumer Valuation of Sustainably Produced Rice in Vietnam. Sustain. Prod. Consum. 2022, 31, 1–12. [Google Scholar] [CrossRef]
- He, A.; Jiang, M.; Nie, L.; Sun, H.; Chen, L.; Man, J. A Preliminary Study of ‘Tidy Field Technology’ to Assess Growth, Development and Weed Control in Direct-Seeded Rice. Field Crops Res. 2022, 277, 108408. [Google Scholar] [CrossRef]
- Ahmed, S.; Alam, M.J.; Hossain, A.; Islam, A.K.M.M.; Awan, T.H.; Soufan, W.; Qahtan, A.A.; Okla, M.K.; El Sabagh, A. Interactive Effect of Weeding Regimes, Rice Cultivars, and Seeding Rates Influence the Rice-Weed Competition under Dry Direct-Seeded Condition. Sustainability 2020, 13, 317. [Google Scholar] [CrossRef]
- Chauhan, B.S.; Awan, T.H.; Abugho, S.B.; Evengelista, G. Effect of Crop Establishment Methods and Weed Control Treatments on Weed Management, and Rice Yield. Field Crops Res. 2015, 172, 72–84. [Google Scholar] [CrossRef]
- Xu, L.; Li, X.; Wang, X.; Xiong, D.; Wang, F. Comparing the Grain Yields of Direct-Seeded and Transplanted Rice: A Meta-Analysis. Agronomy 2019, 9, 767. [Google Scholar] [CrossRef]
- Nazir, A.; Bhat, M.A.; Bhat, T.A.; Bhat, S.F.; Qayoom, S.; Hussain, A.; Lone, B.A.; Jan, B.; Dar, S.A.; John, J. Impact of Crop Establishment Techniques and Weed Management Practices on Oryza sativa L. Growth and Yield. Agron. J. 2023, 115, 1812–1826. [Google Scholar] [CrossRef]
- Mahajan, G.; Kaur, G.; Chauhan, B.S. Seeding Rate and Genotype Effects on Weeds and Yield of Dry-Seeded Rice. Crop Prot. 2017, 96, 68–76. [Google Scholar] [CrossRef]
- Shekhawat, K.; Rathore, S.S.; Chauhan, B.S. Weed Management in Dry Direct-Seeded Rice: A Review on Challenges and Opportunities for Sustainable Rice Production. Agronomy 2020, 10, 1264. [Google Scholar] [CrossRef]
- Chauhan, B.S.; Johnson, D.E. Growth and Reproduction of Junglerice (Echinochloa Colona) in Response to Water Stress. Weed Sci. 2010, 58, 132–135. [Google Scholar] [CrossRef]
- Narayana Rao, A. Echinochloa colona and Echinochloa crus-galli. In Biology and Management of Problematic Crop Weed Species; Elsevier: Amsterdam, The Netherlands, 2021; pp. 197–239. [Google Scholar] [CrossRef]
- Wu, D.; Shen, E.; Jiang, B.; Feng, Y.; Tang, W.; Lao, S.; Jia, L.; Lin, H.Y.; Xie, L.; Weng, X.; et al. Genomic Insights into the Evolution of Echinochloa Species as Weed and Orphan Crop. Nat. Commun. 2022, 13, 689. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Futakuchi, K. Improving Estimation of Weed Suppressive Ability of Upland Rice Varieties Using Substitute Weeds. Field Crops Res. 2014, 162, 1–5. [Google Scholar] [CrossRef]
- Saito, K.; Azoma, K.; Rodenburg, J. Plant Characteristics Associated with Weed Competitiveness of Rice under Upland and Lowland Conditions in West Africa. Field Crops Res. 2010, 116, 308–317. [Google Scholar] [CrossRef]
- Bharamappanavara, M.; Siddaiah, A.M.; Ponnuvel, S.; Ramappa, L.; Patil, B.; Appaiah, M.; Maganti, S.M.; Sundaram, R.M.; Shankarappa, S.K.; Tuti, M.D.; et al. Mapping QTL Hotspots Associated with Weed Competitive Traits in Backcross Population Derived from Oryza sativa L. and O. Glaberrima Steud. Sci. Rep. 2020, 10, 22103. [Google Scholar] [CrossRef]
- Panda, S.; Majhi, P.K.; Anandan, A.; Mahender, A.; Veludandi, S.; Bastia, D.; Guttala, S.B.; Singh, S.K.; Saha, S.; Ali, J. Proofing Direct-Seeded Rice with Better Root Plasticity and Architecture. Int. J. Mol. Sci. 2021, 22, 6058. [Google Scholar] [CrossRef]
- Schumaker, B.C.; Stallworth, S.; De Castro, E.; Fuller, M.G.; Shrestha, S.; Tseng, T.-M. Repeatable Stair-Step Assay to Access the Allelopathic Potential of Weedy Rice (Oryza sativa Ssp.). J. Vis. Exp. 2020, 155, e60764. [Google Scholar] [CrossRef]
- Schumaker, B.; Stallworth, S.; Tucker, A.; Yue, Z.; Tseng, T.-M. Phenotyping of Weedy Rice to Assess Root Characteristics Associated with Allelopathy. Am. J. Plant Sci. 2021, 12, 1210–1221. [Google Scholar] [CrossRef]
- Swanton, C.J.; Nkoa, R.; Blackshaw, R.E. Experimental Methods for Crop–Weed Competition Studies. Weed Sci. 2015, 63, 2–11. [Google Scholar] [CrossRef]
- Chaudhary, A.; Venkatramanan, V.; Kumar Mishra, A.; Sharma, S. Agronomic and Environmental Determinants of Direct Seeded Rice in South Asia. Circ. Econ. Sustain. 2023, 3, 253–290. [Google Scholar] [CrossRef] [PubMed]
- Dimaano, N.G.B.; Ali, J.; Sta. Cruz, P.C.; Baltazar, A.M.; Diaz, M.G.Q.; Acero, B.L.; Li, Z. Performance of Newly Developed Weed-Competitive Rice Cultivars under Lowland and Upland Weedy Conditions. Weed Sci. 2017, 65, 798–817. [Google Scholar] [CrossRef]
- Tonnessen, B.W.; Manosalva, P.; Lang, J.M.; Baraoidan, M.; Bordeos, A.; Mauleon, R.; Oard, J.; Hulbert, S.; Leung, H.; Leach, J.E. Rice Phenylalanine Ammonia-Lyase Gene OsPAL4 Is Associated with Broad Spectrum Disease Resistance. Plant Mol. Biol. 2015, 87, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, J.; Quan, R.; Pan, X.; Wan, L.; Huang, R. EAR Motif Mutation of Rice OsERF3 Alters the Regulation of Ethylene Biosynthesis and Drought Tolerance. Planta 2013, 237, 1443–1451. [Google Scholar] [CrossRef] [PubMed]
- Goldberg-Cavalleri, A.; Onkokesung, N.; Franco-Ortega, S.; Edwards, R. ABC Transporters Linked to Multiple Herbicide Resistance in Blackgrass (Alopecurus Myosuroides). Front. Plant Sci. 2023, 14, 1082761. [Google Scholar] [CrossRef]
- Lu, Y.; Chuan, M.; Wang, H.; Chen, R.; Tao, T.; Zhou, Y.; Xu, Y.; Li, P.; Yao, Y.; Xu, C.; et al. Genetic and Molecular Factors in Determining Grain Number per Panicle of Rice. Front. Plant Sci. 2022, 13, 964246. [Google Scholar] [CrossRef]
- Yu, S.; Ali, J.; Zhang, C.; Li, Z.; Zhang, Q. Genomic Breeding of Green Super Rice Varieties and Their Deployment in Asia and Africa. Theor. Appl. Genet. 2020, 133, 1427–1442. [Google Scholar] [CrossRef]
- Ali, J.; Aslam, U.M.; Tariq, R.; Murugaiyan, V.; Schnable, P.S.; Li, D.; Marfori-Nazarea, C.M.; Hernandez, J.E.; Arif, M.; Xu, J.; et al. Exploiting the Genomic Diversity of Rice (Oryza sativa L.): SNP-Typing in 11 Early-Backcross Introgression-Breeding Populations. Front. Plant Sci. 2018, 9, 849. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chou, H.H. Lucy2: An Interactive DNA Sequence Quality Trimming and Vector Removal Tool. Bioinformatics 2004, 20, 2865–2866. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.D.; Nacu, S. Fast and SNP-Tolerant Detection of Complex Variants and Splicing in Short Reads. Bioinformatics 2010, 26, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping: Integrated Software for Genetic Linkage Map Construction and Quantitative Trait Locus Mapping in Biparental Populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef]
- Pang, Y.; Chen, K.; Wang, X.; Wang, W.; Xu, J.; Ali, J.; Li, Z. Simultaneous Improvement and Genetic Dissection of Salt Tolerance of Rice (Oryza sativa L.) by Designed QTL Pyramiding. Front. Plant Sci. 2017, 8, 1275. [Google Scholar] [CrossRef]
- Murugaiyan, V.; Ali, J.; Mahender, A.; Aslam, U.M.; Jewel, Z.A.; Pang, Y.; Marfori-Nazarea, C.M.; Wu, L.B.; Frei, M.; Li, Z. Mapping of Genomic Regions Associated with Arsenic Toxicity Stress in a Backcross Breeding Populations of Rice (Oryza sativa L.). Rice 2019, 12, 61. [Google Scholar] [CrossRef]
- Manangkil, O.E.; Vu, H.T.T.; Mori, N.; Yoshida, S.; Nakamura, C. Mapping of Quantitative Trait Loci Controlling Seedling Vigor in Rice (Oryza sativa L.) under Submergence. Euphytica 2013, 192, 63–75. [Google Scholar] [CrossRef]
- Teixeira, S.B.; Pires, S.N.; Ávila, G.E.; Silva, B.E.P.; Schmitz, V.N.; Deuner, C.; da Silva Armesto, R.; da Silva Moura, D.; Deuner, S. Application of Vigor Indexes to Evaluate the Cold Tolerance in Rice Seeds Germination Conditioned in Plant Extract. Sci. Rep. 2021, 11, 11038. [Google Scholar] [CrossRef]
- Mahajan, G.; Chauhan, B.S. The Role of Cultivars in Managing Weeds in Dry-Seeded Rice Production Systems. Crop Prot. 2013, 49, 52–57. [Google Scholar] [CrossRef]
- Mennan, H.; Ngouajio, M.; Sahin, M.; Isık, D.; Altop, E.K. Competitiveness of Rice (Oryza sativa L.) Cultivars against Echinochloa crus-galli (L.) Beauv. in Water-Seeded Production Systems. Crop Prot. 2012, 41, 1–9. [Google Scholar] [CrossRef]
- Fageria, N.K. Yield Physiology of Rice. J. Plant Nutr. 2007, 30, 843–879. [Google Scholar] [CrossRef]
- Flint, J. Mapping Quantitative Traits and Strategies to Find Quantitative Trait Genes. Methods 2011, 53, 163–174. [Google Scholar] [CrossRef]
- Dole, J.; Weber, D.F. Detection of Quantitative Trait Loci Influencing Recombination Using Recombinant Inbred Lines. Genetics 2007, 177, 2309–2319. [Google Scholar] [CrossRef]
- Ott, A.; Liu, S.; Schnable, J.C.; Yeh, C.-T.E.; Wang, K.-S.; Schnable, P.S. TGBS® Genotyping-by-Sequencing Enables Reliable Genotyping of Heterozygous Loci. Nucleic Acids Res. 2017, 45, e178. [Google Scholar] [CrossRef]
- Yang, J.; Yang, G.; Yang, M.; Su, L.; Xia, A.; Li, D.; Huang, C.; Zhou, D.; Liu, Y.; Wang, H.; et al. Quantitative Trait Locus Analysis of Seed Germination and Early Seedling Growth in Rice. Front. Plant Sci. 2019, 10, 467540. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Fei, Y.; Wang, Y.; Zhao, W.; Hou, L.; Cao, Y.; Wu, M.; Wu, H. Identification of a Seed Vigor–Related QTL Cluster Associated with Weed Competitive Ability in Direct–Seeded Rice (Oryza sativa L.). Rice 2023, 16, 45. [Google Scholar] [CrossRef] [PubMed]
- Singh, U.M.; Yadav, S.; Dixit, S.; Ramayya, P.J.; Devi, M.N.; Raman, K.A.; Kumar, A. QTL Hotspots for Early Vigor and Related Traits under Dry Direct-Seeded System in Rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 229771. [Google Scholar] [CrossRef]
- Dimaano, N.G.B.; Ali, J.; Mahender, A.; Sta. Cruz, P.C.; Baltazar, A.M.; Diaz, M.G.Q.; Pang, Y.L.; Acero, B.L.; Li, Z. Identification of Quantitative Trait Loci Governing Early Germination and Seedling Vigor Traits Related to Weed Competitive Ability in Rice. Euphytica 2020, 216, 159. [Google Scholar] [CrossRef]
- Park, S.I.; Kim, J.J.; Shin, S.Y.; Kim, Y.S.; Yoon, H.S. ASR Enhances Environmental Stress Tolerance and Improves Grain Yield by Modulating Stomatal Closure in Rice. Front. Plant Sci. 2020, 10, 487076. [Google Scholar] [CrossRef]
- Todaka, D.; Nakashima, K.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Toward Understanding Transcriptional Regulatory Networks in Abiotic Stress Responses and Tolerance in Rice. Rice 2012, 5, 6. [Google Scholar] [CrossRef]
- Zou, X.; Sun, H. DOF Transcription Factors: Specific Regulators of Plant Biological Processes. Front. Plant Sci. 2023, 14, 1044918. [Google Scholar] [CrossRef]
- Schrick, K.; Bruno, M.; Khosla, A.; Cox, P.N.; Marlatt, S.A.; Roque, R.A.; Nguyen, H.C.; He, C.; Snyder, M.P.; Singh, D.; et al. Shared Functions of Plant and Mammalian StAR-Related Lipid Transfer (START) Domains in Modulating Transcription Factor Activity. BMC Med. 2014, 12, 70. [Google Scholar] [CrossRef] [PubMed]
- Zhai, R.; Ye, S.; Zhu, G.; Lu, Y.; Ye, J.; Yu, F.; Chu, Q.; Zhang, X. Identification and Integrated Analysis of Glyphosate Stress-Responsive MicroRNAs, LncRNAs, and MRNAs in Rice Using Genome-Wide High-Throughput Sequencing. BMC Genom. 2020, 21, 238. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Trivedi, P.K. Glutathione S-Transferases: Role in Combating Abiotic Stresses Including Arsenic Detoxification in Plants. Front. Plant Sci. 2018, 9, 751. [Google Scholar] [CrossRef]
- Dasari, S.; Ganjayi, M.S.; Yellanurkonda, P.; Basha, S.; Meriga, B. Role of Glutathione S-Transferases in Detoxification of a Polycyclic Aromatic Hydrocarbon, Methylcholanthrene. Chem. Biol. Interact. 2018, 294, 81–90. [Google Scholar] [CrossRef]
- Kusano, M.; Fukushima, A.; Tabuchi-Kobayashi, M.; Funayama, K.; Kojima, S.; Maruyama, K.; Yamamoto, Y.Y.; Nishizawa, T.; Kobayashi, M.; Wakazaki, M.; et al. Cytosolic GLUTAMINE SYNTHETASE1;1 Modulates Metabolism and chloroplast Development in Roots. Plant Physiol. 2020, 182, 1894–1909. [Google Scholar] [CrossRef] [PubMed]
- Hundertmark, M.; Hincha, D.K. LEA (Late Embryogenesis Abundant) Proteins and Their Encoding Genes in Arabidopsis thaliana. BMC Genom. 2008, 9, 118. [Google Scholar] [CrossRef]
- Dirk, L.M.A.; Abdel, C.G.; Ahmad, I.; Neta, I.C.S.; Pereira, C.C.; Pereira, F.E.C.B.; Unêda-Trevisoli, S.H.; Pinheiro, D.G.; Downie, A.B. Late Embryogenesis Abundant Protein–Client Protein Interactions. Plants 2020, 9, 814. [Google Scholar] [CrossRef]

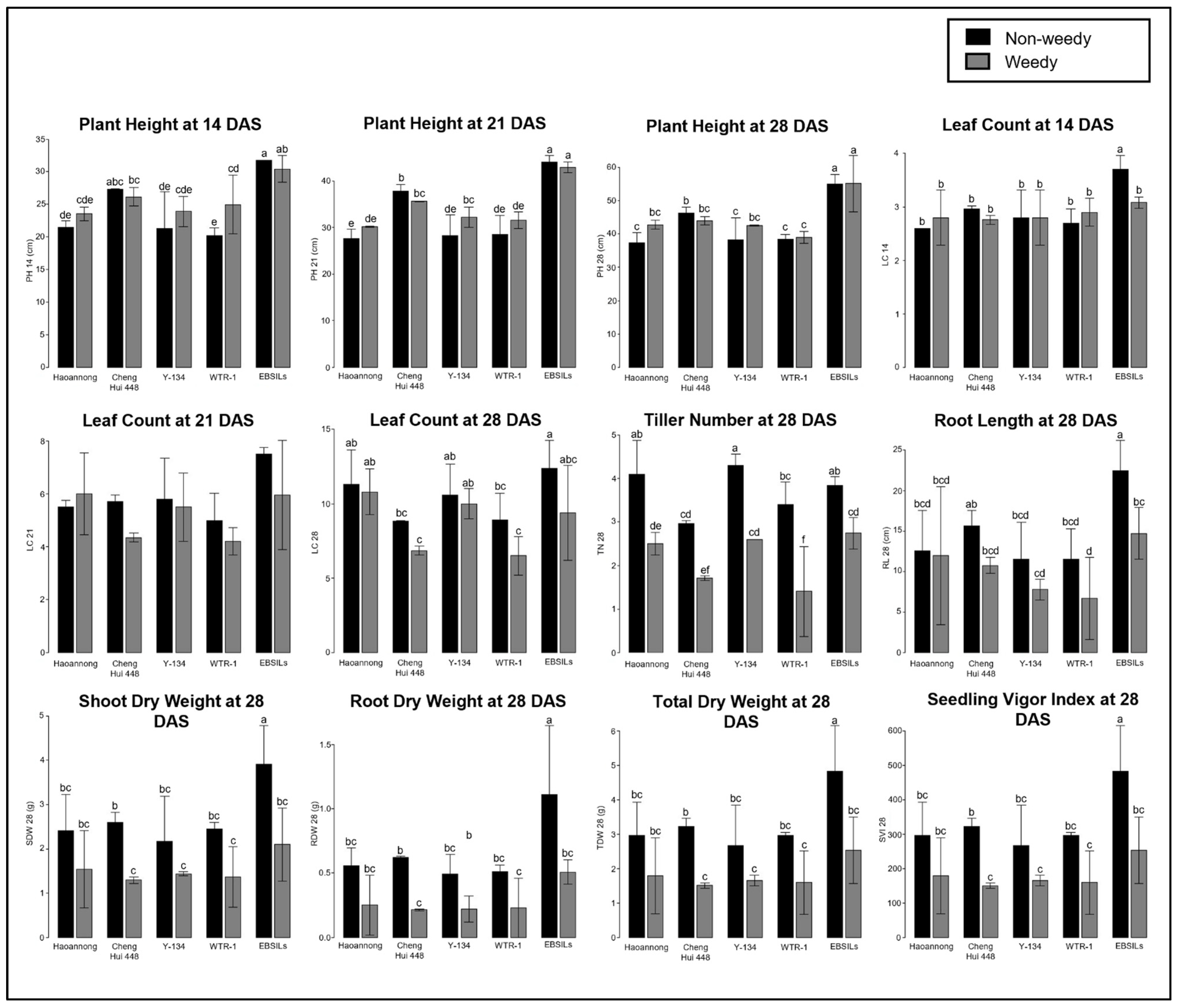

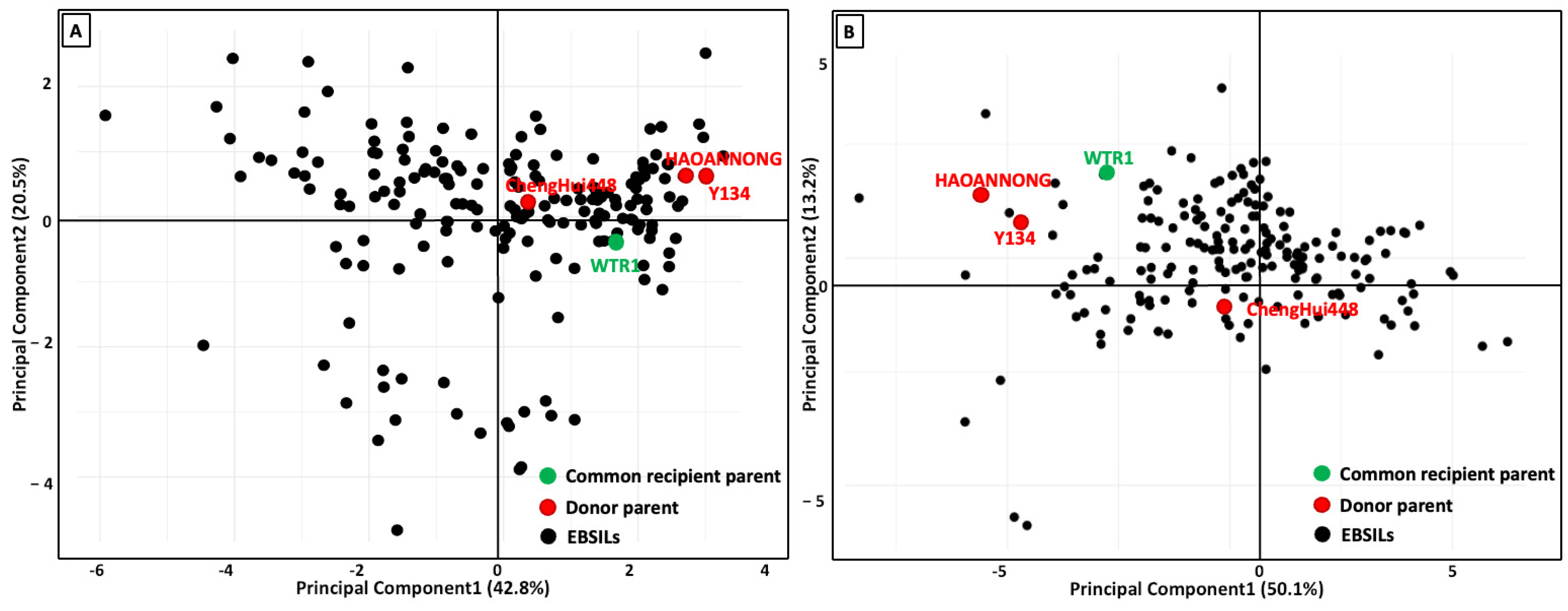

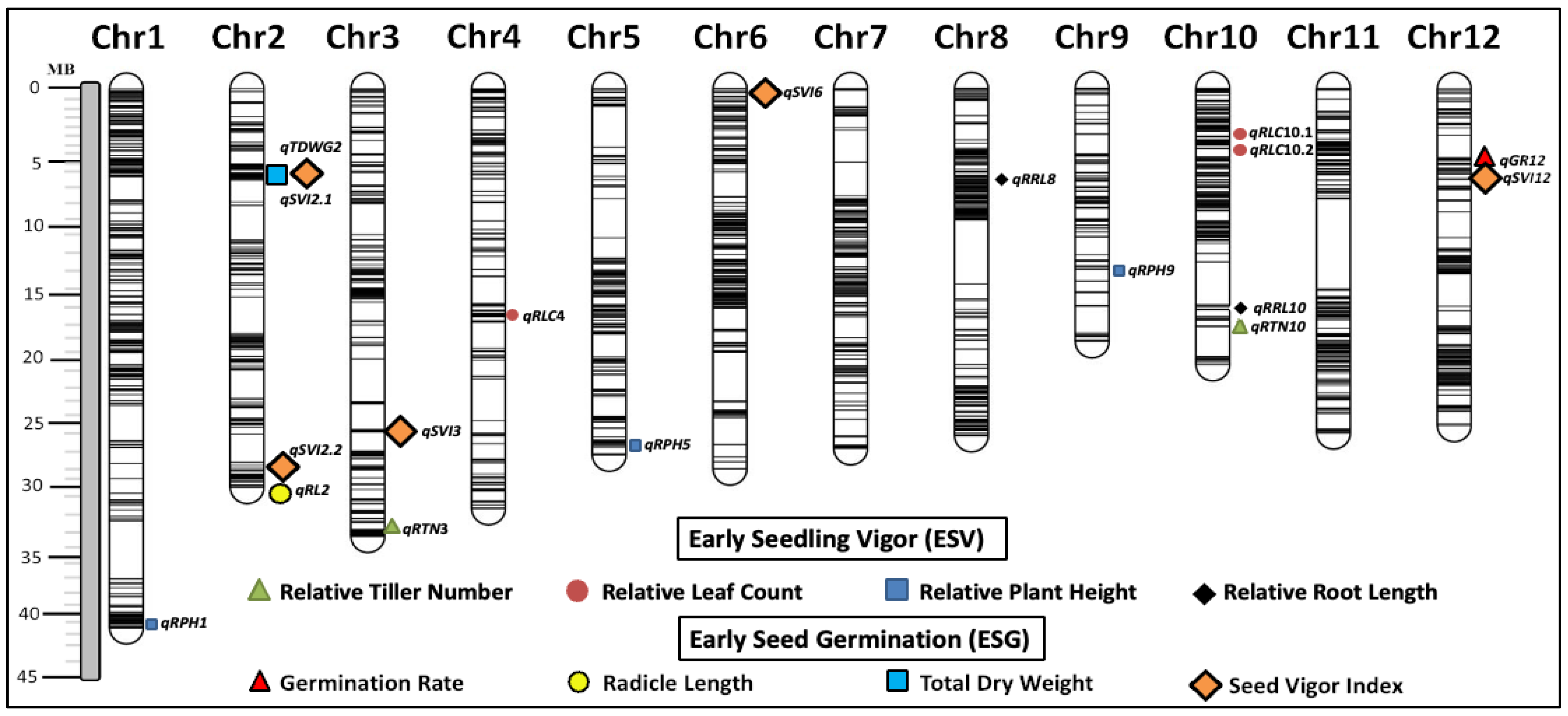
| Traits | Min. | Max. | Mean | Sum of Squares | Mean Square | F-Value | p-Value |
|---|---|---|---|---|---|---|---|
| Second-Day Germination Count | 3 | 25 | 19.01 | 5671 | 30.821 | 7.082 | 0.0000 *** |
| Seventh-Day Germination Count | 19 | 25 | 23.77 | 356.9 | 1.940 | 1.553 | 0.00147 ** |
| Germination Rate (%) | 15 | 100 | 79.77 | 88,891 | 483.1 | 8.34 | 0.0000 *** |
| Coleoptile Length (cm) | 3.16 | 9.79 | 6.536 | 876.7 | 4.765 | 8.927 | 0.0000 *** |
| Radicle Length (cm) | 1.12 | 15.4 | 5.24 | 3274 | 17.80 | 5.614 | 0.0000 *** |
| Total Dry Weight of Germinated Seeds (g) | 0.13 | 0.69 | 0.3953 | 0.6862 | 0.0037 | 2.145 | 0.0000 *** |
| Average Dry Weight of Germinated Seeds (g) | 0.0054 | 0.03 | 0.0166 | 0.0010 | 5.597 × 10−6 | 2.032 | 0.0000 *** |
| Seed Vigor Index | 5.25 | 63 | 31.68 | 21560 | 117.2 | 4.986 | 0.0000 *** |
| Traits | Non-Weedy | Weedy | ANOVA Result | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Min. | Max. | Mean | Min. | Max. | Mean | G | T | G*T | |
| Plant Height at 14 DAS (cm) | 13.2 | 41.2 | 25.89 | 13 | 36.5 | 24.79 | *** | *** | *** |
| Plant Height at 21 DAS (cm) | 22.2 | 54.9 | 36.68 | 17.2 | 51.4 | 34.1 | *** | *** | *** |
| Plant Height at 28 DAS (cm) | 23 | 65.6 | 45.79 | 20 | 90 | 42.88 | *** | *** | *** |
| Leaf Count at 14 DAS | 2 | 5 | 3 | 1 | 5 | 2.86 | *** | *** | *** |
| Leaf Count at 21 DAS | 2 | 10 | 5.87 | 2 | 8 | 4.44 | *** | *** | *** |
| Leaf Count at 28 DAS | 3 | 17 | 9.5 | 3 | 16 | 6.56 | *** | *** | *** |
| Number of Tiller at 28 das | 1 | 6 | 2.99 | 1 | 5 | 1.71 | *** | *** | *** |
| Seedling Vigor Index | 141 | 655 | 352.37 | 26 | 419 | 149.44 | *** | *** | *** |
| Shoot Dry Weight (g) | 1.2 | 4.87 | 2.85 | 0.13 | 3.15 | 1.29 | *** | *** | *** |
| Root Dry Weight (g) | 0.06 | 1.68 | 0.67 | 0.01 | 1.04 | 0.21 | ** | *** | ** |
| Total Dry Weight (g) | 1.41 | 6.55 | 3.52 | 0.26 | 4.19 | 1.5 | *** | *** | *** |
| Root Length (cm) | 4.5 | 37 | 16.33 | 2.8 | 24.9 | 9.64 | *** | *** | *** |
| No. | QTL a | Trait | Chr. | Position b | Associated Marker c | LOD d | PVE e | Additive Effect f | Tolerance Allele g |
|---|---|---|---|---|---|---|---|---|---|
| 1 | qGR12 | Germination Rate | 12 | 5720211–6950257 | S12_6548722 | 4.51 | 10.57 | −6.15 | Haoannong |
| 2 | qRL2 | Radicle Length | 2 | 34379631–34576493 | S2_34576493 | 6.68 | 15.25 | 0.55 | ChengHui448 |
| 3 | qTDWG2 | Total Dry Weight | 2 | 8474520–8752801 | S2_8699045 | 6.08 | 13.98 | −0.02 | Y134 |
| 4 | qSVI2.1 | Seed Vigor Index | 2 | 8474495–8752801 | S2_8474495 | 7.03 | 15.97 | −4.04 | Y134 |
| 5 | qSVI2.2 | 2 | 30478421–30791659 | S2_30791659 | 7.17 | 16.26 | −3.62 | Y134 | |
| 6 | qSVI3 | 3 | 25401607–25452773 | S3_25401672 | 4.74 | 11.08 | −3.25 | Y134 | |
| 7 | qSVI6 | 6 | 1542513–1877725 | S6_1698496 | 5.26 | 12.2 | −2.81 | Y134 | |
| 8 | qSVI12 | 12 | 6950207–7011126 | S12_6950257 | 5.76 | 13.29 | −3.45 | Y134 | |
| 9 | qRPH1 | Relative Plant Height at 14 DAS | 1 | 42171596–42617013 | S1_42549502 | 4.84 | 11.29 | −4.27 | WTR1 |
| 10 | qRPH5 | Relative Plant Height at 21 DAS | 5 | 28497478–28567356 | S5_28525048 | 4.29 | 10.10 | −4.61 | Y134 |
| 11 | qRPH9 | 9 | 14626500–14826499 | S9_14725794 | 4.64 | 10.87 | −5.19 | Y134 | |
| 12 | qRPH1 | Relative Plant Height at 28 DAS | 1 | 42171596–42617013 | S1_42171596 | 4.23 | 9.94 | −3.47 | Y134 |
| 13 | qRLC10.1 | Relative Leaf Count at 28 DAS | 10 | 416500–516499 | S10_466091 | 4.32 | 10.15 | −4.32 | ChengHui448 |
| 14 | qRLC10.2 | 10 | 1393500–1493499 | S10_1441265 | 4.27 | 10.04 | −4.61 | ChengHui448 | |
| 15 | qRLC4 | 4 | 1753300–1753386 | S4_1753338 | 4.86 | 11.34 | 4.42 | ChengHui448 | |
| 16 | qRTN3 | Relative Tiller Number at 28 DAS | 3 | 35948030–35965394 | S3_35948030 | 6.47 | 14.80 | −6.18 | Haoannong |
| 17 | qRTN10 | 10 | 18051000–18164000 | S10_18105284 | 4.36 | 10.24 | −5.46 | Haoannong | |
| 18 | qRRL8 | Relative Root Length at 28 DAS | 8 | 7492455–7777214 | S8_7541070 | 4.52 | 10.60 | −4.42 | Haoannong |
| 19 | qRRL10 | 10 | 18060329–18452140 | S10_18228061 | 4.41 | 10.34 | −4.88 | Haoannong |
| S.No | Stage | Locus | Annotation | SNPs in 3K RGP | Haoannong | ChengHui448 | Y134 | WTR-1 | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P-SNPs | NS-SNPs | P-SNPs | NS-SNPs | P-SNPs | NS-SNPs | P-SNPs | NS-SNPs | |||||
| 1 | Early Seedling Vigor | LOC_Os01g73250 | Abscisic stress-ripening, putative, expressed (ASR4) | 146 | 5 | 1 | 10 | 2 | 23 | 5 | 8 | 2 |
| 2 | LOC_Os09g24560 | No apical meristem protein, putative, expressed | 39 | 0 | 0 | 5 | 1 | 3 | 1 | 5 | 1 | |
| 3 | LOC_Os09g24800 | MYB family transcription factor, putative, expressed | 52 | 1 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | |
| 4 | LOC_Os09g24820 | ZF-HD protein dimerization region containing protein expressed | 267 | 2 | 0 | 27 | 1 | 34 | 3 | 37 | 3 | |
| 5 | LOC_Os10g33940 | Auxin response factor 22, putative, expressed (ARF22) | 251 | 27 | 4 | 26 | 4 | 23 | 4 | 23 | 4 | |
| 6 | LOC_Os10g33960 | START domain-containing protein expressed (OSHB2) | 325 | 20 | 1 | 24 | 2 | 25 | 2 | 25 | 2 | |
| 7 | LOC_Os10g34020 | Glutathione S-transferase, putative, expressed (GSTU47) | 354 | 24 | 0 | 13 | 2 | 18 | 1 | 31 | 1 | |
| 8 | LOC_Os10g34430 | Dicer, putative, expressed (DCL3B) | 469 | 21 | 8 | 22 | 8 | 26 | 9 | 26 | 9 | |
| 9 | Early Seed Germination | LOC_Os12g10720 | Glutathione S-transferase, putative, expressed (GSTZ1) | 258 | 2 | 0 | 7 | 0 | 0 | 0 | 11 | 1 |
| 10 | LOC_Os12g10730 | Glutathione S-transferase, putative, expressed (GSTZ2) | 199 | 0 | 0 | 6 | 1 | 0 | 0 | 7 | 1 | |
| 11 | LOC_Os12g12580 | NADP-dependent oxidoreductase, putative, expressed (CLPC2) | 306 | 18 | 1 | 65 | 11 | 6 | 0 | 69 | 13 | |
| 12 | LOC_Os02g15250 | Late embryogenesis abundant domain-containing protein, putative, expressed (LEA15) | 88 | 0 | 0 | 1 | 1 | 11 | 6 | 10 | 5 | |
| 13 | LOC_Os02g15340 | No apical meristem protein, putative, expressed | 113 | 0 | 0 | 0 | 0 | 12 | 5 | 12 | 4 | |
| 14 | LOC_Os02g15350 | dof zinc finger domain-containing protein, putative, expressed (RPBF) | 267 | 3 | 1 | 3 | 0 | 14 | 3 | 52 | 3 | |
| 15 | LOC_Os02g50240 | Glutamine synthetase, a catalytic domain-containing protein, expressed (GLN1;1) | 194 | 2 | 0 | 8 | 1 | 10 | 1 | 10 | 1 | |
| 16 | LOC_Os02g50330 | RNA-dependent RNA polymerase, putative, expressed (RDR1) | 344 | 6 | 0 | 17 | 1 | 18 | 1 | 6 | 0 | |
| 17 | LOC_Os06g04070 | pyridoxal-dependent decarboxylase protein, putative, expressed (ACD1) | 343 | 2 | 1 | 0 | 0 | 6 | 3 | 0 | 0 | |
| 18 | LOC_Os06g04200 | Starch synthase, putative, expressed (WX1) | 403 | 6 | 0 | 0 | 0 | 28 | 1 | 2 | 0 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nocito, K.D.; Murugaiyan, V.; Ali, J.; Pandey, A.; Casal, C., Jr.; De Asis, E.J.; Dimaano, N.G. Genome-Wide Dissection of Novel QTLs and Genes Associated with Weed Competitiveness in Early-Backcross Selective Introgression-Breeding Populations of Rice (Oryza sativa L.). Biology 2025, 14, 413. https://doi.org/10.3390/biology14040413
Nocito KD, Murugaiyan V, Ali J, Pandey A, Casal C Jr., De Asis EJ, Dimaano NG. Genome-Wide Dissection of Novel QTLs and Genes Associated with Weed Competitiveness in Early-Backcross Selective Introgression-Breeding Populations of Rice (Oryza sativa L.). Biology. 2025; 14(4):413. https://doi.org/10.3390/biology14040413
Chicago/Turabian StyleNocito, Kim Diane, Varunseelan Murugaiyan, Jauhar Ali, Ambika Pandey, Carlos Casal, Jr., Erik Jon De Asis, and Niña Gracel Dimaano. 2025. "Genome-Wide Dissection of Novel QTLs and Genes Associated with Weed Competitiveness in Early-Backcross Selective Introgression-Breeding Populations of Rice (Oryza sativa L.)" Biology 14, no. 4: 413. https://doi.org/10.3390/biology14040413
APA StyleNocito, K. D., Murugaiyan, V., Ali, J., Pandey, A., Casal, C., Jr., De Asis, E. J., & Dimaano, N. G. (2025). Genome-Wide Dissection of Novel QTLs and Genes Associated with Weed Competitiveness in Early-Backcross Selective Introgression-Breeding Populations of Rice (Oryza sativa L.). Biology, 14(4), 413. https://doi.org/10.3390/biology14040413









