Transcriptome Profiling Revealed Light-Mediated Gene Expression Patterns of Plants in Forest Vertical Structures
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site and Plant Material
2.2. RNA Extraction and RNA-Seq
2.3. Transcriptome Assembly, Gene Annotation, and Differential Expression Analysis
2.4. Statistical Analysis
3. Results
3.1. Expression Pattern of Photosynthetic and Photorespiratory Genes
3.2. Expression Pattern of Photoreceptors
3.3. Expression Pattern of Photoprotection Gene and Photolyases
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kohyama, T.S.; Takada, T. One-sided competition for light promotes coexistence of forest trees that share the same adult height. J. Ecol. 2012, 100, 1501–1511. [Google Scholar] [CrossRef]
- Onoda, Y.; Salunga, J.B.; Akutsu, K.; Aiba, S.; Yahara, T.; Anten, N.P.R. Trade-off between light interception efficiency and light use efficiency: Implications for species coexistence in one-sided light competition. J. Ecol. 2014, 102, 167–175. [Google Scholar] [CrossRef]
- Sercu, B.K.; Baeten, L.; van Coillie, F.; Martel, A.; Lens, L.; Verheyen, K.; Bonte, D. How tree species identity and diversity affect light transmittance to the understory in mature temperate forests. Ecol. Evol. 2017, 7, 10861–10870. [Google Scholar] [CrossRef]
- Tang, H.; Dubayah, R. Light-driven growth in Amazon evergreen forests explained by seasonal variations of vertical canopy structure. Proc. Natl. Acad. Sci. USA 2017, 114, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Casal, J.J. Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 2013, 64, 403–427. [Google Scholar] [CrossRef]
- Sheerin, D.J.; Hiltbrunner, A. Molecular mechanisms and ecological function of far-red light signalling. Plant Cell Environ. 2017, 40, 2509–2529. [Google Scholar] [CrossRef] [PubMed]
- Moretti, A.P.; Olguin, F.Y.; Pinazo, M.A.; Graciano, C. Water and light stresses drive acclimation during the establishment of a timber tree under different intensities of rainforest canopy coverage. CERNE 2019, 25, 93–104. [Google Scholar] [CrossRef]
- Nelson, N.; Yocum, C.F. Structure and function of photosystems I and II. Annu. Rev. Plant Biol. 2006, 57, 521–565. [Google Scholar] [CrossRef]
- Klimmek, F.; Sjodin, A.; Noutsos, C.; Leister, D.; Jansson, S. Abundantly and rarely expressed Lhc protein genes exhibit distinct regulation patterns in plants. Plant Physiol. 2006, 140, 793–804. [Google Scholar] [CrossRef]
- Pietrzykowska, M.; Suorsa, M.; Semchonok, D.A.; Tikkanen, M.; Boekema, E.J.; Aro, E.M.; Jansson, S. The light-harvesting chlorophyll a/b binding proteins Lhcb1 and Lhcb2 play complementary roles during state transitions in Arabidopsis. Plant Cell 2014, 26, 3646–3660. [Google Scholar] [CrossRef]
- Benson, S.L.; Maheswaran, P.; Ware, M.A.; Hunter, C.N.; Horton, P.; Jansson, S.; Ruban, A.V.; Johnson, M.P. An intact light harvesting complex I antenna system is required for complete state transitions in Arabidopsis. Nat. Plants 2015, 1, 15176. [Google Scholar] [CrossRef] [PubMed]
- Mauser, H.; King, W.A.; Gready, J.E.; Andrews, T.J. CO2 fixation by Rubisco: Computational dissection of the key steps of carboxylation, hydration, and C-C bond cleavage. J. Am. Chem. Soc. 2001, 123, 10821–10829. [Google Scholar] [CrossRef]
- Feller, U.; Anders, I.; Mae, T. Rubiscolytics: Fate of Rubisco after its enzymatic function in a cell is terminated. J. Exp. Bot. 2008, 9, 1615–1624. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Davydov, I.I.; Besnard, G.; Salamin, N. Duplication history and molecular evolution of the rbcS multigene family in angiosperms. J. Exp. Bot. 2019, 70, 6127–6139. [Google Scholar] [CrossRef] [PubMed]
- Busch, F.A. Photorespiration in the context of Rubisco biochemistry, CO2 diffusion and metabolism. Plant J. 2020, 101, 919–939. [Google Scholar] [CrossRef]
- Voss, I.; Sunil, B.; Scheibe, R.; Raghavendra, A.S. Emerging concept for the role of photorespiration as an important part of abiotic stress response. Plant Biol. 2013, 15, 713–722. [Google Scholar] [CrossRef]
- Timm, S.; Woitschach, F.; Heise, C.; Hagemann, M.; Bauwe, H. Faster removal of 2-phosphoglycolate through photorespiration improves abiotic stress tolerance of Arabidopsis. Plants 2019, 8, 563. [Google Scholar] [CrossRef]
- Ruban, A.V. Plants in light. Commun. Integr. Biol. 2009, 2, 50–55. [Google Scholar] [CrossRef]
- Kong, S.G.; Okajima, K. Diverse photoreceptors and light responses in plants. J. Plant Res. 2016, 12, 111–114. [Google Scholar] [CrossRef]
- Somers, D.E.; Devlin, P.F.; Kay, S.A. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 1998, 282, 1488–1490. [Google Scholar] [CrossRef]
- Christie, J.M. Phototropin blue-light receptors. Annu. Rev. Plant Biol. 2007, 58, 21–45. [Google Scholar] [CrossRef]
- Sharrock, R.A. The phytochrome red/far-red photoreceptor superfamily. Genome Biol. 2008, 9, 230. [Google Scholar] [CrossRef] [PubMed]
- Kianianmomeni, A. More light behind gene expression. Trends Plant Sci. 2014, 19, 488–490. [Google Scholar] [CrossRef] [PubMed]
- Fankhauser, C.; Batschauer, A. Shadow on the plant: A strategy to exit. Cell 2016, 164, 15–17. [Google Scholar] [CrossRef]
- Courbier, S.; Pierik, R. Canopy light quality modulates stress responses in plants. iScience 2019, 22, 441–452. [Google Scholar] [CrossRef]
- Kasahara, M.; Kagawa, T.; Oikawa, K.; Suetsugu, N.; Miyao, M.; Wada, M. Chloroplast avoidance movement reduces photodamage in plants. Nature 2002, 420, 829–832. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Badger, M.R. Photoprotection in plants: A new light on photosystem II damage. Trends Plant Sci. 2011, 16, 53–60. [Google Scholar] [CrossRef]
- Roach, T.; Krieger-Liszkay, A. The role of the PsbS protein in the protection of photosystems I and II against high light in Arabidopsis thaliana. Biochim. Biophys. Acta Bioenerg. 2012, 1817, 2158–2165. [Google Scholar] [CrossRef]
- Croce, R. PsbS is the plants’ pick for sun protection. Nat. Struct. Mol. Biol. 2015, 22, 650–652. [Google Scholar] [CrossRef]
- Gill, S.S.; Anjum, N.A.; Gill, R.; Jha, M.; Tuteja, N. DNA damage and repair in plants under ultraviolet and ionizing radiations. Sci. World J. 2015, 2015, 250158. [Google Scholar] [CrossRef]
- Mei, Q.; Dvornyk, V. Evolutionary history of the photolyase/cryptochrome superfamily in eukaryotes. PLoS ONE 2015, 10, e0135940. [Google Scholar] [CrossRef]
- Kavakli, I.H.; Baris, I.; Tardu, M.; Gul, S.; Oner, H.; Cal, S.; Bulut, S.; Yarparvar, D.; Berkel, C.; Ustaoglu, P.; et al. The Photolyase/cryptochrome family of proteins as DNA repair enzymes and transcriptional repressors. Photochem. Photobiol. 2017, 93, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Violle, C.; Navas, M.L.; Vile, D.; Kazakou, E.; Fortunel, C.; Hummel, I.; Garnier, E. Let the concept of trait be functional! Oikos 2007, 116, 882–892. [Google Scholar] [CrossRef]
- Ellsworth, D.S.; Reich, P.B. Canopy structure and vertical patterns of photosynthesis and related leaf traits in a deciduous forest. Oecologia 1993, 96, 169–178. [Google Scholar] [CrossRef]
- Mo, Q.F.; Lin, Q.; Wang, W.J.; Zhou, Q.; Peng, Z.T.; Chen, Y.Q.; Wang, Y.Y.; Zhao, Q. Assessments of foliar functional traits of understory shrubs in two 13-year reforested plantations in subtropical China. Glob. Ecol. Conserv. 2020, 23, e01202. [Google Scholar] [CrossRef]
- Alvarez, M.; Schrey, A.W.; Richards, C.L. Ten years of transcriptomics in wild populations: What have we learned about their ecology and evolution? Mol. Ecol. 2015, 24, 710–725. [Google Scholar] [CrossRef]
- Todd, E.V.; Black, M.A.; Gemmell, N.J. The power and promise of RNA-seq in ecology and evolution. Mol. Ecol. 2016, 25, 1224–1241. [Google Scholar] [CrossRef]
- Han, B.; Umaña, M.N.; Mi, X.; Liu, X.; Chen, L.; Wang, Y.; Liang, Y.; Wei, W.; Ma, K. The role of transcriptomes linked with responses to light environment on seedling mortality in a subtropical forest, China. J. Ecol. 2017, 105, 592–601. [Google Scholar] [CrossRef]
- Swenson, N.G.; Iida, Y.; Howe, R.; Wolf, A.; Umana, M.N.; Petprakob, K.; Turner, B.L.; Ma, K. Tree co-occurrence and transcriptomic response to drought. Nat. Commun. 2017, 8, 1996. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 2018, 7, 1–6. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Pertea, G.; Huang, X.Q.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Parvizi, B.; et al. TIGR Gene Indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Mount, D.W. Using the basic local alignment search tool (BLAST). CSH Protoc. 2007, 2007, pdb.top17. [Google Scholar] [CrossRef]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011, 12, 323. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 1 December 2021).
- Bates, D.; Maechler, M.; Bolker, B.; Walkerm, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Kuznetsova, A.; Brockhoff, P.B.; Christensen, R.H.B. lmerTest package: Tests in linear mixed effects models. J. Stat. Softw. 2017, 82, 1–26. [Google Scholar] [CrossRef]
- Lin, C. Blue light receptors and signal transduction. Plant Cell 2002, 14 (Suppl. 1), S207–S225. [Google Scholar] [CrossRef]
- Redekop, P.; Rothhausen, N.; Rothhausen, N.; Melzer, M.; Mosebach, L.; Dulger, E.; Bovdilova, A.; Caffarri, S.; Hippler, M.; Jahns, P. PsbS contributes to photoprotection in Chlamydomonas reinhardtii independently of energy dissipation. Biochim. Biophys. Acta Bioenerg. 2020, 1861, 148183. [Google Scholar] [CrossRef]
- Feng, C.M.; Qiu, Y.; Van Buskirk, E.K.; Yang, E.J.; Chen, M. Light-regulated gene repositioning in Arabidopsis. Nat. Commun. 2014, 5, 3027. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Kihara-Doim, T.; Kawazu, T.; Miyake, C.; Makino, A. Differences in Rubisco content and its synthesis in leaves at different positions in Eucalyptus globulus seedlings. Plant Cell Environ. 2010, 33, 1314–1323. [Google Scholar] [CrossRef]
- Craine, J.M.; Reich, P.B. Leaf-level light compensation points in shade-tolerant woody seedlings. New Phytol. 2005, 166, 710–713. [Google Scholar] [CrossRef]
- Sefcik, L.T.; Zak, D.R.; Ellsworth, D.S. Photosynthetic responses to understory shade and elevated carbon dioxide concentration in four northern hardwood tree species. Tree Physiol. 2006, 26, 1589–1599. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K.; Buschmann, C.; Doll, M.; Fietz, H.J.; Bach, T.; Kozel, U.; Meier, D.; Rahmsdorf, U. Photosynthetic activity, chloroplast ultrastructure, and leaf characteristics of high-light and low-light plants and of sun and shade leaves. Photosynth. Res. 1981, 2, 115–141. [Google Scholar] [CrossRef] [PubMed]
- Laisk, A.; Eichelmann, H.; Oja, V.; Rasulov, B.; Padu, E.; Bichele, I.; Pettai, H.; Kull, O. Adjustment of leaf photosynthesis to shade in a natural canopy: Rate parameters. Plant Cell Environ. 2005, 28, 375–388. [Google Scholar] [CrossRef]
- Walters, R.G. Towards an understanding of photosynthetic acclimation. J. Exp. Bot. 2005, 56, 435–447. [Google Scholar] [CrossRef]
- McKim, S.M.; Durnford, D.G. Translational regulation of light-harvesting complex expression during photoacclimation to high-light in Chlamydomonas reinhardtii. Plant Physiol. Biochem. 2006, 44, 857–865. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Lou, Y.; Sun, H.; Li, L.; Wang, L.; Dong, L.; Gao, Z. Transcriptome and comparative gene expression analysis of Phyllostachys edulis in response to high light. BMC Plant Biol. 2016, 16, 34. [Google Scholar] [CrossRef]
- Chukhutsina, V.U.; Liu, X.; Xu, P.; Croce, R. Light-harvesting complex II is an antenna of photosystem I in dark-adapted plants. Nat. Plants 2020, 6, 860–868. [Google Scholar] [CrossRef]
- Ganeteg, U.; Klimmek, F.; Jansson, S. Lhca5--an LHC-type protein associated with photosystem I. Plant Mol. Biol. 2004, 54, 641–651. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Fukao, Y.; Fujiwara, M.; Takami, T.; Shikanai, T. Efficient operation of NAD(P)H dehydrogenase requires supercomplex formation with photosystem I via minor LHCI in Arabidopsis. Plant Cell 2009, 21, 3623–3640. [Google Scholar] [CrossRef]
- Peterson, R.B.; Schultes, N.P. Light-harvesting complex B7 shifts the irradiance response of photosynthetic light-harvesting regulation in leaves of Arabidopsis thaliana. J. Plant Physiol. 2014, 171, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Allorent, G.; Petroutsos, D. Photoreceptor-dependent regulation of photoprotection. Curr. Opin. Plant Biol. 2017, 37, 102–108. [Google Scholar] [CrossRef]
- Hwang, S.; Herrin, D.L. Control of lhc gene transcription by the circadian clock in Chlamydomonas reinhardtii. Plant Mol. Biol. 1994, 26, 557–569. [Google Scholar] [CrossRef]
- Piechulla, B. Circadian expression of the light-harvesting complex protein genes in plants. Chronobiol. Int. 1999, 16, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Im, C.S.; Eberhard, S.; Huang, K.; Beck, C.F.; Grossman, A.R. Phototropin involvement in the expression of genes encoding chlorophyll and carotenoid biosynthesis enzymes and LHC apoproteins in Chlamydomonas reinhardtii. Plant J. 2006, 48, 1–16. [Google Scholar] [CrossRef]
- Reed, J.W.; Nagatani, A.; Elich, T.D.; Fagan, M.; Chory, J. Phytochrome-A and phytochrome-B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 1994, 104, 1139–1149. [Google Scholar] [CrossRef]
- Ohgishi, M.; Saji, K.; Okada, K.; Sakai, T. Functional analysis of each blue light receptor, cry1, cry2, phot1, and phot2, by using combinatorial multiple mutants in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 2223–2228. [Google Scholar] [CrossRef]
- Kagawa, T.; Kimura, M.; Wada, M. Blue light-induced phototropism of inflorescence stems and petioles is mediated by phototropin family members phot1 and phot2. Plant Cell Physiol. 2009, 50, 1774–1785. [Google Scholar] [CrossRef]
- He, D.; Yan, E.R. Size-dependent variations in individual traits and trait scaling relationships within a shade-tolerant evergreen tree species. Am. J. Bot. 2018, 105, 1165–1174. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.R.; Paul, N.D. Seduced by the dark side: Integrating molecular and ecological perspectives on the influence of light on plant defence against pests and pathogens. New Phytol. 2006, 170, 677–699. [Google Scholar] [CrossRef] [PubMed]
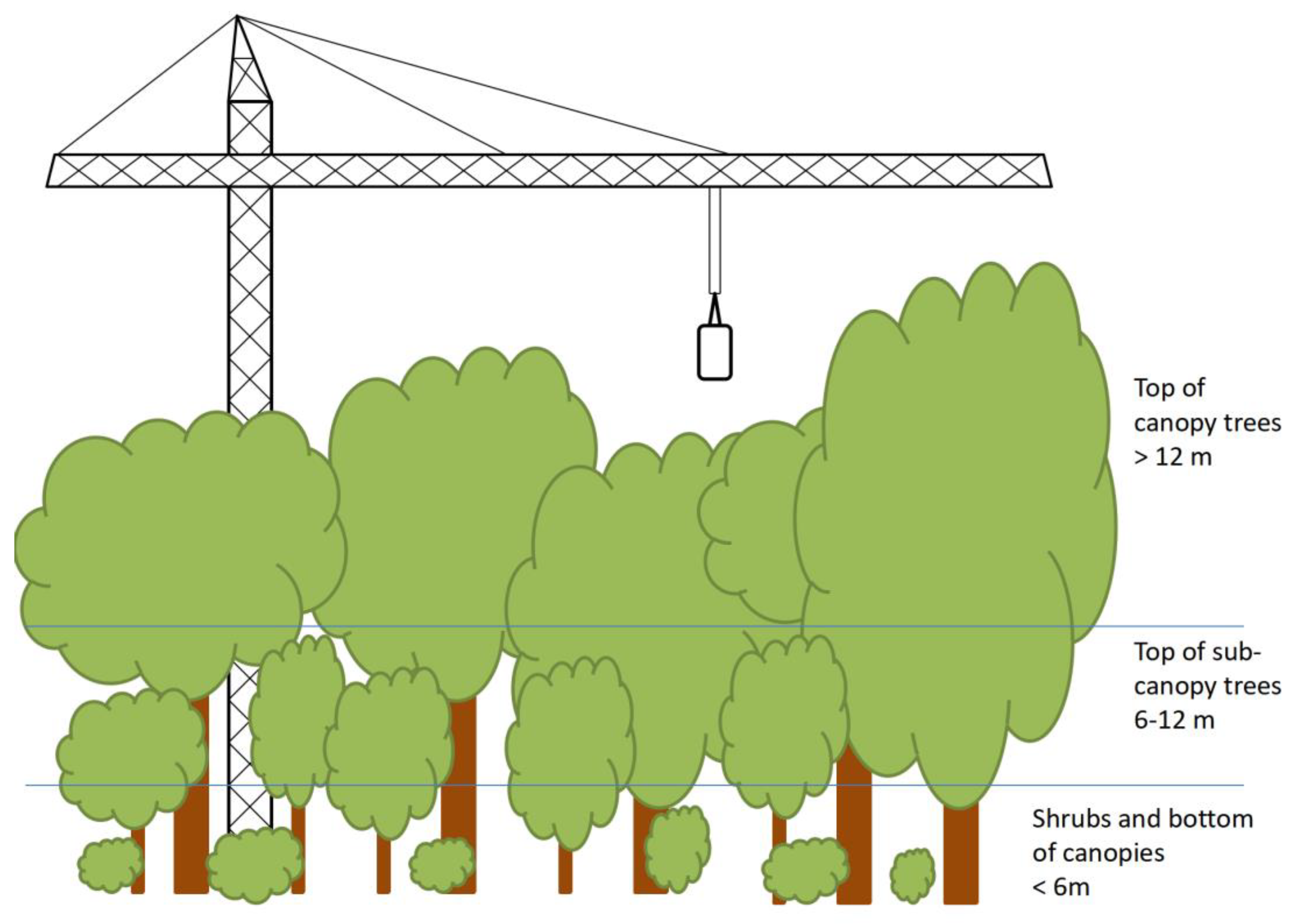

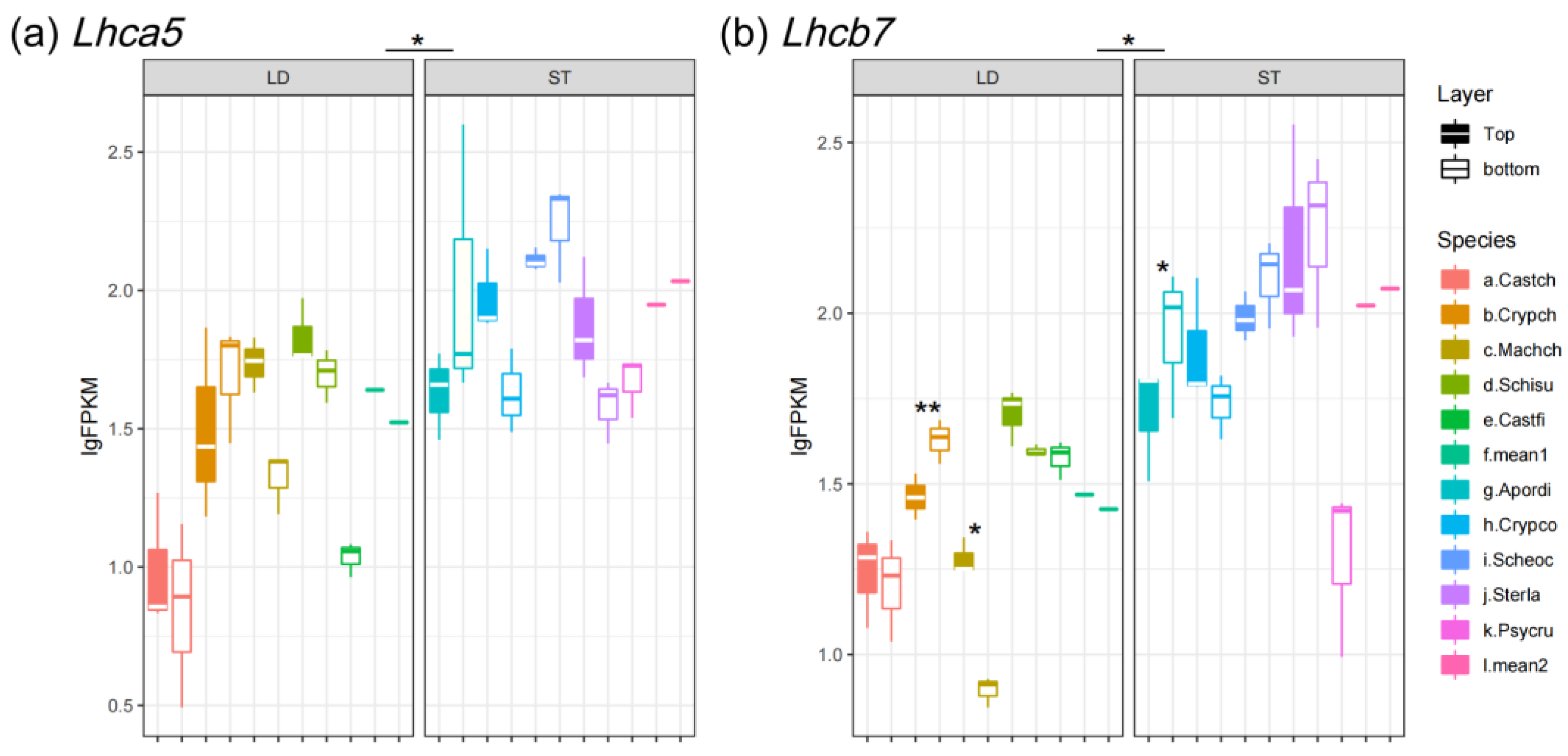
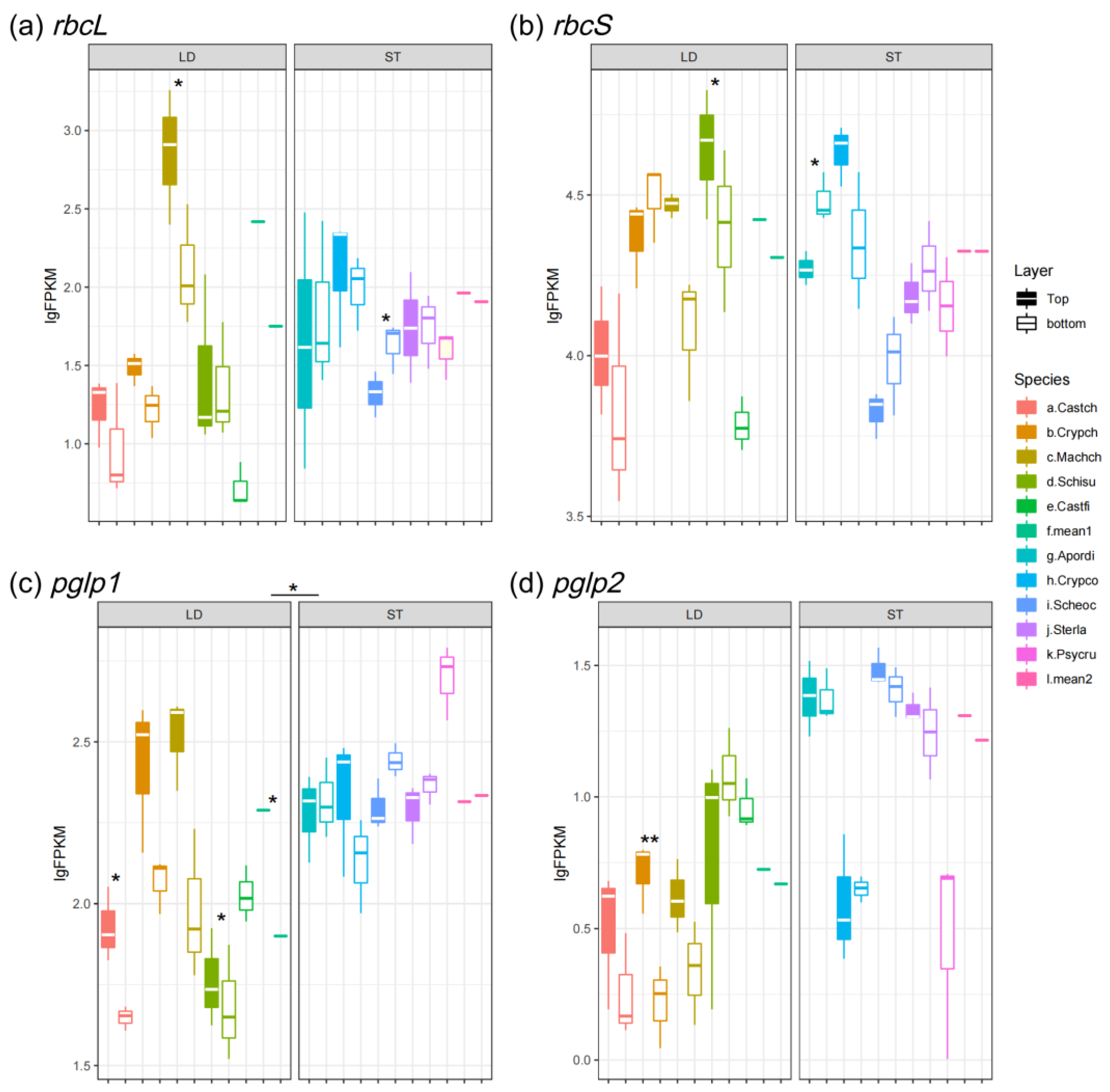



| Genes | Function | Expression Level in Light-Demanding Species | Expression Level in Shade-Tolerant Species |
|---|---|---|---|
| Abundantly expressed LHCs (Lhca1/2 and Lhcb1/2) | Light-harvesting of photosynthesis | Higher | Lower |
| Rarely expressed LHCs (Lhca5 and Lhcb7) | Light-harvesting of photosynthesis and photoprotection | Lower | Higher |
| RuBisCO (rbcL and rbcS) | Carbon fixation and photorespiration | Similar | Similar |
| 2-PG phosphatase (pglp1/2) | Photorespiration | Lower | Higher |
| Cryptochromes (cry1/2) | Blue- and UV-A-light receptors | Similar | Similar |
| Phototropins (pot1/2) | Blue-light receptors | Lower | Higher |
| Phytochromes (phyA/B) | Red- and far-red-light receptors | Lower | Higher |
| NPQ (PsbS) | Photoprotection | Lower | Higher |
| Photolyases (CPD Phr, (6-4) Phr and cry-DASH) | Repairing UV-induced damage | Lower | Higher |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mei, Q.; Zheng, Y.; Feng, J.; Wang, Z.; Cao, H.; Lian, J. Transcriptome Profiling Revealed Light-Mediated Gene Expression Patterns of Plants in Forest Vertical Structures. Biology 2025, 14, 434. https://doi.org/10.3390/biology14040434
Mei Q, Zheng Y, Feng J, Wang Z, Cao H, Lian J. Transcriptome Profiling Revealed Light-Mediated Gene Expression Patterns of Plants in Forest Vertical Structures. Biology. 2025; 14(4):434. https://doi.org/10.3390/biology14040434
Chicago/Turabian StyleMei, Qiming, Yi Zheng, Jiayi Feng, Zhengfeng Wang, Honglin Cao, and Juyu Lian. 2025. "Transcriptome Profiling Revealed Light-Mediated Gene Expression Patterns of Plants in Forest Vertical Structures" Biology 14, no. 4: 434. https://doi.org/10.3390/biology14040434
APA StyleMei, Q., Zheng, Y., Feng, J., Wang, Z., Cao, H., & Lian, J. (2025). Transcriptome Profiling Revealed Light-Mediated Gene Expression Patterns of Plants in Forest Vertical Structures. Biology, 14(4), 434. https://doi.org/10.3390/biology14040434





