Transcriptome and Metabolome Analyses Reveal High-Altitude Adaptation in the Qinghai Toad-Headed Lizard Phrynocephalus vlangalii
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Collection and Treatment
2.2. Resting Metabolic Rate (RMR) Measurement and Data Analysis
2.3. Transcriptome Analysis
2.4. Metabolome Analysis
2.5. Association Analysis of Metabolome and Transcriptome
3. Results
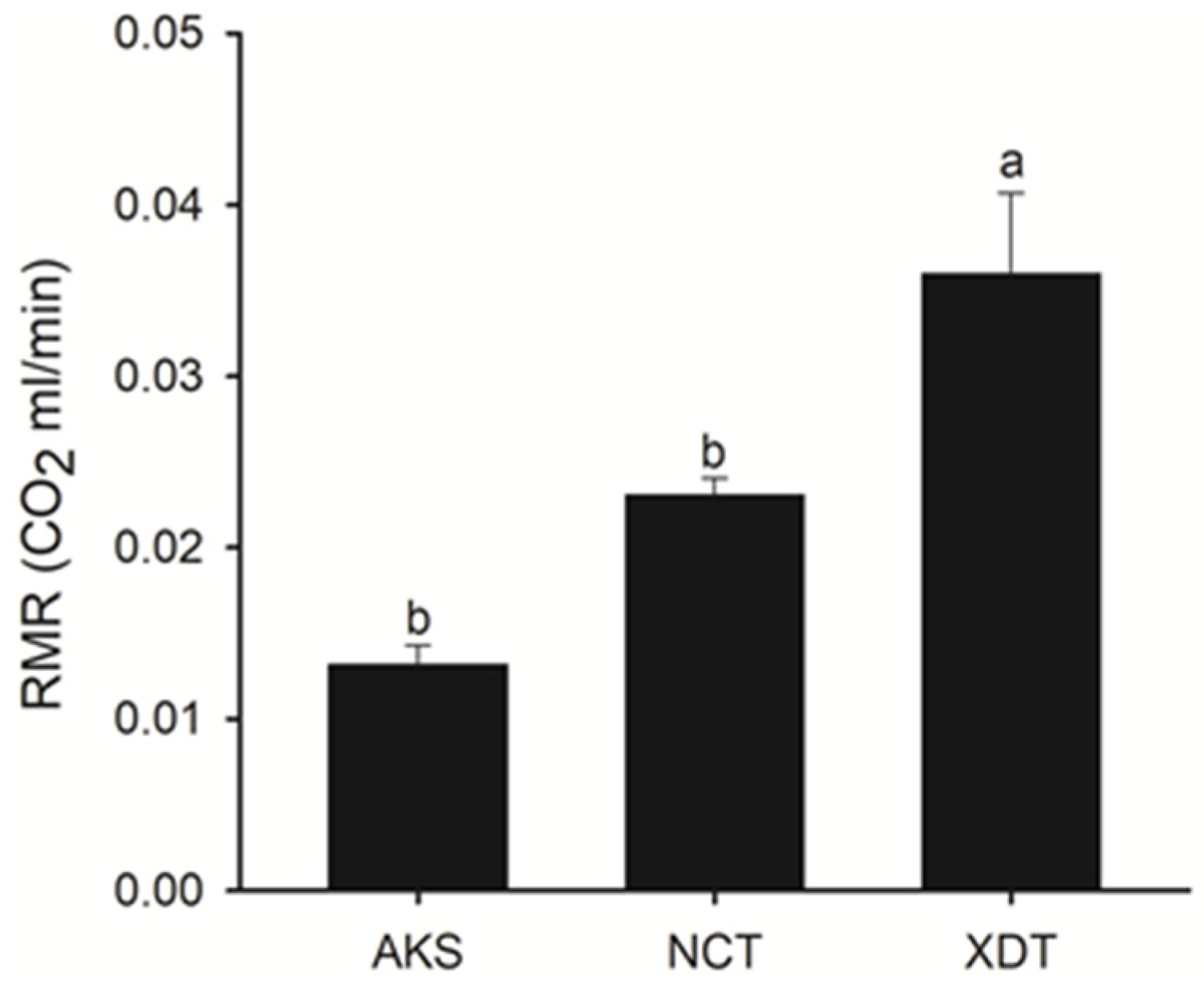
3.1. Altitudinal Trend of Resting Metabolic Rate (RMR)
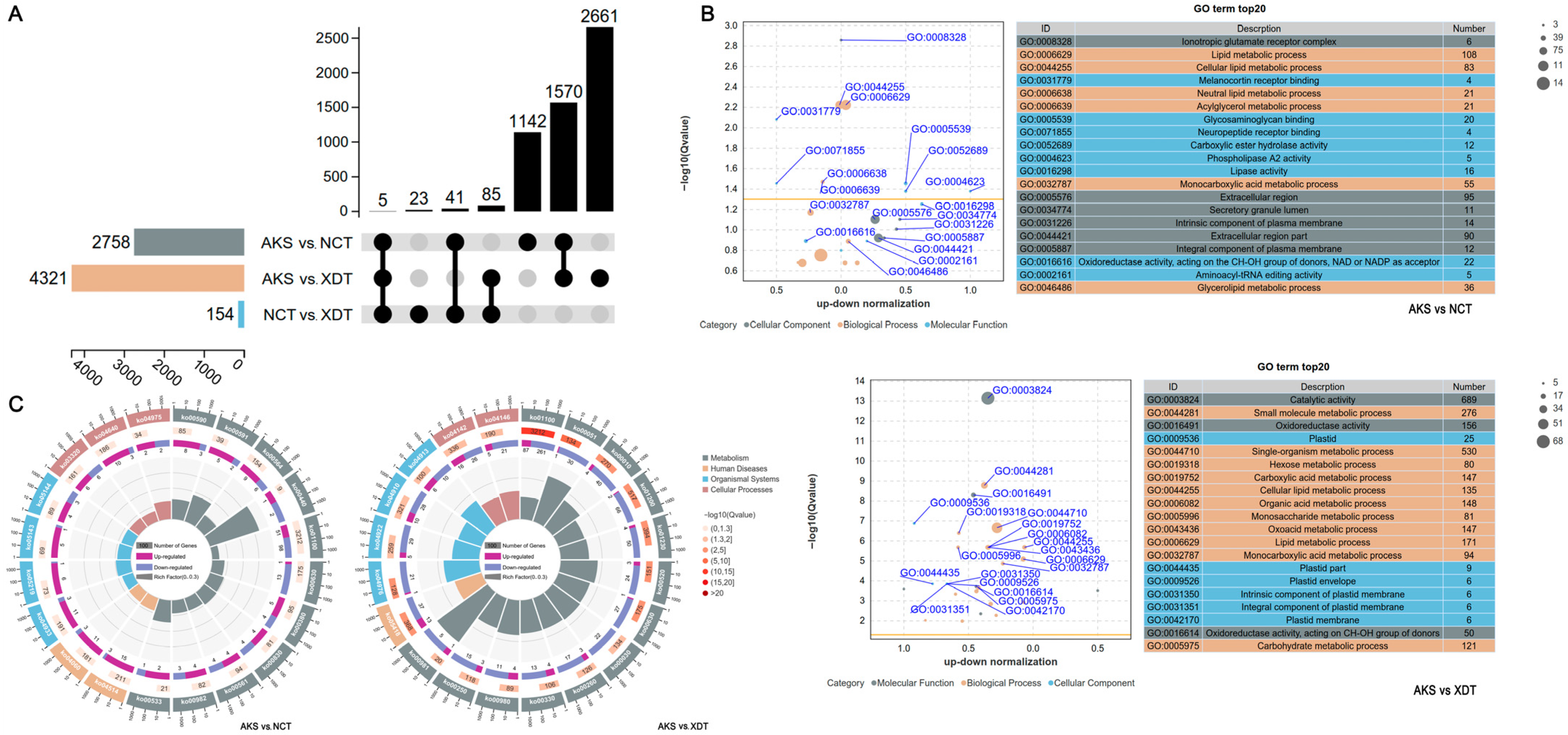
3.2. Differently Expressed Genes (DEGs) Revealed the Metabolism Change with Altitude
3.3. Metabolomic Data Revealed the Glycogen and Lipid Metabolism Change with Altitude
3.4. Joint Analysis of Transcriptome and Metabolome Changes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DEGs | Differentially expressed genes |
| DAMs | Differential metabolites |
| RMR | Resting metabolic rate |
| MS | Metabolites |
| HOAD | β-hydroxyacyl-CoA dehydrogenase |
| LDH | Lactate dehydrogenase |
| CS | Citrate synthase |
| PDK | Pyruvate dehydrogenase kinase |
References
- Storz, J.F.; Moriyama, H. Mechanisms of hemoglobin adaptation to high-altitude hypoxia. High Alt. Med. Biol. 2008, 9, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Root, T.L.; Price, J.T.; Hall, K.R.; Schneider, S.H.; Rosenzweig, C.; Pounds, J.A. Fingerprints of global warming on wild animals and plants. Nature 2003, 421, 57–60. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, R.; Lenoir, J.; Piedallu, C.; Riofrio-Dillon, G.; de Ruffray, P.; Vidal, C.; Pierrat, J.C.; Gegout, J.C. Changes in plant community composition lag behind climate warming in lowland forests. Nature 2011, 479, 517–520. [Google Scholar] [CrossRef]
- Zhang, K.-L.; Yao, L.-J.; Meng, J.-S.; Tao, J. Maxent modeling for predicting the potential geographical distribution of two peony species under climate change. Sci. Total Environ. 2018, 634, 1326–1334. [Google Scholar] [CrossRef] [PubMed]
- Storz, J.F.; Scott, G.R.; Cheviron, Z.A. Phenotypic plasticity and genetic adaptation to high-altitude hypoxia in vertebrates. J. Exp. Biol. 2010, 213, 4125–4136. [Google Scholar] [CrossRef]
- Ren, Y.; Liu, P.-F.; Zhu, W.-L.; Zhang, H.; Cai, J.-H. Higher altitude and lower temperature regulate the body mass and energy metabolism in male Eothenomys miletus. Pak. J. Zool. 2020, 52, 139–146. [Google Scholar] [CrossRef]
- Arias-Reyes, C.; Soliz, J.; Joseph, V. Mice and rats display different ventilatory, hematological, and metabolic features of acclimatization to hypoxia. Front. Physiol. 2021, 12, 647822. [Google Scholar] [CrossRef]
- Brummelen, A.C.V.; Olszewski, K.L.; Wilinski, D.; Llinas, M.; Louw, A.I.; Birkholtz, L.M. Co-inhibition of Plasmodium falciparum S-adenosylmethionine decarboxylase/ornithine decarboxylase reveals perturbation-specific compensatory mechanisms by transcriptome, proteome, and metabolome analyses. J. Biol. Chem. 2009, 284, 4635–4646. [Google Scholar] [CrossRef]
- Liang, S.-W.; Li, W.-X.; Zhang, Y.; Tang, X.-L.; He, J.-Z.; Bai, Y.-C.; Li, D.-Q.; Wang, Y.; Chen, Q. Seasonal variation of metabolism in lizard Phrynocephalus vlangalii at high-altitude. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2017, 203, 341–347. [Google Scholar] [CrossRef]
- Chicco, A.J.; Le, C.H.; Gnaiger, E.; Dreyer, H.C.; Muyskens, J.B.; D’Alessandro, A.; Nemkov, T.; Hocker, A.D.; Prenni, J.E.; Wolfe, L.M.; et al. Adaptive remodeling of skeletal muscle energy metabolism in high-altitude hypoxia: Lessons from AltitudeOmics. J. Biol. Chem. 2018, 293, 6659–6671. [Google Scholar] [CrossRef]
- Brandl, S.J.; Lefcheck, J.S.; Bates, A.E.; Rasher, D.B.; Norin, T. Can metabolic traits explain animal community assembly and functioning? Biol. Rev. 2023, 98, 1–18. [Google Scholar] [CrossRef]
- Toledo, L.F.; Brito, S.P.; Milsom, W.K.; Abe, A.S.; Andrade, D.V. Effects of season, temperature, and body mass on the standard metabolic rate of tegu lizards (Tupinambis merianae). Physiol. Biochem. Zool. 2008, 81, 158–164. [Google Scholar] [CrossRef]
- Cheviron, Z.A.; Bachman, G.C.; Connaty, A.D.; McClelland, G.B.; Storz, J.F. Regulatory changes contribute to the adaptive enhancement of thermogenic capacity in high-altitude deer mice. Proc. Natl. Acad. Sci. USA 2012, 109, 8635–8640. [Google Scholar] [CrossRef] [PubMed]
- Storz, J.F.; Cheviron, Z.A. Physiological genomics of adaptation to high-altitude hypoxia. Annu. Rev. Anim. Biosci. 2021, 9, 149–171. [Google Scholar] [CrossRef] [PubMed]
- Dawson, N.J.; Scott, G.R. Adaptive increases in respiratory capacity and O2 affinity of subsarcolemmal mitochondria from skeletal muscle of high-altitude deer mice. FASEB J. 2022, 36, e22391. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Chen, B.-C.; Langda, S.; Pu, P.; Zhu, X.-J.; Zhou, S.-W.; Kalds, P.; Zhang, K.; Bhati, M.; Leonard, A.; et al. Multi-omic analyses shed light on the genetic control of high-altitude adaptation in sheep. Genom. Proteom. Bioinform. 2024, 22, qzae030. [Google Scholar] [CrossRef]
- Harris, P.; Castillo, Y.; Gibson, K.; Heath, D.; Arias-Stella, J. Succinic and lactic dehydrogenase activity in myocardial homogenates from animals at high and low altitude. J. Mol. Cell. Cardiol. 1970, 1, 189–193. [Google Scholar] [CrossRef]
- Sheafor, B.A. Metabolic enzyme activities across an altitudinal gradient: An examination of pikas (genus Ochotona). J. Exp. Biol. 2003, 206, 1241–1249. [Google Scholar] [CrossRef]
- Seebacher, F.; Sparrow, J.; Thompson, M.B. Turtles (Chelodina longicollis) regulate muscle metabolic enzyme activity in response to seasonal variation in body temperature. J. Comp. Physiol. B 2004, 174, 205–210. [Google Scholar] [CrossRef]
- Qi, Y.; Zhang, T.; Wu, Y.-Y.; Yao, Z.-Y.; Qiu, X.; Pu, P.; Tang, X.-L.; Fu, J.-Z.; Yang, W.-Z. A multilevel assessment of plasticity in response to high-altitude environment for Agama lizards. Front. Ecol. Evol. 2022, 10, 845072. [Google Scholar] [CrossRef]
- Kuang, L.-D.; Zheng, Y.-C.; Lin, Y.-Q.; Xu, Y.-O.; Jin, S.-Y.; Li, Y.-P.; Dong, F.; Jiang, Z.-Y. High-altitude adaptation of yak based on genetic variants and activity of lactate dehydrogenase-1. Biochem. Genet. 2010, 48, 418–427. [Google Scholar] [CrossRef]
- Lyons, S.A.; Tate, K.B.; Welch, K.C., Jr.; McClelland, G.B. Lipid oxidation during thermogenesis in high-altitude deer mice (Peromyscus maniculatus). Am. J. Physiol. Regul. Integr. Comp. Physiol. 2021, 320, R735–R746. [Google Scholar] [CrossRef]
- Raben, N.; Nagaraju, K.; Lee, E.; Kessler, P.; Byrne, B.; Lee, L.; Lamarca, M.; King, C.; Ward, J.; Sauer, B.; et al. Targeted disruption of the acid α-glucosidase gene in mice causes an illness with critical features of both infantile and adult human glycogen storage disease type II. J. Biol. Chem. 1998, 273, 19086–19092. [Google Scholar] [CrossRef] [PubMed]
- Fuson, A.L.; Cowan, D.F.; Kanatous, S.B.; Polasek, L.K.; Davis, R.W. Adaptations to diving hypoxia in the heart, kidneys and splanchnic organs of harbor seals (Phoca vitulina). J. Exp. Biol. 2003, 206, 4139–4154. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.N.; Vats, P.; Kumria, M.M.L.; Ranganathan, S.; Shyam, R.; Arora, M.P.; Jain, C.L.; Sridharan, K. Effect of high-altitude (7620 m) exposure on glutathione and related metabolism in rats. Eur. J. Appl. Physiol. 2001, 84, 233–237. [Google Scholar] [CrossRef]
- Dawson, N.J.; Alza, L.; Nandal, G.; Scott, G.R.; McCracken, K.G. Convergent changes in muscle metabolism depend on duration of high-altitude ancestry across Andean waterfowl. eLife 2020, 9, e56259. [Google Scholar] [CrossRef] [PubMed]
- Xin, J.W.; Chai, Z.X.; Zhang, C.F.; Yang, Y.M.; Zhang, Q.; Zhu, Y.; Cao, H.W.; Ji, C.D.Y.; Zhong, J.C.; Ji, Q.M. Transcriptome analysis identified long non-coding RNAs involved in the adaption of yak to high-altitude environments. R. Soc. Open Sci. 2020, 7, 200625. [Google Scholar] [CrossRef]
- Li, J.-Y.; Huang, N.-T.; Zhang, X.; Sun, C.; Chen, J.-R.; Wei, Q. Changes of collagen content in lung tissues of plateau yak and its mechanism of adaptation to hypoxia. PeerJ 2024, 12, e18250. [Google Scholar] [CrossRef]
- Yang, W.-Z.; Qi, Y.; Fu, J.-Z. Genetic signals of high-altitude adaptation in amphibians: A comparative transcriptome analysis. BMC Genet. 2016, 17, 134. [Google Scholar] [CrossRef]
- Guo, K.; Yuan, S.-J.; Wang, H.; Zhong, J.; Wu, Y.-Q.; Chen, W.; Hu, C.-C.; Chang, Q. Species distribution models for predicting the habitat suitability of Chinese fire-bellied newt Cynops orientalis under climate change. Ecol. Evol. 2021, 11, 10147–10154. [Google Scholar] [CrossRef]
- Zhang, Y.; Liang, S.-W.; He, J.-Z.; Bai, Y.-C.; Niu, Y.-G.; Tang, X.-L.; Li, D.-Q.; Chen, Q. Oxidative stress and antioxidant status in a lizard Phrynocephalus vlangalii at different altitudes or acclimated to hypoxia. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2015, 190, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Li, W.-X.; Liang, S.-W.; Wang, H.-H.; Xin, Y.; Lu, S.-S.; Tang, X.-L.; Chen, Q. The effects of chronic hypoxia on thermoregulation and metabolism in Phrynocephalus vlangalii. Asian Herpetol. Res. 2016, 7, 103–111. [Google Scholar]
- Han, J.-M.; Guo, R.-H.; Li, J.-Q.; Guan, C.; Chen, Y.; Zhao, W. Organ mass variation in a toad headed lizard Phrynocephalus vlangalii in response to hypoxia and low temperature in the Qinghai-Tibet Plateau, China. PLoS ONE 2016, 11, e0162572. [Google Scholar] [CrossRef]
- Zhao, E.-M.; Adler, K. Herpetology of China; Society for the Study of Amphibians and Reptiles: Oxford, OH, USA, 1993. [Google Scholar]
- Yao, Y.-T.; Du, Y.; Fang, M.-C.; Lin, L.-H.; Ji, X. Developmental stage does not affect resting metabolic rate in the monitor lizard, Varanus salvator. Anim. Biol. 2019, 69, 199–212. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.-D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; Mccue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Zhang, J.; Geng, X.-Q.; Zhang, Y.-H.; Zhao, X.-L.; Zhang, P.-W.; Sun, G.-R.; Li, W.-T.; Li, D.-H.; Han, R.-L.; Li, G.-X.; et al. Interaction between cecal metabolites and liver lipid metabolism pathways during induced molting in laying hens. Front. Physiol. 2022, 13, 862721. [Google Scholar] [CrossRef]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- Li, Q.-F.; Sun, R.-Y.; Huang, C.-X.; Wang, Z.-K.; Liu, X.-T.; Hou, J.-J.; Liu, J.-S.; Cai, L.-Q.; Li, N.; Zhang, S.-Z.; et al. Cold adaptive thermogenesis in small mammals from different geographical zones of China. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2001, 129, 949–961. [Google Scholar] [CrossRef]
- Thompson, L.J.; Downs, C.T. Altitudinal variation in metabolic parameters of a small Afrotropical bird. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2017, 212, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Polymeropoulos, E.T.; Elliott, N.G.; Frappell, P.B. Hypoxic acclimation leads to metabolic compensation after reoxygenation in Atlantic salmon yolk-sac alevins. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. A 2017, 213, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Woods, A.L.; Sharma, A.P.; Garvican-Lewis, L.A.; Saunders, P.U.; Rice, A.J.; Thompson, K.G. Four weeks of classical altitude training increases resting metabolic rate in highly trained middle-distance runners. Int. J. Sport Nutr. Exerc. Metab. 2017, 27, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Plasman, M.; Bautista, A.; McCue, M.D.; De La Vega-Perez, A.H. Resting metabolic rates increase with altitude in a mountain-dwelling lizard. Integr. Zool. 2020, 15, 363–374. [Google Scholar] [CrossRef]
- Hodkinson, I.D. Terrestrial insects along elevation gradients: Species and community responses to altitude. Biol. Rev. 2005, 80, 489–513. [Google Scholar] [CrossRef]
- Lau, D.S.; Connaty, A.D.; Mahalingam, S.; Wall, N.; Cheviron, Z.A.; Storz, J.F.; Scott, G.R.; McClelland, G.B. Acclimation to hypoxia increases carbohydrate use during exercise in high-altitude deer mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2017, 312, R400–R411. [Google Scholar] [CrossRef]
- Liu, G.-W.; Li, Y.-H.; Liao, N.; Shang, X.-Z.; Xu, F.-Q.; Yin, D.-C.; Shao, D.-Y.; Jiang, C.-M.; Shi, J.-L. Energy metabolic mechanisms for high-altitude sickness: Downregulation of glycolysis and upregulation of the lactic acid/amino acid-pyruvate-TCA pathways and fatty acid oxidation. Sci. Total Environ. 2023, 894, 164998. [Google Scholar] [CrossRef]
- Ding, X.-Z.; Liang, C.-N.; Guo, X.; Wu, X.-Y.; Wang, H.-B.; Johnson, K.A.; Yan, P. Physiological insight into the high-altitude adaptations in domesticated yaks (Bos grunniens) along the Qinghai-Tibetan Plateau altitudinal gradient. Livest. Sci. 2014, 162, 233–239. [Google Scholar] [CrossRef]
- Ma, J.; Zhang, T.-L.; Wang, W.-X.; Chen, Y.; Cai, W.-T.; Zhu, B.; Xu, L.-Y.; Gao, H.-J.; Zhang, L.-P.; Li, J.-Y.; et al. Comparative transcriptome analysis of gayal (Bos frontalis), yak (Bos grunniens), and cattle (Bos taurus) reveal the high-altitude adaptation. Front. Genet. 2022, 12, 778788. [Google Scholar] [CrossRef]
- Yang, W.Z.; Qi, Y.; Lu, B.; Qiao, L.; Wu, Y.Y.; Fu, J.Z. Gene expression variations in high-altitude adaptation: A case study of the Asiatic toad (Bufo gargarizans). BMC Genet. 2017, 18, 62. [Google Scholar] [CrossRef]
- Chen, Y.; Tan, S.; Fu, J.-Z. Modified metabolism and response to UV radiation: Gene expression variations along an elevational gradient in the Asiatic toad (Bufo gargarizans). J. Mol. Evol. 2022, 90, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Yomano, L.P.; Scopes, R.K.; Ingram, L.O. Cloning, sequencing, and expression of the Zymomonas mobilis phosphoglycerate mutase gene (pgm) in Escherichia coli. J. Bacteriol. 1993, 175, 3926–3933. [Google Scholar] [CrossRef]
- Reddy, G.K.; Wendisch, V.F. Characterization of 3-phosphoglycerate kinase from Corynebacterium glutamicum and its impact on amino acid production. BMC Microbiol. 2014, 14, 54. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-D.; Oeser, J.K.; Yang, C.-M.; Sarkar, S.; Hackl, S.I.; Hasty, A.H.; McGuinness, O.P.; Paradee, W.; Hutton, J.C.; Powell, D.R.; et al. Deletion of the gene encoding the ubiquitously expressed glucose-6-phosphatase catalytic subunit-related protein (UGRP)/glucose-6-phosphatase catalytic subunit-β results in lowered plasma cholesterol and elevated glucagon. J. Biol. Chem. 2006, 281, 39982–39989. [Google Scholar] [CrossRef]
- Kim, D.-G.; Yoo, J.-C.; Kim, E.; Lee, Y.-S.; Yarishkin, O.V.; Lee, D.-Y.; Lee, K.-H.; Hong, S.-G.; Hwang, E.-M.; Park, J.Y. A novel cytosolic isoform of mitochondrial trans-2-enoyl-coa reductase enhances peroxisome proliferator-activated receptor α activity. Endocrinol. Metab. 2014, 29, 185–194. [Google Scholar] [CrossRef]
- Qu, S.; Altomonte, J.; Perdomo, G.; He, J.; Fan, Y.; Kamagate, A.; Meseck, M.; Dong, H.H. Aberrant forkhead box O1 function is associated with impaired hepatic metabolism. Endocrinology 2006, 147, 5641–5652. [Google Scholar] [CrossRef]
- Matsumoto, M.; Pocai, A.; Rossetti, L.; DePinho, R.A.; Accili, D. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor FoxO1 in liver. Cell Metab. 2007, 6, 208–216. [Google Scholar] [CrossRef]
- Xiong, X.-W.; Tao, R.-Y.; DePinho, R.A.; Dong, X.-C. Deletion of hepatic FoxO1/3/4 genes in mice significantly impacts on glucose metabolism through downregulation of gluconeogenesis and upregulation of glycolysis. PLoS ONE 2013, 8, e74340. [Google Scholar] [CrossRef]
- Wilhelm, K.; Happel, K.; Eelen, G.; Schoors, S.; Oellerich, M.F.; Lim, R.; Zimmermann, B.; Aspalter, I.M.; Franco, C.A.; Boettger, T.; et al. FOXO1 couples metabolic activity and growth state in the vascular endothelium. Nature 2016, 529, 216–220. [Google Scholar] [CrossRef]
- Yan, D.; Cai, Y.; Luo, J.-R.; Liu, J.-J.; Li, X.; Ying, F.; Xie, X.; Xu, A.-M.; Ma, X.-S.; Xia, Z.-Y. FOXO1 contributes to diabetic cardiomyopathy via inducing imbalanced oxidative metabolism in type 1 diabetes. J. Cell. Mol. Med. 2020, 24, 7850–7861. [Google Scholar] [CrossRef]
- Wu, Y.-L.; Guo, Y.-Y.; Wang, Q. USP21 accelerates the proliferation and glycolysis of esophageal cancer cells by regulating the STAT3/FOXO1 pathway. Tissue Cell 2022, 79, 101916. [Google Scholar] [CrossRef] [PubMed]
- Piao, L.; Sidhu, V.K.; Fang, Y.-H.; Ryan, J.J.; Parikh, K.S.; Hong, Z.-G.; Toth, P.T.; Morrow, E.; Kutty, S.; Lopaschuk, G.D.; et al. FOXO1-mediated upregulation of pyruvate dehydrogenase kinase-4 (PDK4) decreases glucose oxidation and impairs right ventricular function in pulmonary hypertension: Therapeutic benefits of dichloroacetate. J. Mol. Med. 2013, 91, 333–346. [Google Scholar] [CrossRef]
- Kim, D.H.; Ha, S.; Choi, Y.J.; Dong, H.H.; Yu, B.P.; Chung, H.Y. Altered FoxO1 and PPARγ interaction in age-related ER stress-induced hepatic steatosis. Aging 2019, 11, 4125–4144. [Google Scholar] [CrossRef]
- Yu, S.; Matsusue, K.; Kashireddy, P.; Cao, W.Q.; Yeldandi, V.; Yeldandi, A.V.; Rao, M.S.; Gonzalez, F.J.; Reddy, J.K. Adipocyte-specific gene expression and adipogenic steatosis in the mouse liver due to peroxisome proliferator-activated receptor γ1 (PPARγ1) overexpression. J. Biol. Chem. 2003, 278, 498–505. [Google Scholar] [CrossRef]
- Lei, M.-M.; Li, Y.-X.; Li, J.-Y.; Liu, J.; Dai, Z.-C.; Chen, R.; Zhu, H.-X. Low testosterone and high leptin activate PPAR signaling to induce adipogenesis and promote fat deposition in caponized ganders. Int. J. Mol. Sci. 2024, 25, 8686. [Google Scholar] [CrossRef]
- Zhu, Y.-P.; Jing, L.; Li, X.-Y.; Zheng, D.; Zhou, G.-Q.; Zhang, Y.; Sang, Y.-J.; Shi, Z.-X.; Sun, Z.-W.; Zhou, X.-Q. Decabromodiphenyl ether disturbs hepatic glycolipid metabolism by regulating the PI3K/AKT/GLUT4 and mTOR/PPARγ/RXRα pathway in mice and L02 cells. Sci. Total Environ. 2021, 763, 142936. [Google Scholar] [CrossRef] [PubMed]
- Moran-Salvador, E.; Lopez-Parra, M.; Garcia Alonso, V.; Titos, E.; Martinez-Clemente, M.; Gonzalez-Periz, A.; Barak, Y.; Arroyo, V.; Claria, J. Role for PPARγ in obesity-induced hepatic steatosis as determined by hepatocyte- and macrophage-specific conditional knockouts. FASEB J. 2011, 25, 2538–2550. [Google Scholar] [CrossRef] [PubMed]
- Angilletta, M.J.; Winters, R.S.; Dunham, A.E. Thermal effects on the energetics of lizard embryos: Implications for hatchling phenotypes. Ecology 2000, 81, 2957–2968. [Google Scholar] [CrossRef]
- Hou, D.-M.; Jia, T.; Ren, Y.; Zhu, W.-L.; Liu, P.-F. Phenotypic trait variations in the frog Nanorana parkeri: Differing adaptive strategies to altitude between sexes. J. Vertebr. Biol. 2023, 72, 23008.1–23008.11. [Google Scholar] [CrossRef]
- Nishimura, T.; Arima, H.; Koirala, S.; Ito, H.; Yamamoto, T. Individual variations and sex differences in hemodynamics and percutaneous arterial oxygen saturation (SpO2) in Tibetan highlanders of Tsarang in the Mustang district of Nepal. J. Physiol. Anthropol. 2022, 41, 9. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, J.; Chen, J.; Lu, Y.-H.; Huang, Y.-F.; Hong, M.-S.; Ji, X. Transcriptome and Metabolome Analyses Reveal High-Altitude Adaptation in the Qinghai Toad-Headed Lizard Phrynocephalus vlangalii. Biology 2025, 14, 459. https://doi.org/10.3390/biology14050459
Zhong J, Chen J, Lu Y-H, Huang Y-F, Hong M-S, Ji X. Transcriptome and Metabolome Analyses Reveal High-Altitude Adaptation in the Qinghai Toad-Headed Lizard Phrynocephalus vlangalii. Biology. 2025; 14(5):459. https://doi.org/10.3390/biology14050459
Chicago/Turabian StyleZhong, Jun, Jian Chen, Yu-Hong Lu, Yu-Fei Huang, Ming-Sheng Hong, and Xiang Ji. 2025. "Transcriptome and Metabolome Analyses Reveal High-Altitude Adaptation in the Qinghai Toad-Headed Lizard Phrynocephalus vlangalii" Biology 14, no. 5: 459. https://doi.org/10.3390/biology14050459
APA StyleZhong, J., Chen, J., Lu, Y.-H., Huang, Y.-F., Hong, M.-S., & Ji, X. (2025). Transcriptome and Metabolome Analyses Reveal High-Altitude Adaptation in the Qinghai Toad-Headed Lizard Phrynocephalus vlangalii. Biology, 14(5), 459. https://doi.org/10.3390/biology14050459









